Abstract
Coenzyme Q (ubiquinone or Q) is a crucial mitochondrial lipid required for respiratory electron transport in eukaryotes. 4-Hydroxybenozoate (4HB) is an aromatic ring precursor that forms the benzoquinone ring of Q and is used extensively to examine Q biosynthesis. However, the direct precursor compounds and enzymatic steps for synthesis of 4HB in yeast are unknown. Here we show that para-aminobenzoic acid (pABA), a well known precursor of folate, also functions as a precursor for Q biosynthesis. A hexaprenylated form of pABA (prenyl-pABA) is normally present in wild-type yeast crude lipid extracts but is absent in yeast abz1 mutants starved for pABA. A stable 13C6-isotope of pABA (p- amino[aromatic-13C6]benzoic acid ([13C6]pABA)), is prenylated in either wild-type or abz1 mutant yeast to form prenyl-[13C6]pABA. We demonstrate by HPLC and mass spectrometry that yeast incubated with either [13C6]pABA or [13C6]4HB generate both 13C6-demethoxy-Q (DMQ), a late stage Q biosynthetic intermediate, as well as the final product 13C6-coenzyme Q. Pulse-labeling analyses show that formation of prenyl-pABA occurs within minutes and precedes the synthesis of Q. Yeast utilizing pABA as a ring precursor produce another nitrogen containing intermediate, 4-imino-DMQ6. This intermediate is produced in small quantities in wild-type yeast cultured in standard media and in abz1 mutants supplemented with pABA. We suggest a mechanism where Schiff base-mediated deimination forms DMQ6 quinone, thereby eliminating the nitrogen contributed by pABA. This scheme results in the convergence of the 4HB and pABA pathways in eukaryotic Q biosynthesis and has implications regarding the action of pABA-based antifolates.
Keywords: Folate Metabolism, Lipid Synthesis, Mass Spectrometry (MS), Mitochondrial Metabolism, Yeast Metabolism, Coenzyme Q Metabolism
Introduction
Coenzyme Q (Q)2 is an essential polyprenylated benzoquinone lipid in cellular energy metabolism. The prenyl tail anchors Q in cellular membranes, whereas the redox chemistry of the benzoquinone ring plays a crucial role in respiratory electron transport, in catabolic and biosynthetic metabolism (1), as a co-antioxidant able to recycle vitamin E, and as a chain-terminating antioxidant (2). In these reactions the quinone ring of Q thus cycles between oxidized and reduced (QH2, or hydroquinone) states.
Cells rely on de novo synthesis for an adequate supply of Q. Studies in Escherichia coli, Schizosaccharomyces pombe, and Saccharomyces cerevisiae have made use of Q-deficient mutants to elucidate the biosynthetic pathway (3–5). In S. cerevisiae, nine COQ genes are required, and each of the yeast coq mutants (coq1 through coq9) lack Q6 and are unable to grow on media containing non-fermentable carbon sources such as glycerol or ethanol. The dedicated precursors in the biosynthesis of Q are polyisoprene diphosphate, which provides the tail (S. cerevisiae synthesizes Q6, with a tail containing six isoprene units), and 4-hydroxybenzoic acid (4HB) (6, 7). Studies in animal cells and in E. coli indicate that different metabolic pathways are used to produce 4HB. Animals (e.g. rats and humans) generate 4HB from the essential dietary amino acid tyrosine (6–8). Phenylalanine also acts as a precursor for 4HB, however, the incorporation is thought to proceed primarily following its conversion to tyrosine via phenylalanine hydroxylase (8). The biosynthetic steps leading from 4-hydroxyphenylpyruvate to 4HB in animal cells are not yet characterized (see Fig. 1). E. coli relies on shikimate biosynthesis, the formation of chorismate, and chorismate pyruvate lyase (encoded by the ubiC gene) to synthesize 4HB (9, 10). E. coli ubiC mutants lack Q unless 4HB is provided in the growth media (9). E. coli mutants lacking shikimate or chorismate also require exogenous 4HB to synthesize Q (11). Thus, E. coli cells are unable to convert tyrosine or phenylalanine to Q and rely exclusively on the de novo synthesis of 4HB from chorismate.
FIGURE 1.
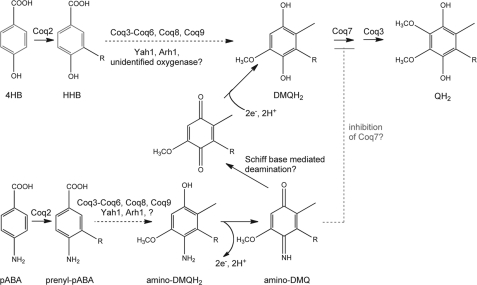
Yeast aromatic ring precursors involved in Q biosynthesis. We propose that yeast generate aromatic precursors for Q biosynthesis by at least two pathways. One branches from chorismate to produce pABA, a new aromatic ring precursor in Q biosynthesis. The other pathway also branches from chorismate to produce the typical aromatic ring precursor 4HB, via unknown gene products from tyrosine or 4-hydroxyphenylpyruvate. Both 4HB and pABA are prenylated by Coq2. Yeast lack a homolog of E. coli UbiC (chorismate pyruvate lyase), which directly forms 4HB from chorismate (dotted line). Animals lack the ability to produce shikimate, and rely on Tyr (or Phe) for production of 4HB. The steps involved in converting 4-hdroxyphenylpyruvate to 4HB are not known in yeast or animal cells.
In contrast, S. cerevisiae may utilize either shikimate or tyrosine to synthesize the aromatic ring precursor of Q (6, 12). Yeast preferentially utilize shikimate to produce Q, and tyrosine is utilized only when the synthesis of shikimate is blocked (12). Thus, yeast aro1C mutants (unable to synthesize shikimate), and yeast aro2 mutants (unable to synthesize chorismate) still synthesize Q de novo, because they are able to utilize exogenously added tyrosine (Fig. 1). The steps producing 4HB from tyrosine have not been identified, although the pathway may be similar to that described for the catabolism of p-coumarate to 4HB in Acinetobacter bayli (13). Although it has been assumed that yeast may generate 4HB via chorismate pyruvate lyase activity, S. cerevisiae lack a homolog of UbiC. This raises the question: how do yeast utilize chorismate to produce a ring precursor of Q?
Here we describe our surprising findings that para-aminobenzoic acid (pABA), a known precursor of folates, is also an aromatic precursor for Q biosynthesis, via the synthesis of 3-hexaprenyl-4-aminobenzoic acid (prenyl-pABA). These pathways are described in Fig. 1. The biosynthetic steps in yeast necessary for the production of pABA are catalyzed by the ABZ1 and ABZ2 gene products. Abz1 amidates chorismate to make the 4-aminodeoxychorismate intermediate (14, 15), and the Abz2 lyase forms free pABA (16). Import of pABA into the mitochondria is necessary for further folate synthesis; the FOL1 gene product is required for this import and also performs multiple enzymatic functions in pteroglutamoyl synthesis (17). Immunogold particle labeling and a Fol1-GFP fusion localized the tri-functional polypeptide Fol1p in yeast to mitochondrial membranes (17).
We recently became aware of similar work identifying pABA and prenyl-pABA as Q biosynthetic precursors (18). These authors identified pABA as a Q precursor in their search for iron-mediated effects on the function of the Coq7 monooxygenase in Q biosynthesis. Our studies independently determined that pABA is a novel coenzyme Q precursor, and we show prenyl-pABA is an endogenously synthesized intermediate in the Q biosynthetic pathway. We further demonstrate the relative contributions of the 13C6-isotope of 4HB and pABA under competition conditions with the alternative unlabeled ring precursor. In addition we identify 4-imino-DMQ6 in wild-type yeast and in pABA-supplemented abz1 null mutants. Based on our identification of this intermediate, we suggest a possible mechanism for the removal of the nitrogen donated by pABA, and its replacement with an oxygen atom to form the 1,4-quinone moiety in DMQ via Schiff base chemistry.
EXPERIMENTAL PROCEDURES
Yeast Growth Analysis
Yeast strains used in this work are described in Table 1. The abz1 null mutant (W303Δabz1) was generated as described (19). Yeast colonies from YPD (2% glucose, 1% yeast extract, 2% peptone, 2% agar) plates were inoculated into 18- × 100-mm culture tubes containing 5 ml of Drop Out Galactose (Dogal media): 2% galactose, 0.1% dextrose, and 6.8 g/liter Bio101 yeast nitrogen base minus pABA minus folate with ammonium sulfate (MP Biomedicals) and 5.83 mm sodium monophosphate (pH adjusted to 6.0 with NaOH). Amino acids and nucleotides were included at the following final concentrations (milligrams/liter): adenine hemisulfate, 80; arginine hydrochloride, 40; aspartic acid, 100; cysteine hydrochloride, 80; glutamic acid, 100; histidine hydrochloride, 80; isoleucine, 60; leucine, 120; lysine hydrochloride, 60; methionine, 80; phenylalanine, 80; serine, 60; threonine, 400; tryptophan, 200; tyrosine, 40; uracil, 80; and valine, 150. Following overnight incubation, yeast cultures were diluted 1:100 into fresh Dogal minimal media to deplete intrinsic stores of pABA and folate. Alternatively, cultures were diluted into Drop Out Glycerol Ethanol media (Doge; made as above, except galactose was replaced with 3% glycerol, 2% ethanol). Solid plate media were made by adding 2.5 g/liter Gelrite (Sigma). When noted media were supplemented with folinic acid, 0.4 μg/ml; 4HB, 2 μg/ml; or pABA (Sigma) 2 μg/ml.
