Abstract
Chaperonins are large protein complexes consisting of two stacked multisubunit rings, which open and close in an ATP-dependent manner to create a protected environment for protein folding. Here, we describe the first crystal structure of a group II chaperonin in an open conformation. We have obtained structures of the archaeal chaperonin from Methanococcus maripaludis in both a peptide acceptor (open) state and a protein folding (closed) state. In contrast with group I chaperonins, in which the equatorial domains share a similar conformation between the open and closed states and the largest motions occurs at the intermediate and apical domains, the three domains of the archaeal chaperonin subunit reorient as a single rigid body. The large rotation observed from the open state to the closed state results in a 65% decrease of the folding chamber volume and creates a highly hydrophilic surface inside the cage. These results suggest a completely distinct closing mechanism in the group II chaperonins as compared with the group I chaperonins.
Keywords: Alzheimer Disease, Amyloid, Chaperone Chaperonin, Protein Folding, X-ray Crystallography
Introduction
Chaperonins play an essential role in ensuring efficient folding of newly translated or stress-denatured proteins (1). The cellular accumulation of misfolded protein has been associated with several diseases, including amyloid diseases and cancer (2, 3). This strong relationship between incorrectly folded protein and pathological states emphasizes the importance of understanding in vivo folding mechanisms (1).
Chaperonins consist of two stacked rings of seven to nine subunits each, creating a large cylindrical protein complex of ∼1 MDa. The subunit architecture comprises three distinct domains: an equatorial domain that includes the ATP binding site, an apical domain involved with substrate binding, and an intermediate domain that connects the equatorial and apical domains via two hinge regions (4, 5) (see Fig. 1). This subunit architecture is conserved highly among the two classes of chaperonins, which otherwise show distinct structural differences. Group I chaperonins, such as GroEL from Escherichia coli, are found in prokaryotes and eukaryotic organelles (6) and generally consist of double homoheptameric ring arrangements. In contrast, group II chaperonins exist in archaeal and eukaryotic cytosol (7) and consist of a double hetero (or homo)-octameric or nonameric ring (8). Moreover, group I chaperonins require a ring-shaped GroES-like cofactor to fully close the protein folding chamber in the presence of ATP, whereas group II chaperonins contain a “built-in” lid formed by a protrusion at the tip of the apical domains and lack the GroES-like cofactor (4, 5).
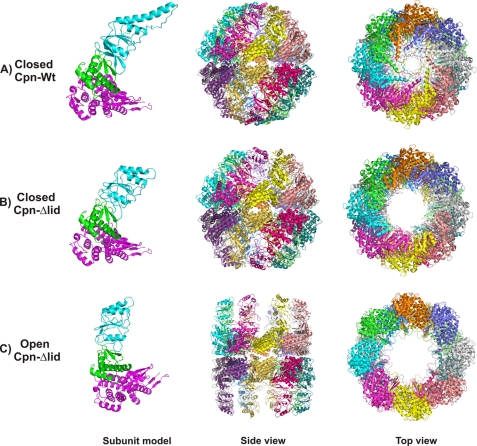
FIGURE 1.
Overall architecture of closed Cpn-WT (A), closed Cpn-Δlid (B), and open Cpn-Δlid (C) structures. The single subunit shows in different colors for the equatorial domain (purple), intermediate domain (green), and apical domain (cyan). The double octamer-ring complex of Cpn is represented by a schematic in both side and top views.
Several conformational states of GroEL associated with its protein folding mechanism have been characterized using electron microscopy (9–11) and x-ray crystallography (12–17). The crystal structure of the GroEL14-GroES7-ADP7 complex showed that the character of the folding chamber changes from hydrophobic in the peptide acceptor state (open) to hydrophilic in the protein folding state (closed) (17). In the first step of GroEL-mediated protein folding, non-native protein binds to the open ring state of GroEL via hydrophobic interactions (18, 19). ATP and GroES then bind to this ring, creating the so-called cis-ring complex. The non-native protein binding site of GroEL is located in the same region as the GroES binding site; consequently, binding of GroES induces the release of substrate inside the folding chamber, and the cofactor acts as a lid for the cis-ring complex (20–22). The closure mechanism of GroEL is characterized as the downward en bloc rotation of the intermediate domains followed by an upward rotation of the apical domains (17). ATP hydrolysis in the cis-ring weakens the interaction between the GroEL and GroES. In a mechanism that is not yet well understood, conditions within the chamber facilitate folding of the non-native protein. Finally, binding of ATP to the adjacent ring (trans-ring) induces release of the folded protein and GroES cofactor (22–24). The highly allosteric nature of protein folding by the group I chaperonins (25–27) has been observed in group II chaperonins as well (25, 28–31). ATP binding to one subunit enhances ATP association with other subunits in the same ring, whereas ATP binding to one ring inhibits the ATP association with the subunits of the adjacent ring. This positive intra-ring and negative inter-ring cooperativity allow chaperonins to function as “two-stroke” machines (4).