TABLE 1.
Genotypes and sources of S. cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothsteina |
| W303ΔCOQ2 | MATaade2-1 his3-1,15 leu2-3,112 trp 1-1 ura3-1 coq2::HIS3 | (49) |
| W303ΔABZ1 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 abz1::KANMX4 | This work |
| NM101 | MATaleu2-3,112, ura3-52, coq7-1 | (29) |
| E2–249 | MATamet6, coq3 | (50) |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | (51)b |
| BY4741Δabz1 | MATaabz1Δ::kanMX4 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | (52)b |
a Dept. of Human Genetics, Columbia University, New York, NY.
b European Saccharomyces cerevisiae Archive for Functional Analysis (EUROSCARF), available on-line.
Radioactive and Stable Isotope Labeling
Radioactive compounds included p-hydroxy[U-14C]benzoic acid (450 mCi/mmol, 0.1 mCi/ml, American Radiolabeled Chemicals, Inc., St. Louis, MO), and p-amino[aromatic-14C]benzoic acid (57 mCi/mmol, 0.1 mCi/ml, Moravek Biochemicals, Brea CA). p-Amino[aromatic-13C6]benzoic acid ([13C6]pABA) or p-hydroxy[aromatic-13C6]benzoic acid ([13C6]4HB) were obtained from Cambridge Isotope Laboratories (Andover, MA). The manufacturers' analyses of the pure 13C-labeled aromatic ring compounds by GC-MS and NMR verified a better than 98% chemical purity with 99% isotopic enrichment. 13C6- and 14C-labeled aromatic ring precursors were also examined for purity by HPLC. Yeast cells were grown as described to deplete pABA and folate. Optical density (A) measurements (600 nm) were recorded for each sample at time of harvest, and the total A600 nm values were used to normalize lipid content. For radioactive labeling studies, the cells were suspended in 300–600 ml of fresh media as above (Dogal or Doge) and grown to an A600 nm of 0.6, then collected by centrifugation (1000 × g, 5 min) and resuspended in small volumes of fresh media (∼2 ml), containing 4 μCi, of the designated 14C-labeled ring precursor. Cells were collected by centrifugation as before, and pellets were stored at −20 °C. Stable isotopic labeling was essentially as above except that individual amounts of [13C6]pABA or [13C6]4HB were weighed and dissolved in DMSO (Sigma), and then added to a final concentration of 2–10 μg/ml.
Competition and Pulse Experiments
Time course and competition experiments with normal and stable isotope forms of precursors were conducted in 18- × 100-mm culture tubes. Cells were collected from a larger volume culture, and equivalent optical density amounts of yeast cells (between 20 and 100 A600 nm) were re-suspended in 5 ml of fresh media as described, which contained either the DMSO (vehicle control), [13C6]pABA, or [13C6]4HB at 10 μg/ml, or each of these plus an additional 10 μg/ml of the competing unlabeled precursor 4HB or pABA. These samples were prepared in duplicate or triplicate, and analyzed via mass spectrometry two or three times (n = 4–9). For pulse analyses cells were grown in large volume as described above, re-suspended in pre-warmed media (30 °C) to a total volume of 16 ml in a 125-ml flask, and incubated with shaking (250 rpm, 30 °C). Prior to addition of labeled ring precursors, two (1 ml) aliquots were removed, to represent a “no-label” control. [13C6]pABA was added (final concentration, 10 μg/ml), and further 1-ml aliquots were collected in duplicate at the time points as described. The time zero point was defined by removing two aliquots prior to addition of labeled precursors. The first time point (termed 2 min) represented the addition of label with mixing and immediate removal of two sequential samples, effectively allowing the label to be present for ∼1 min and 3 min, respectively.
Lipid Extraction
Yeast cell pellets were thawed on ice, and solvents were added as follows: 100 μl of H2O, 1 ml of methanol, and 2 ml of petroleum ether. Samples were vortexed for 30 s each. This was repeated, and then samples were centrifuged at 1000 × g to separate layers. The upper layer was moved to a new tube, 2 ml of petroleum ether was added to the lower phase, and the sample was vortexed. This upper phase was added to the previous upper organic phase, and the solvent was evaporated under N2 gas. Samples were routinely analyzed immediately after extraction. When Q or other intermediates were quantified, Q4 (Sigma) was added in a known amount (expected final concentration, 1 pmol/μl upon analysis) as internal standard to all samples, and to a simultaneously prepared and extracted calibration curve. Typical standard curve final concentrations were 0.2 fmol/μl, 1 fmol/μl, 25 fmol/μl, 200 fmol/μl, 1 pmol/μl, and 5 pmol/μl. The petroleum ether extracts were dried under nitrogen gas and resuspended in 200 μl of ethanol (USP, Aaper Alcohol and Chemical Co., Shelbyville, KY), in sample vials compatible for use with HPLC. Lipid extractions for the pulse experiments were similar, except that the cells were collected onto glass microfiber filter disks (Whatman) placed on a vacuum apparatus, and the collected cells and disks were immersed in ice-cold methanol (2 ml), containing 125 μl of 0.1% bromcresol green. Samples were stored in methanol at −20 °C. Q4 was added as an internal standard as described above, and samples were kept on ice during the extraction. Re-extraction with petroleum ether (3 ml) was repeated two times. For all quantitative analyses, the standard curve was prepared and analyzed along side the samples, with freshly prepared internal standard and analytes. Routinely, the low end of the standard curve was monitored for recovery of the analyte by comparison of these concentrations a non-extracted standard curve. To ensure that analytes in low concentration gave a reproducible response, a signal to noise threshold ratio was set at 5. If measured analytes did not meet these criteria, or if the standard integrations did not identify the peak of interest accurately, the data were not used.
RP-HPLC and Detection by Scintillation Counting
Detection of radioactive coenzyme Q and intermediates used a Β-ram model 2 (IN/US Systems, Inc., Tampa, FL) with a 500-μl flow cell. The column eluate was combined with Safety Solve (Research Products International Corp., Mount Prospect, IL) at a 2:1 ratio with a dwell setting of 1. Data from the device were collected into the Chem Station software supplied with the Agilent 1090 HPLC system. Simultaneous UV data were collected from the intrinsic diode array detector, 274 ± 4 nm, and 250 ± 4 nm. The system contained a Peltier cooled sample chamber maintained at 4 °C and a column oven set to 40 °C. A binary HPLC solvent delivery system was used with a phenyl-hexyl column (Luna 5μ, 100 × 4.60 mm, 5 μm, Phenomenex). The mobile phase consisted of Solvent A (methanol:isopropanol, 95:5, with 2.5 mm ammonium formate) to Solvent B (isopropanol, 2.5 mm ammonium formate) beginning at 100% Solvent A and over 8 min decreasing to 95%. The total run time was 11 min. Sample volumes for injection ranged from 40 to 70 μl, and the software was set to respond at 1 mV = 1 cpm.
RP-HPLC with Detection by MS
A 4000 QTRAP linear MS/MS spectrometer from Applied Biosystems (Foster City, CA) was used in combination with an Agilent Technologies 1200 HPLC system consisting of a PAL autosampler with thermostatted tray holders and Stack (LEAP Technologies, Carrboro, NC). Nitrogen was used for all gases in the mass spectrometer; the nitrogen gas was provided as boil-off from a bulk liquid nitrogen storage tank. Applied Biosystems software, Analyst version 1.4.2, was used for data acquisition and processing. Infusion experiments for tuning and optimization were performed with a model 11 plus syringe infusion pump (Harvard Apparatus, Inc., South Natick, MA). RP-HPLC separation was performed as described above. The 4000QT spectrometer was operated in Turbo electrospray positive mode. Q1 and Q3 were operated in unit resolution. For multiple reaction monitoring detection, the precursor-to-product ion transitions in multiple reaction monitoring mode were used to quantify Q and intermediates (m/z): 591.4/197 (Q6), 455.2/197 (Q4), 561.4/167.0 (DMQ6), and 546.4/150 (prenyl-pABA). Optimum positive turbospray conditions for coenzyme Q compounds: nebulizer gas, 50 psig; turbo gas, 60 psig; curtain gas, 20 psig; collision gas set to “medium”; nebulizer current, 20; and temperature, 450 °C. Optimal settings for compound-dependent parameters are in volts, and dwell is in milliseconds (data are declustering potential, entrance potential, collision energy, collision cell exit potential, and dwell): Q4 (75, 10, 29, 12, and 125), Q6 (111, 10, 37, 10, and 125), DMQ6 (96, 9, 37, 11, and 125), and prenyl-pABA (96, 9, 33, 11, and 125). The same settings were used for 13C-labeled forms. Settings as described are theoretical and based on differences required for the analyses of farnesylated standards (20), with a compensation for increased isoprene length. For ion trap detection, similar gases as above were used for Q and Q intermediates: nebulizer gas, 45 psig; turbo gas, 55 psig; curtain gas, 25 psig; collision gas, high; nebulizer current, 35; and temperature, 450 °C. The enhanced product ion scan used default dynamic settings for trap filling and other parameters. The mass spectrometer detection conditions also included an enhanced resolution scan with standard parameters, between m/z 520 and m/z 620. The injection volume was 10 or 20 μl. Stock solutions of the Q4 and Q6 (Sigma) were prepared in hexanes and stored under argon gas at −20 °C. Aliquots added to ethanol and the concentrations were then determined spectrophotometrically with a molar extinction coefficient of E = 14,900 at 275 nm (21). Integration of peak areas was performed with Analyst software, with a bunching factor of 1 and 3 smoothing events. Area ratios were constructed in Microsoft Excel for the calibration curve and experimental samples. The slope was calculated with a linear curve forced through zero. Standard deviations represent duplicate/triplicate samples, independently extracted with duplicate/triplicate injections (n = 4–6).
RESULTS
Prenyl-pABA Is a Naturally Occurring Lipid Component of Yeast Cells
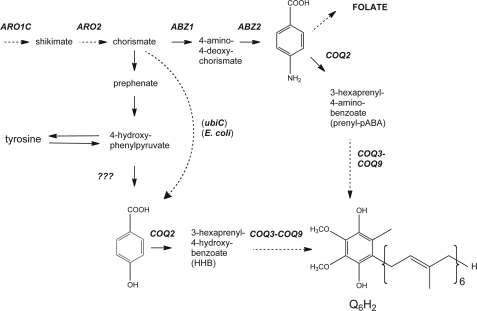
Neutral lipid extracts prepared from wild-type yeast cells cultured in standard rich media contain a lipid that we have identified as prenyl-pABA. The identification of prenyl-pABA was based on the presence of an HPLC peak with an elution similar to that for HHB, a previously characterized yeast Q intermediate (22). The precursor ion [M+H]+ of 546, and predominant tropylium [m/z = 150] and chromenylium [m/z = 190] product ions, detected in ion-trap analyses, were consistent with a ring amino replacing the ring hydroxyl present in HHB (Fig. 2A). The tropylium-like ion is a transition ion generated from prenylated aromatic and benzoquinone rings and is formed under dissociation conditions by incorporation of a methylene remnant (produced by fragmentation of the prenyl tail after the first carbon) to form a 7-membered ring (23). The chromenylium-like ion is larger in mass by +40 (C3H4) under these electrospray ionization conditions and is derived by fragmentation and cyclization to include the first four prenyl tail carbons (23).
FIGURE 2.
Detection of prenyl-pABA, DMQ6, and Q6 in lipid extracts of wild-type yeast cultured in the absence or presence of 13C6-pABA. Wild-type yeast (W303-1A) were pre-cultured in Dogal media (minus pABA and folate) as described under “Experimental Procedures.” Cells were harvested during mid-log from large cultures (300–600 ml of media; A600 nm = 0.6), resuspended in 2 ml of fresh media or fresh media containing 2–10 μg/ml [13C6]pABA (final concentration), incubated at 30 °C and harvested after 16 h. Lipid extracts were prepared and quinones and prenylated intermediates were subjected to RP-HPLC-ESI-MS/MS as described. A–F show product ion spectra: A, prenyl-pABA [M+H]+ precursor ion (C37H55NO2+; exact mass, 546.4) and the prenyl-pABA tropylium ion [M]+ (C8H8N1O2+; exact mass, 150.0); B, prenyl-[13C6]pABA [M+H]+ precursor ion (13C612C31H55NO2+; exact mass, 552.4) and the 13C6-prenyl-pABA tropylium ion [M]+ (13C612C2H8NO2+; exact mass, 156.0); C, DMQ6 [M+H]+ precursor ion (C38H56O3+; exact mass, 561.4) and the DMQ6 tropylium ion [M]+ (C9H11O3+; exact mass, 167.1); D, [13C6]DMQ6 [M+H]+ precursor ion (13C612C32H56O3+; exact mass, 567.4) and the [13C6]DMQ6 tropylium ion [M]+ (13C612C3H11O3+; exact mass, 173.1); E, Q6 [M+H]+ precursor ion (C39H58O4+; exact mass, 591.4) and the Q6 tropylium ion [M]+ (C10H13O4+; exact mass, 197.1); F, [13C6]Q6 [M+H]+ precursor ion (13C612C33H58O4+; exact mass, 597.4) and the [13C6]Q6 tropylium ion [M]+ (13C612C4H13O4+; exact mass, 203.1). Q6 (Sigma) and the lipid extract prepared from NM101 (coq7-1) yeast strains were used to establish the retention times of Q6 and DMQ6, respectively (29). A, C, and E show the structures of the compounds detected and indicate the identity of the predicted tropylium-like ion of the base peak (tallest peak) formed under collision associated dissociative conditions.
To confirm the identity of prenyl-pABA, wild-type yeast (W303-1A) were pre-cultured in Dogal media (minus pABA and folate) to deplete cellular stores of pABA (16). Either dextrose or galactose can be used as a fermentable carbon source in minimal media; galactose is used because it is non-repressing to aerobic respiration (24). Yeast cells were transferred to fresh Dogal medium plus folate, and then cultured in the presence of [13C6]pABA as described under “Experimental Procedures.” Product ion analyses from the crude lipids of yeast grown in the presence of [13C6]pABA show that yeast incorporate the ring carbons into prenyl-pABA to generate prenyl-[13C6]pABA (13C612C31H55NO2) (Fig. 2B). Although prenyl-pABA is readily detectable in lipid extracts of wild-type yeast (W303-1A harvested at 1.8 A in YPGal media contain 89.0 ± 5.6 fmol/A), it is much less abundant than Q6 (84.6 ± 4.3 pmol) under standard log phase growth conditions in rich media.
Yeast coq3 Mutants Cultured with 13C6-4HB Produce 13C6-labeled HHB
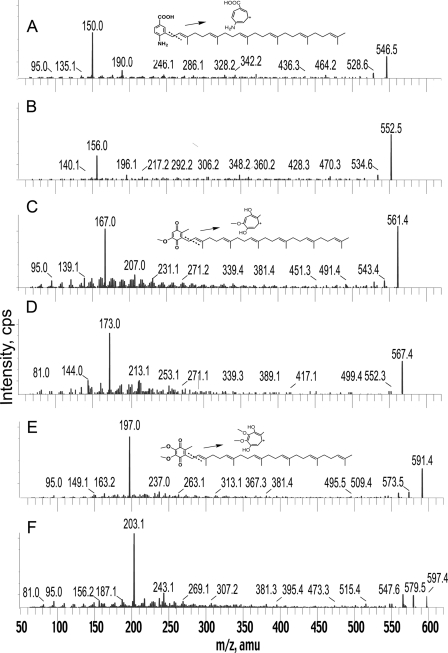
Previous work has shown that yeast coq mutants grown in the presence of 4HB produce HHB (25). However, this intermediate is unstable and difficult to detect without derivatization (22). For purposes of comparison, we wished to generate both the normal and 13C-labeled form of HHB. To do this, a coq3 yeast mutant was cultured in the presence of [13C6]4HB. The E2–249 coq3 mutant is Q-deficient and is a member of the G31 complementation group defined by Dieckmann and Tzagoloff (26, 27). As shown in Fig. 3 both the normal isotopic form of HHB and [13C6]HHB are detected in lipid extracts prepared from the coq3 mutant. Our product ion spectra match that described previously (28); of particular note is the fragmentation pattern showing the shift in mass from the precursor molecule (Fig. 3A) with an analogous series of fragments from the same compound in the separated crude lipids of [13C6]4HB-labeled coq3 point mutant (Fig. 3B). Both HHB compounds elute with exactly the same retention time. These results demonstrate that the precursor and product ions of prenyl-pABA are each one mass unit less than for HHB, the intermediate formed via the 4HB pathway; HHB [M+H]+ of 547; tropylium ion [m/z = 151] (compare Figs. 2A and 2B with 3A and 3B).