In contrast with group I, crystal structures of group II chaperonins have previously only been solved in the closed conformation (8, 32). This paper describes for the first time a crystallographic model for the open state of a group II chaperonin. Comparison with the closed state suggests a closing mechanism that is distinct from the group I chaperonins; the equatorial, intermediate, and apical domains undergo a large conformational change between the open and closed states, possibly rotating as a single unit. In the group I chaperonins, the equatorial domains are relatively stationary between the open and closed states, and chamber closing is due to the motion of the intermediate and apical domains (17). Differences in the closing mechanism between the two classes of chaperonins are not entirely unexpected, because no external cofactor is required for the group II chaperonins, and protein folding depends exclusively on ATP hydrolysis. The structures of the closed and open states of group II chaperonin from Methanococcus maripaludis (MmCpn) provide new insight into the folding mechanism of group II chaperonins.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Cloning, expression, and purification of Cpn-WT3 and Cpn-Δlid have been described by Reissmann et al. (4). Both the Cpn-WT and Cpn-Δlid protein were concentrated and dialyzed against 20 mm Hepes buffer, pH 7.4, containing 50 mm NaCl, 5 mm MgCl2, and 10% glycerol. The final concentration of Cpn-WT and Cpn-Δlid used for crystallization trials were 10 mg·ml−1 and 12 mg·ml−1, respectively.
Crystallization of Cpn-WT and Cpn-Δlid
The protein solution of Cpn-WT was brought to 5.0 mm of ATP analogue (adenosine 5′-[β,γ-imido]triphosphate) and the Cpn-Δlid protein was complexed with 5 mm ADP (adenosine 5′-diphosphate) or non- hydrolyzable ATP analogue (adenosine 5′-[γ-thio]triphosphate) prior to crystallization. Cpn-WT and Cpn-Δlid protein were screened using the sparse matrix method (33) with a Phoenix Robot (Art Robbins Instruments, Sunnyvale, CA) using the following crystallization screens: Crystal Screen I and II, PEG/Ion, SaltRx, and Index (Hampton Research, Aliso Viejo, CA). The optimum condition for the crystals of Cpn-WT was found in 200 mm sodium fluoride and 20% PEG 3350, and the optimum condition for the crystals of Cpn-Δlid were found in 0.2 m Li2SO4 and 20% PEG 3350 for closed state and 0.1 m MES buffer pH 6.5, 5 mm spermidine, and 30% of 2-methyl-2,4-pentanediol for the open state. Crystals were obtained after 1 day by the sitting-drop vapor-diffusion method with the drops consisting of a mixture of 0.2 μl of protein solution and 0.2 μl of reservoir solution.
X-ray Data Collection
Crystals of Cpn-WT and Cpn-Δlid were placed in a reservoir solution containing 100% of paratone oil and 20% (v/v) glycerol, respectively, and then flash-frozen in liquid nitrogen. The data sets for the Cpn-WT and Cpn-Δlid were collected at the Berkeley Center for Structural Biology beamlines 5.0.2 and 8.2.2 at the Advanced Light Source, Lawrence Berkeley National Laboratory. The data sets were processed using the program HKL-2000 (34).