FIGURE 3.
Detection of HHB in lipid extracts of a yeast coq3 mutant cultured in the absence (A) or presence (B) of 13C6-4HB. The yeast coq3 mutant E2-249 was pre-cultured in Dogal media as described previously, harvested, and incubated in fresh media or fresh media with 10 μg/ml [13C6]4HB as described in Fig. 2. Lipid extracts were prepared and subjected to RP-HPLC-ESI-MS/MS as described. Product ion spectra are shown: A, HHB [M+ H]+ precursor ion (C37H54O3+; exact mass, 547.4) and the HHB tropylium ion [M]+ (C8H7O3+; exact mass, 151.0); B, [13C6]HHB [M+H]+ precursor ion (13C612C31H54O3+; exact mass, 553.4) and the [13C6]HHB tropylium ion (13C612C2H7O3+; exact mass, 157.0).
Yeast Cultured with [U-14C]pABA Produce 14C-Labeled Q6 and DMQ6
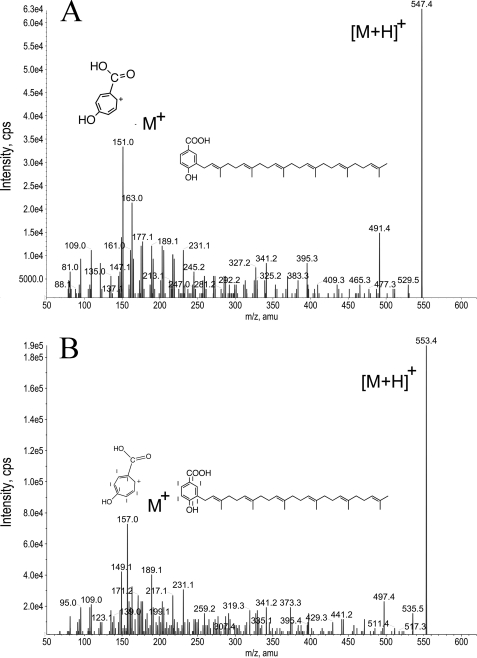
The identification of prenyl-pABA in yeast neutral lipid extracts led us to investigate whether pABA might serve as a ring precursor in yeast Q biosynthesis. We obtained [14C]4HB and [14C]pABA and determined that [ring-14C]pABA is free of detectable 4HB and vice versa (“Experimental Procedures”). S. cerevisiae wild-type cells (W303-1A), coq7-1 mutants (NM101), or coq2 mutants (W303ΔCOQ2), were pre-cultured in Dogal media (minus pABA and folate) to deplete cellular stores of pABA (16). Yeast cells were transferred to fresh Dogal medium plus folate, with the addition of either [14C]4HB or [14C]pABA (in each case the specific activity was adjusted to 50 Ci/mmol; 800 nm final concentration). Cells were incubated 24 h, lipid extracts were prepared and subjected to RP-HPLC, and the radioactivity was detected as described under “Experimental Procedures.” Yeast lipid extracts prepared from wild-type cells contained two peaks of 14C-radiolabeled material, consistent with the elution of Q6 and Q6H2 (the hydroquinone) (Fig. 4). Similarly, the yeast coq7-1 mutant NM101, when incubated with either [14C]4HB or [14C]pABA, produced radiolabeled material slightly more polar than Q6, previously identified as DMQ6 (29). The synthesis of [14C]Q6 under both experimental conditions requires the yeast COQ2 gene product (Fig. 4). These results indicate that DMQ6 and Q6 may be synthesized in yeast from either pABA or 4HB aromatic ring precursors, and both ring precursors require Coq2p for prenyl tail attachment.
FIGURE 4.
Yeast cultured with 14C-pABA or 14C-4HB produce 14C-labeled Q6 and DMQ6. Wild-type yeast (W303-1A), coq7-1 mutants (NM101), or coq2 mutants (W303ΔCOQ2), were pre-cultured in Dogal media (minus pABA, minus folate) to deplete cellular stores of pABA (16). Yeast cells were transferred to fresh Dogal medium plus folate, with the addition of either [14C]4HB or [14C]pABA (in each case specific activity was adjusted to 50 Ci/mmol; 800 nm final concentration). Cells were incubated 24 h, and lipid extracts were prepared and subjected to RP-HPLC and the radioactivity detected as described under “Experimental Procedures.” The bottom blue trace indicates the Q6 standard (12.2 min, 274 nm). Green and red traces show elution of 14C-radiolabeled material present in lipid extracts of wild-type yeast cultured (24 h) with either [14C]4HB or [14C]pABA, as indicated. The elution of [14C]Q6 at 12.35 min includes a time delay of 0.15 min between the UV detector and the BetaRam (Model 2 in-line scintillation) detector. Reduced [14C]Q6H2 eluted 2 min earlier at 11.3 min. Olive and Pink traces identify 14C-radiolabeled material eluting at 12.1 min as DMQ6, because the coq7-1 yeast mutant lacks Q6 and contains DMQ6 (29). The top two traces (dark green and purple) indicate the lack of incorporation of 14C precursors into coq2 null cells. The arrow designates the 14C-material co-migrating with DMQ6, present in the NM101 extracts. Chromatograms are not normalized; the amounts of [14C]DMQ6 produced in NM101 is 5- to 20-fold lower than the amount of [14C]Q6 present in wild-type cells.
Yeast Cultured with [13C6]pABA produce 13C6-labeled Prenyl-pABA, DMQ6, and Q6
The results obtained with the 14C-labeled precursors strongly suggest that pABA functions as a ring precursor in yeast Q6 biosynthesis. Metabolic labeling studies with stable isotopes provide a definitive test, because the ring carbons can be detected in both precursor and product ions by mass spectrometry. Wild-type yeast were first depleted of pABA and folate as described above and then cultured in the presence of [13C6]pABA as described under “Experimental Procedures.” The identification of hexaprenylated compounds in the crude lipids of wild-type yeast is represented by the spectra in Fig. 2. The normal (unlabeled) precursor ions monitored included: prenyl-pABA (Fig. 2A), demethoxy-Q6 (Fig. 2C), and Q6 (Fig. 2E). Product ion analyses from the crude lipids of yeast grown in the presence of [13C6]pABA show that the ring carbons of this compound are incorporated into demethoxy-Q6 and Q6 and alter the average isotopic masses of the tropylium-like ion and molecular ion by 6 mass units. This results in the following 13C6-labeled compounds in Fig. 2: prenyl-[13C6]pABA (13C612C31H55NO2) (Fig. 2B); 13C6-demethoxy-Q6 (13C612C32H56O3) (Fig. 2D); and [13C6]Q6 (13C612C33H58O4) (Fig. 2F). The incorporation of [13C6]pABA into the penultimate Q intermediate, DMQ6, was readily detected in wild-type yeast cell lipid extracts (Fig. 2D). Wild-type yeast cultured in the presence of [13C6]4HB also generate the expected +6 isotopically labeled forms of Q6 and DMQ6, however, under these labeling conditions, it was difficult to detect the +6 form of HHB (data not shown).
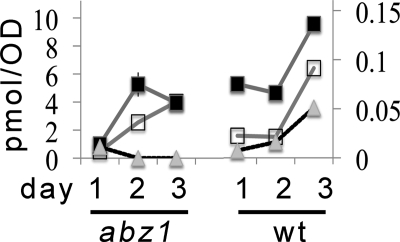
Prenyl-pABA Is Absent in Yeast abz1 Mutants Starved for pABA, Yet Content of Q Is Similar to That of Wild-type Yeast
Yeast produce pABA from chorismate by a two-step process that requires the ABZ1 and ABZ2 gene products (14, 16). Thus, it seemed likely that the production of prenyl-pABA would depend on the supply of pABA from this biosynthetic pathway, or from the pABA supplied in the media. To test this idea, wild-type yeast, or abz1 yeast mutants were serially cultured under conditions where the exogenous supply of pABA was eliminated (16). As shown in Fig. 5, abz1 mutants starved for pABA show dramatic decline in the content of prenyl-pABA, whereas the content of prenyl-pABA in wild-type cells remains unchanged or increased. These results indicate that a combination of the abz1 deletion and nutritional depletion of pABA results in the depletion of prenyl-pABA. Under these pABA-depleted conditions, the abz1 mutants are still able to produce DMQ6 and Q6. This is consistent with the presence of at least two pathways in yeast able to supply aromatic ring precursors for Q biosynthesis (Fig. 1).
FIGURE 5.
Nutritional and genetic depletion of pABA eliminate the formation of prenyl-pABA. BY4741 wild-type yeast and BY4741Δabz1 yeast mutants were grown overnight in YPD, and then diluted 1:100 (v/v) into fresh Dogal media minus pABA and folate and incubated for 1 day. The day 1 culture was used to inoculate fresh Dogal media (minus pABA and folate), and the process was repeated to generate the day2 and day 3 cultures. The serial dilution into pABA minus media exhausts endogenous stores of pABA (16). Cells were harvested, and lipid extracts were examined for content of Q6 (filled squares; left y axis), DMQ6 (open squares; right y axis), and prenyl-pABA (triangles; right y axis).
What Are the Relative Contributions of the pABA and 4HB Ring Precursors to Q Biosynthesis?
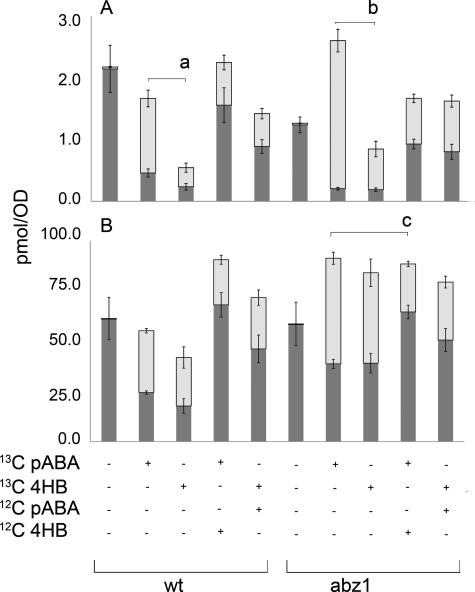
Once we recognized that [13C6]pABA could serve as a precursor to [13C6]Q6, we decided to investigate whether cells demonstrated a preference for pABA or 4HB as an aromatic ring precursor for Q. As expected, incubation of wild-type or abz1 mutant cells labeled for 3.5 h with a single designated precursor show that either [13C6]pABA or [13C6]4HB serve as ring precursors in biosynthesis of DMQ6 (Fig. 6A) and Q6 (Fig. 6B); the darkly shaded lower section of each column designates the amount of the [12C]quinone, and the upper light gray bar shows the amount of [13C6]quinone. We then performed competition experiments to examine the ability of the normal isotopic 12C form of the alternative precursor to diminish the incorporation of the 13C-form into [13C6]DMQ6 or [13C6]Q6 (Fig. 6). For these competitor experiments, cells were incubated (3.5 h) in the presence of equivalent concentrations of the alternative ring precursor (10 μg/ml). In wild-type cells, the presence of the competitor did not substantially affect the amount of [13C6]Q6 content. Indeed, total levels of Q6 were slightly higher in wild-type cells incubated with both aromatic ring precursors concurrently. In abz1 mutant cells, the total content of Q6 was increased (relative to the no-addition control) by addition of any combination of aromatic ring precursors, and the presence of added 12C competitor significantly decreased the amount of [13C6]Q6 produced from the 13C-aromatic ring precursor. It seems likely that the presence of both ring precursors better satisfies the requirement for both folate and Q.
FIGURE 6.
4HB and pABA are mutually incorporated into Q6, but differentially incorporated into DMQ6. Wild-type (W303-1A) or abz1 null (W303ΔABZ1) cells were serially cultured to deplete them of pABA and folinic acid as described. The depleted cells were harvested and used to inoculate a 4-ml culture. This was divided to supply duplicate samples of cells to five different media conditions. The media contained either no addition or 10 μg/ml of the following: [13C6]pABA, [13C6]4HB, or similar cultures in the presence of equivalent concentrations of the alternative unlabeled (12C) ring precursor (10 μg/ml). Independent replicates were made of each condition. The amount of the competing 12C form of precursor (ratio of 13C:12C) was 1:1. After incubation for 3.5 h, cells were harvested, and lipid extracts were subjected to analyses by RP-HPLC-ESI/MS-MS. A, DMQ6 content; B, Q6 content. The content of [12C]DMQ6 or [12C]Q6 is designated by the darkly shaded lower section of each bar (in A and B, respectively), and [13C6]DMQ6 and [13C6]Q6 by the upper light gray bar (in A and B, respectively). Each condition was cultured as an independent replicate, and each lipid extract was injected twice (n = 4). This experiment is representative of three others, conducted independently. The brackets with letters denote the values being compared: Content of [13C6]DMQ6 is significantly different in wild-type cells cultured in [13C]pABA as compared with [13C]4HB (a, p = 0.0002); content of [13C6]DMQ6 is significantly different in abz1 mutant cells cultured in [13C]pABA as compared with [13C]4HB (b, p = 0.0002); and content of [13C6]Q6 is significantly different in in abz1 mutant cells cultured in [13C]pABA as compared with [13C]pABA plus unlabeled 4HB (c, p = 0.0001).
Although the different ring precursors had only modest effects on the amount of Q6 formed, they had dramatic effects on the content of DMQ6. For example, both wild-type yeast and abz1 mutants had increased content of DMQ6 when incubated with pABA as compared with 4HB. In fact abz1 mutants incubated with [13C6]pABA produced a high content of DMQ6 of which almost all is [13C6]DMQ6 (Fig. 6A). We speculate that prenyl-pABA, or more likely a subsequent intermediate derived from prenyl-pABA, might act to inhibit the hydroxylation of DMQ.
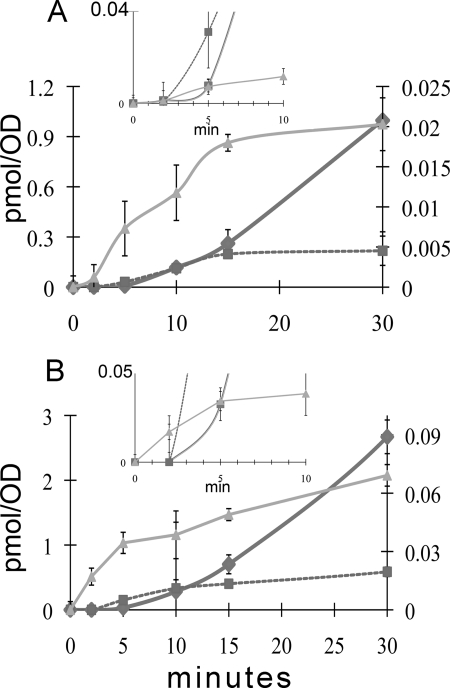
Prenylation of pABA Precedes Biosynthesis of DMQ and Q from pABA
A pulse-labeling experiment was conducted to determine whether the incorporation of 13C6-ring carbons into 13C6-prenyl-pABA preceded the formation of [13C6]DMQ6 and [13C6]Q6. Wild-type cells and abz1 yeast mutants were pre-cultured as described in Fig. 6 and incubated with [13C6]pABA over a time course from 0 to 30 min, as described in Fig. 7 and “Experimental Procedures.” Samples were removed from the incubation at stated times, collected by filtration, and quenched, and lipid extracts were analyzed to determine the amounts of prenyl-pABA, DMQ6, and Q6 by HPLC-MS/MS and multiple reaction monitoring. In both wild-type and abz1 mutant yeast, prenyl-[13C6]pABA is detectable within a minute of label addition (see inset, Fig. 7), and its de novo synthesis precedes that of [13C6]DMQ6 and [13C6]Q6, consistent with the notion of a precursor-product relationship.
FIGURE 7.
Time course assays with 13C6-pABA. A, wild-type (W303-1A) or B, abz1 null (W303ΔABZ1) cells were pre-cultured as described to deplete endogenous pABA, resuspended in pre-warmed Dogal media (30 °C) to a total volume of 16 ml in a 125-ml flask, and incubated with shaking (250 rpm, 30 °C). Prior to addition of ring precursors, two (1 ml) aliquots were removed, to represent a “no-label or zero time” control. [13C6]pABA was added (final concentration, 10 μg/ml), and further 1-ml aliquots were collected in duplicate at the time points indicated on the glass filter disks for lipid extraction: no label control (defines the “zero” time point), 2, 5, 10, 15, or 30 min. Lipid extracts were subjected to analyses by RP-HPLC-ESI/MS-MS as described, and the amount of de novo prenyl-[13C6]pABA (triangles), [13C6]DMQ6 (squares), and [13C6]Q6 (diamonds) is depicted. The left y axis represents the picomoles/A (for plots of [13C6]DMQ6 and [13C6]Q6), whereas the right y axis shows the content in picomoles/A for prenyl-[13C6]pABA. Values are plotted as average ± S.D. (n = 4, wild type; n = 6 abz1 null). The insets in each panel depict early time points (in minutes) for all three components plotted on the same scale (pmol/A).
pABA-replete Yeast Produce 4-Imino-DMQ6
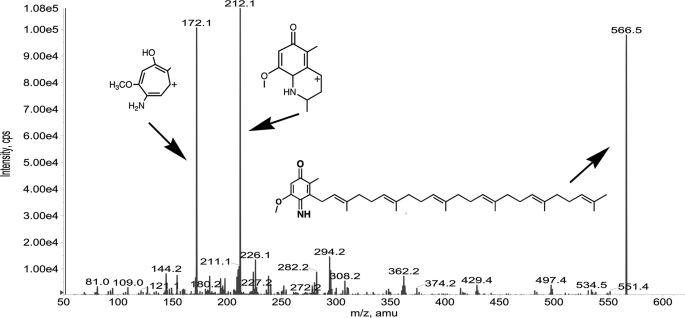
In the crude lipid extracts of wild-type cells grown in media supplemented with [13C6]pABA, we identify what appears to be a nitrogen containing form of 4-imino-DMQ6 (Fig. 8). The tropylium, chromenylium, and molecular ion are all shifted in accordance with M+6 (m/z), as is detected for DMQ6 (Fig. 2D); however, the masses of these ions are reduced by 1 Da relative to the fragment ion masses of DMQ6, consistent with an imino nitrogen rather than a quinone oxygen atom in the para ring position. The chromenylium ion fragment of 4-imino-DMQ6 is abundant, because it is likely to be more stable to dissociative conditions due to the interaction of the first four carbons of the prenyl tail with the imino nitrogen forming a second ring (23). The normal isotopic form of 4-imino-DMQ6 is present in crude lipid extracts of strains grown in YPD (data not shown).
FIGURE 8.
Identification of 4-imino-DMQ6 in lipid extracts of wild-type cells. An imino form of [13C6]DMQ6 is present in wild-type yeast cultured in the presence of [13C6]pABA and is consistent with the structures as shown. The imino substituent forms a more stable interaction with isoprene proximal to the ring, creating more stable and abundant chromenylium ion upon collision associated dissociation: m/z 212. Masses detected at m/z 497.4, 429.4, 362.2 and 294.2 are characteristic of the loss of successive isoprene units (m/z 68). This compound can be detected in wild-type strains grown in rich media and appears to be a normally accumulating form of demethoxy-Q6 (see Fig. 9). [13C6]imino-DMQ6 [M+H]+ precursor ion (13C612C32H56NO2+; exact mass, 566.4), the [13C6]imino-DMQ6 tropylium ion [M]+ (13C612C3H11NO2+; exact mass, 172.2), and the [13C6]imino-DMQ6 chromenylium ion [M]+ (13C612C6H16NO2+; exact mass, 212.3).
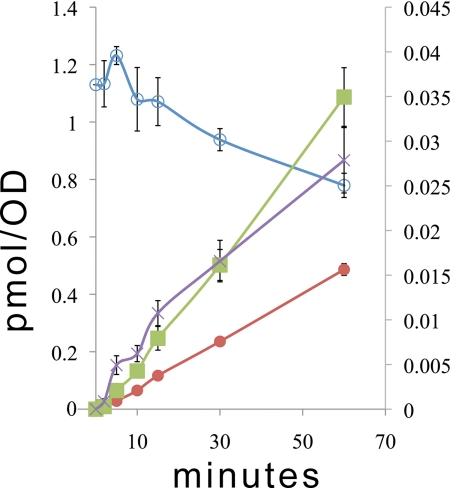
The detection of 4-imino-DMQ6 led us to test whether it might function as an intermediate in Q6 biosynthesis. To simplify the precursor-product relationship between 4-imino-DMQ6 and DMQ6, we examined the synthesis of both as a function of time in NM101 yeast, a strain known to be blocked in Q biosynthesis and to produce DMQ6 due to a mutation in COQ7 (29). The pulse labeling with [13C6]pABA was performed over a time course of 0 to 60 min, and the content of 13C-labeled intermediates (prenyl-[13C6]pABA, [13C6]imino-DMQ6, and [13C6]DMQ6) were determined (Fig. 9). The rate of formation of prenyl-[13C6]pABA and [13C6]imino-DMQ6 were very similar, suggesting that both serve as biosynthetic precursors in the synthesis of DMQ6. We note that DMQ6 is relatively unstable in the coq7-1 mutant, as is evident from the rate of loss of the [12C]DMQ6. This rate of loss seems to be compensated by the formation of [13C6]DMQ6. These results suggest that the nitrogen substituent of pABA may be retained up to the step generating 4-imino-DMQ6, and are consistent with a possible mechanism where oxygen from a water-based hydroxyl could replace the nitrogen imino via Schiff base chemistry (Fig. 10).
FIGURE 9.
Time course assays with 13C6-pABA examine the formation novel Q intermediates in NM101 (coq7-1) yeast. NM101 yeast cells were pre-cultured and analyzed by pulse-labeling with [13C6]pABA as described in Fig. 7. Lipid extracts were subjected to analyses by RP-HPLC-ESI/MS-MS as described, and the amount of de novo prenyl-[13C6]pABA (green squares), [13C6]DMQ6 (red circles), and [13C6]imino-DMQ6 (purple cross) is depicted. Also depicted is the decay of non-isotopically labeled DMQ6 (blue open circles). The right y axis represents the pmol/A (for plots of prenyl-[13C6]pABA and [13C6]imino-DMQ6), whereas the left y axis shows the content in pmol/A for DMQ6 and [13C6]DMQ6. Values are plotted as average ± standard deviations (n = 4).
FIGURE 10.
Scheme for generation of 4-imino-DMQ6 and loss of nitrogen generating DMQ6. The pulse-labeling studies in Figs. 7 and 9 suggest that prenyl-pABA and 4-imino-DMQ6 are bona fide precursors in yeast Q synthesis from pABA. A possible mechanism for replacement of the imino nitrogen with a hydroxy substituent via Schiff base chemistry is proposed. The new steps of oxidation, de-imination, and reduction allow the two pathways to converge at formation of DMQ6H2.
DISCUSSION
Schemes of Q biosynthesis in E. coli, yeast, and animals universally depict 4HB as the aromatic ring precursor. In each of these species, isoprenylation of 4HB is thought to represent a committed step in Q biosynthesis. Here we show that S. cerevisiae can also utilize pABA as a ring precursor in Q biosynthesis. This is a surprising finding, because pABA is a crucial intermediate in folate biosynthesis. It is also surprising because the addition of pABA to either E. coli or human cells causes a concentration-dependent inhibition of Q biosynthesis (30–33). In E. coli, rat, and human cells, the pABA ring competes with 4HB at the ring:polyprenyltransferase step (catalyzed by Coq2), and the product prenyl-pABA appears to be a dead-end product. Recently another aromatic ring inhibitor, 4-nitrobenzoic acid, was shown to inhibit Q biosynthesis in mammalian cells through its competition with 4HB for Coq2 (34). Thus it appears that several benzoic acid ring analogs function as competitive inhibitors of Q biosynthesis in mammalian cells (33, 34).
In contrast, our studies identify prenyl-pABA, a normal metabolite present in lipid extracts of wild-type yeast, as a Q intermediate. The synthesis of prenyl-pABA depends on Coq2 (Fig. 4), and we show that prenyl-pABA is a normal metabolite present in lipid extracts of wild-type yeast cultured in standard yeast media (Fig. 2). An interesting independent confirmation of prenyl-pABA in yeast neutral lipid extracts was recently published, found in lipid extracts of yeast with defects in ferredoxin (YAH1) and ferredoxin reductase (ARH1) (18). These authors discovered the role of pABA in Q biosynthesis through their analyses of iron chaperones required for the activity of Coq7, the Coq diiron enzyme required for the last ring hydroxylation in Q biosynthesis (29, 35).
Our work clearly explores the biochemical relationship of prenyl-pABA in yeast Q biosynthesis, demonstrating prenyl-pABA is a bona fide biosynthetic precursor. pABA can be prenylated immediately following its addition to cells. Pulse labeling studies with [13C6]pABA show that pABA-deficient abz1 null yeast mutants synthesize prenyl-[13C6]pABA within 1 min of incubation (Fig. 7). This rapid incorporation of the isotope into prenyl-pABA precedes the formation of [13C6]DMQ6 and [13C6]Q6 and is consistent with a precursor-product relationship. Yeast abz1 mutants, known to be defective in pABA biosynthesis (15, 16), become depleted in prenyl-pABA when cultured in pABA-free media (Fig. 5). However, under these culture conditions Q is still produced, reflecting use of an alternate ring precursor, 4HB (Fig. 1). Prenylation of 4HB by Coq2 is the “classic” arm of the pathway. We also identify for the first time the underivatized forms of HHB, detected in a coq3 yeast mutant.
Identification of 4-Imino-DMQ6 and a Model for Nitrogen Loss
Both wild-type and abz1 yeast null mutants when pre-cultured under conditions to deplete pABA, followed by growth with pABA supplementation, have a higher DMQ6 content as compared with wild-type yeast or abz1 mutants supplemented with 4HB (Fig. 6). In isotopic labeling studies with the abz1 mutant >90% of the DMQ6 detected contains the 13C6-aromatic ring from pABA. The preferential incorporation of pABA into DMQ6 is particularly intriguing in light of our finding that 4-imino-DMQ6, a lipid derived from prenyl-pABA, is present in the pABA-supplemented abz1 mutant and in normally cultured wild-type cells (Fig. 8). The rate of formation of 4-imino-DMQ6 is similar to that of prenyl-pABA (Fig. 9), identifying it as a new Q intermediate and indicating that the nitrogen of pABA can potentially be retained up to the step preceding DMQ6.
If this is the case, what is the fate of 4-imino-DMQ6? We speculate 4-imino-DMQ6 is produced from the two-electron oxidation of 4-amino-DMQ6H2 (Fig. 10). Once the imino-quinone is formed, the potential for loss of the ring nitrogen as ammonia and its replacement by oxygen from a water or hydroxide ion to form the quinone could reasonably occur by Schiff base chemistry. Although we designate the ring nitrogen loss step as immediately preceding DMQ6, it is possible the loss could occur earlier. However, we favor the depiction in Fig. 10, because DMQ6 is a relatively abundant Q intermediate, detected even in wild-type cells (36, 37), and is also a component of a Q biosynthetic complex (38). It seems possible that the enhanced accumulation of DMQ6 in cells first starved for and then supplemented with pABA may be due to inhibition of the Coq7 monooxygenase step (Fig. 10). However, the effect may also be indirect, as the influx of pABA would also replete folate synthesis. We note that the redox state of mitochondria might very well affect the relative rates of oxidation and reduction of amino-DMQ6H2 to imino-DMQ6, and DMQ6 to DMQ6H2.
Which Ring Precursor, 4HB or pABA, Is Normally Preferred?
Our labeling with 13C-aromatic ring precursors is accomplished by lowering the supply of pABA and folate to cells with deficient media prior to isotopic incorporation. Thus, at present we cannot differentiate the relative importance of each ring precursor (4HB or pABA) for Q6 production in wild-type cells, under physiological conditions. It remains possible that pABA could be converted to 4HB prior to lipidation in mitochondria. However, the competition experiments (Fig. 6) suggest that, when one 13C-ring precursor is provided to yeast cells in a one to one molar ratio with the other unlabeled ring, pABA and 4HB are indistinguishable for [13C]Q synthesis. In addition, if [13C6]pABA was converted into [13C6]4HB prior to its prenylation by mitochondrial Coq2, then the enhanced accumulation of [13C6]DMQ6 from the [13C6]pABA labeling relative to that of [13C6]4HB (Fig. 6A) would be unlikely. Although the abz1 null avidly incorporates and synthesizes [13C]demethoxy from [13C]pABA, our data do not suggest that pABA is a better source for demethoxy Q6 in normal yeast. Media conditions, for example carbon source and nitrogen source(s), may radically alter ring precursor preference. Finally, we note that our conclusions regarding the incorporation of 4HB and pABA into Q6 are valid only if the uptake of 4HB and pABA are identical.
It is likely but uncertain that 4HB and pABA may share some mechanisms of uptake and transport of the free form into mitochondria. pABA and 4HB are weakly ionic compounds (pABA: pKa 4.9; 4HB: pKa 4.67 (39)), and their uptake and retention has a pH-dependent component. Uptake is favored at low pH, and the formation of the carboxylate anion once imported into the cell favors retention. Although the pH of our media is 6.0, which has been shown to relieve pABA growth inhibition in S. cerevisiae (40), other work has shown pABA uptake cannot be saturated (41). The studies of inhibition of yeast growth by pABA are intriguing (40), and are different from E. coli growth inhibition by pABA. E. coli have ubiC encoded chorismate lyase, which directly converts chorismate to 4HB (9) (Fig. 1), whereas yeast lack this homolog. E. coli can be growth inhibited by excess pABA, and relief is accomplished by large concentrations of 4HB (31). However, rescue of growth inhibition by pABA in S. cerevisiae requires aromatic amino acids, with phenylalanine and tyrosine in combination to best resume growth (40).
What Are the Pathway(s) to 4HB Production?
Radiolabeled cinnamic and coumaroyl acids produce radiolabeled Q when fed to bakers' yeast (42), with coumarate being the best precursor, and both these compounds are shown to precede 4HB. Other work describes the ability of radiolabeled aromatic amino acids to donate their carbons to Q in yeast (43), however the intermediary compounds have not been described. In other microorganisms the direct precursors for 4HB have been examined more recently and thoroughly. An alternative is described in plants (44), where phenylalanine is a probable precursor, although S. cerevisiae lacks an identifiable phenylalanine ammonia lyase homolog. The Acinetobactor baylyi system describes the production of 4HB precursors from the catabolism of primarily plant cell wall components into hydroxycinnamate precursors (13).
Do Anti-folates Target Both Folate and Q?
Studies of chorismate synthesis have been stimulated by investigations of drug resistance in microorganisms and pABA metabolism. Depletion of folate is difficult, because it is recycled as a cofactor and yeast carry reserves of both folate and pABA. Yeast cells must be serially cultured in media depleted of these nutrients to elicit a pABA or folate growth deficiency (16). A pABA deficiency is also accomplished by inclusion of sulfanilamide in the media, because sulfanilamide acts as a competitive pABA analog (14). The discovery of sulfanilamide antibiotics hinges on the ability of this class of compound to interfere with the condensation of pABA to pteroglutamoyl for the synthesis of folate (19). The enzyme dihydropteroate synthase is the target of the pABA analog sulfamethoxazole, within the “sulfa” drug class. Previous work by Macreadie's group on resistance to this drug showed resistance depended on expression of the dihydropteroate synthase homolog in yeast (FOL1), as long as pABA was supplied in the media (45).
This report describes a novel and physiologically relevant, lipidated form of pABA in yeast, establishing a possible linkage between Q biosynthesis and folate metabolism. Although direct relationships between coenzyme Q biosynthesis and folate metabolism have not been characterized until now, relationships are known to exist between folate synthesis and sulfa drug resistance. A storage form of pABA in microorganisms satisfies the logistics for folate synthesis, just as a glucosylated form of pABA functions in plants (41, 45, 46). It would be undesirable for a crucial metabolic intermediate such as pABA to fall into low supply, but large quantities of the free acid may act as uncouplers to the electrochemical gradient (47). Larger amounts of pABA may also act as a substrate inhibitor of dihydropteroate synthase, the enzyme that couples pABA to a pterin moiety during folate synthesis. Although inhibition occurs at micromolar levels of pABA for the bacterial dihydropteroate synthase homolog (48), its inhibition by high amounts is not complete and it retains much lower but steady activity.
In summary, our analyses document pABA and 4HB as two aromatic rings that serve as precursors for DMQ6 and Q6. The observation of a normally produced imino form of DMQ found in the crude lipid extract of wild-type cells (Fig. 8), and its similar rate of formation as compared with prenyl-pABA in a coq7-1 point mutant (Fig. 9), suggest that both are novel Q intermediates. Based on the 4-imino-DMQ6 intermediate, we suggest a mechanism for the loss of the pABA-derived nitrogen. Finally the findings presented here suggest an intimate relationship may exist between synthesis of folic acid, necessary for many cellular essential functions and cellular respiration in S. cerevisiae, coordinated through the overlapping substrate prenyl-pABA.
Acknowledgments
We thank John Greaves, University of California, Irvine, for his insight and advice about the presence of nitrogen in our unknown that was 1 Da less than HHB. We also thank Jennifer Shepherd, Gonzaga University, for discussions regarding aromatic compounds containing nitrogen substituents, and Jane Strouse, Chemistry and Biochemistry at UCLA, for assistance with mass spectrometry. We thank the C.F. Clarke laboratory group members for discussions and input.
This work was supported, in whole or in part, by National Institutes of Health Grant GM4592 (to C. F. C.). This work was also supported by National Science Foundation Grant 0919609 (to C. F. C. and B. M.). UCLA mass spectrometry resources were funded by a NIH grant 4000QT for mass spectrometry resources and Grant S10RR024605 from the National Center For Research Resources.
- Q
- coenzyme Q
- [13C6]4HB
- p-hydroxy[aromatic-13C6]benzoic acid
- [13C6]pABA
- p-amino[aromatic-13C6]benzoic acid
- DMQ
- demethoxy-Q
- ESI-MS/MS
- electrospray ionization mass spectrometry
- HHB
- 3-hexaprenyl-4-hydroxybenzoic acid
- 4HB
- 4-hydroxybenzoic acid
- 4-imino-DMQ6
- 3-hexaprenyl-4-imino-5-methoxy-2-methylcyclohexa-2,5-dienone
- pABA
- 4-aminobenzoic acid
- prenyl-pABA
- 3-hexaprenyl-4-aminobenzoic acid
- psig
- pound-force per square inch gauge
- Q6
- coenzyme Q6 with a tail of 6 isoprene units
- Q6H2
- reduced or hydroquinone form of Q6
- RP
- reverse phase.
REFERENCES
- 1.Lenaz G., De Santis A. (1985) in Coenzyme Q (Lenaz G. ed) pp. 165–199, Wiley & Sons, Chichester, UK [Google Scholar]
- 2.Bentinger M., Brismar K., Dallner G. (2007) Mitochondrion 7, (suppl.) S41–S50 [DOI] [PubMed] [Google Scholar]
- 3.Meganathan R. (2001) FEMS Microbiol. Lett. 203, 131–139 [DOI] [PubMed] [Google Scholar]
- 4.Kawamukai M. (2009) Biotechnol. Appl. Biochem. 53, 217–226 [DOI] [PubMed] [Google Scholar]
- 5.Tran U. C., Clarke C. F. (2007) Mitochondrion 7, (suppl.) S62–S71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson R. E., Rudney H. (1983) Vitam. Horm. 40, 1–43 [DOI] [PubMed] [Google Scholar]
- 7.Pennock J. F., Threlfall D. R. (1983) in Biosynthesis of Isoprenoid Compounds (Porter J. W., Spurgeon S. L. eds) pp. 191–302, John Wiley and Sons, New York [Google Scholar]
- 8.Olson R. E. (1966) Vitam. Horm. 24, 551–574 [DOI] [PubMed] [Google Scholar]
- 9.Siebert M., Severin K., Heide L. (1994) Microbiology 140, 897–904 [DOI] [PubMed] [Google Scholar]
- 10.Dosselaere F., Vanderleyden J. (2001) Crit. Rev. Microbiol. 27, 75–131 [DOI] [PubMed] [Google Scholar]
- 11.Cox G. B., Gibson F., Pittard J. (1968) J. Bacteriol. 95, 1591–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goewert R. R. (1981) PhD thesis, Studies on the Biosynthesis of Ubiquinone: The Identification of 3,4-Dihydroxy-5-hexaprenylbenzoic Acid, 3-Methoxy-4-hydroxy-5-hexaprenylbenzoic Acid and the Regulation of the Conversion of Tyrosine and Chorismic Acid to Ubiquinone in Saccharomyces cerevisiae, University of St. Louis, St. Louis, MO [Google Scholar]
- 13.Young D. M., Parke D., Ornston L. N. (2005) Annu. Rev. Microbiol. 59, 519–551 [DOI] [PubMed] [Google Scholar]
- 14.Edman J. C., Goldstein A. L., Erbe J. G. (1993) Yeast 9, 669–675 [DOI] [PubMed] [Google Scholar]
- 15.Castelli L. A., Nguyen N. P., Macreadie I. G. (2001) FEMS Microbiol. Lett. 199, 181–184 [DOI] [PubMed] [Google Scholar]
- 16.Botet J., Mateos L., Revuelta J. L., Santos M. A. (2007) Eukaryot. Cell 6, 2102–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Güldener U., Koehler G. J., Haussmann C., Bacher A., Kricke J., Becher D., Hegemann J. H. (2004) Mol. Biol. Cell 15, 3811–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Mühlenhoff U., Ozeir M., Lill R., Fontecave M. (2010) Chem. Biol. 17, 449–459 [DOI] [PubMed] [Google Scholar]
- 19.Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brajcich B. C., Iarocci A. L., Johnstone L. A., Morgan R. K., Lonjers Z. T., Hotchko M. J., Muhs J. D., Kieffer A., Reynolds B. J., Mandel S. M., Marbois B. N., Clarke C. F., Shepherd J. N. (2010) J. Bacteriol. 192, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lester R. L., Crane F. L. (1959) J. Biol. Chem. 234, 2169–2175 [PubMed] [Google Scholar]
- 22.Poon W. W., Marbois B. N., Faull K. F., Clarke C. F. (1995) Arch. Biochem. Biophys. 320, 305–314 [DOI] [PubMed] [Google Scholar]
- 23.Elliot W. H., Waller G. R. (1972) in Biochemical Applications of Mass Spectrometry (Waller G. R. ed) pp. 499–536, Wiley-Interscience, New York [Google Scholar]
- 24.Carlson M. (1999) Curr. Opin. Microbiol. 2, 202–207 [DOI] [PubMed] [Google Scholar]
- 25.Poon W. W., Do T. Q., Marbois B. N., Clarke C. F. (1997) Mol. Aspects Med. 18, (suppl.) S121–S127 [DOI] [PubMed] [Google Scholar]
- 26.Tzagoloff A., Dieckmann C. L. (1990) Microbiol. Rev. 54, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashby M. N., Edwards P. A. (1990) J. Biol. Chem. 265, 13157–13164 [PubMed] [Google Scholar]
- 28.Young I. G., Stroobant P., Macdonald C. G., Gibson F. (1973) J. Bacteriol. 114, 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marbois B. N., Clarke C. F. (1996) J. Biol. Chem. 271, 2995–3004 [DOI] [PubMed] [Google Scholar]
- 30.Alam S. S., Nambudiri A. M., Rudney H. (1975) Arch. Biochem. Biophys. 171, 183–190 [DOI] [PubMed] [Google Scholar]
- 31.Davis B. D. (1951) J. Exp. Med. 94, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton J. A., Cox G. B. (1971) Biochem. J. 123, 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turunen M., Olsson J., Dallner G. (2004) Biochim. Biophys. Acta 1660, 171–199 [DOI] [PubMed] [Google Scholar]
- 34.Forsman U., Sjöberg M., Turunen M., Sindelar P. J. (2010) Nat. Chem. Biol. 6, 515–517 [DOI] [PubMed] [Google Scholar]
- 35.Stenmark P., Grünler J., Mattsson J., Sindelar P. J., Nordlund P., Berthold D. A. (2001) J. Biol. Chem. 276, 33297–33300 [DOI] [PubMed] [Google Scholar]
- 36.Trumpower B. L., Aiyar A. S., Opliger C. E., Olson R. E. (1972) J. Biol. Chem. 247, 2499–2511 [PubMed] [Google Scholar]
- 37.Padilla S., Tran U. C., Jiménez-Hidalgo M., López-Martin J. M., Martín-Montalvo A., Clarke C. F., Navas P., Santos-Ocaña C. (2009) Cell Mol. Life Sci. 66, 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marbois B., Gin P., Faull K. F., Poon W. W., Lee P. T., Strahan J., Shepherd J. N., Clarke C. F. (2005) J. Biol. Chem. 280, 20231–20238 [DOI] [PubMed] [Google Scholar]
- 39.Dawson R. M. (1986) Data for Biochemical Research, 3rd ed., pp. 42 and 116, Clarendon Press, Oxford [Google Scholar]
- 40.Reed L. J., Schram A. C., Loveless L. E. (1959) J. Biol. Chem. 234, 904–908 [PubMed] [Google Scholar]
- 41.Quinlivan E. P., Roje S., Basset G., Shachar-Hill Y., Gregory J. F., 3rd, Hanson A. D. (2003) J. Biol. Chem. 278, 20731–20737 [DOI] [PubMed] [Google Scholar]
- 42.Threlfall D. R., Law A., Whistance G. R. (1970) Biochem. J. 118, 55P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parson W. W., Rudney H. (1964) Proc. Natl. Acad. Sci. U.S.A. 51, 444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sircar D., Mitra A. (2009) J. Plant Physiol. 166, 1370–1380 [DOI] [PubMed] [Google Scholar]
- 45.Iliades P., Berglez J., Meshnick S., Macreadie I. (2003) Microb. Drug Resist. 9, 249–255 [DOI] [PubMed] [Google Scholar]
- 46.Eudes A., Bozzo G. G., Waller J. C., Naponelli V., Lim E. K., Bowles D. J., Gregory J. F., 3rd, Hanson A. D. (2008) J. Biol. Chem. 283, 15451–15459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen F. S., Eisenberg M., McLaughlin S. (1977) J. Membr. Biol. 37, 361–396 [DOI] [PubMed] [Google Scholar]
- 48.Hampele I. C., D'Arcy A., Dale G. E., Kostrewa D., Nielsen J., Oefner C., Page M. G., Schonfeld H. J., Stüber D., Then R. L. (1997) J. Mol. Biol. 268, 21–30 [DOI] [PubMed] [Google Scholar]
- 49.Ashby M. N., Kutsunai S. Y., Ackerman S., Tzagoloff A., Edwards P. A. (1992) J. Biol. Chem. 267, 4128–4136 [PubMed] [Google Scholar]
- 50.Tzagoloff A., Akai A., Needleman R. B. (1975) J. Biol. Chem. 250, 8228–8235 [PubMed] [Google Scholar]
- 51.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 52.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Science 285, 901–906 [DOI] [PubMed] [Google Scholar]