Structure Determination
The crystal structure of closed Cpn-Δlid was determined using the molecular-replacement method with the program PHASER (35), using as a search model a single subunit structure of the group II chaperonin from Thermococcus strain KS-1 (Protein Data Bank code 1Q3S) (8). The closed Cpn-WT and the open Cpn-Δlid structures also were solved by molecular replacement, using the closed Cpn-Δlid as a search model. The resolution cut-offs were determined based on analysis of the σA versus resolution plot (36). Previous studies have shown that with maximum likelihood-based refinement algorithms, this is a reasonable method for determining the effective diffraction limit of the data without losing significant information contained in weak reflections (36, 37). The atomic positions obtained from molecular replacement were used to initiate crystallographic refinement and model rebuilding. Structure refinement was performed using PHENIX (38). For the open Cpn-Δlid structure extensive multizone rigid body refinement (39) was performed for all domains simultaneously to determine the position of intermediate and apical domains. Studies of low resolution crystallographic data have indicated that only rigid body refinement is appropriate typically when the highest resolution experimental data available is between 8–5 Å (36). Translation-libration-screw refinement was applied using the equatorial, intermediate, and apical domains of each subunit as separate translation-libration-screw groups. Residual atomic displacement parameters were refined with very tight noncrystallographic symmetry restraints. Tight noncrystallographic symmetry restraints also were applied to the coordinates in the refinement of both closed structures. Manual rebuilding with COOT (40) allowed construction of the final models. 5% of the reflections randomly were selected in each data set for cross-validation prior to starting refinement.
Root-mean-square deviation differences from ideal geometries for bond lengths, angles, and dihedrals were calculated with PHENIX (38). The overall stereochemical quality of the final models for Cpn-WT and Cpn-Δlid were assessed using the MOLPROBITY (41) program. Atomic models were superposed using the programs LSQKAB from CCP4 and COOT (40).
RESULTS AND DISCUSSION
Structure Determination of Chaperonin
Structural studies of MmCpn were performed using the full-length protein (Cpn-WT) and the lidless version (Cpn-Δlid) described previously, in which the apical protrusions (Ile241–Lys267) are replaced with a short linker composed of four amino acid residues (4). The Cpn-Δlid mutant has been shown to be a functionally open state, able to hydrolyze ATP and bind substrate (4).
Crystals of the closed form of Cpn-WT in the presence of nonhydrolyzable ATP analogue (AMP-PNP) diffracted to 3.3 Å and belonged to the monoclinic space group C2. The electron density map showed clear positions for the equatorial, intermediate, and apical domains, which define one subunit. The crystallographic asymmetric unit contained eight copies of the subunit, forming an octameric ring. The upper and lower rings observed for the Cpn-WT structure are related by the crystallography 2-fold symmetry (Fig. 1A).
The Cpn-Δlid was crystallized in the closed form in the presence of ADP, whereas the open form was obtained by addition of the nonhydrolyzable ATP analogue ATP-γ-thio (Fig. 1, B and C). Binding and hydrolysis of ATP in the equatorial domain has been shown to drive the transition from the open to the closed state in TriC and MmCpn (4); however, Group II chaperonins are not stabilized in the closed state by ADP binding alone (42–45). In this case, the closed form of Cpn-Δlid was obtained due to the presence of sulfate ion in the crystallization buffer. Biochemical studies indicate that group II chaperonins adopt the closed conformation in the presence of a high concentration of sulfate ions (32, 46, 8). Moreover, homologous structures of the group II chaperonins from Thermoplasma acidophilum (32) and several mutants from Thermococcus strain KS-1 (8) were solved in a closed conformation in the presence of sulfate ion, despite the fact that some crystallization conditions lacked nucleotides. The superposition of the closed Cpn-Δlid form onto the closed Cpn-WT indicates the position of a sulfate ion corresponds to the γ-phosphate group of the ATP analogue molecule. The closed Cpn-Δlid crystal diffracted to 3.5-Å resolution and belonged to the same C2 space group observed for Cpn-WT. The deletion of the lid results in the loss of intra-ring contacts involving the lid residues and increases flexibility of the apical domains, which is reflected in the diffraction quality of closed Cpn-Δlid crystals.
A similar effect was observed in crystals of the open state Cpn-Δlid. In the open conformation, the Cpn-Δlid loses not only intra-ring contacts at the lid region but also the inter-subunit contacts at the intermediate domain and apical domain, influencing considerably the diffraction quality of the crystals (Fig. 1C). The lack of intersubunit contacts at the intermediate and apical domains for the open state of Cpn from M. maripaludis has been described by low resolution cryo-electron microscopy (cryo-EM) studies and underlies the flexibility of the peptide acceptor state (47, 48). The flexibility of MmCpn structures is reflected not only in the relative diffraction quality of the crystals but also by the distribution of atomic displacement parameters (B-factors). The closed Cpn-WT shows a homogeneous average B-factor for the equatorial, intermediate, and apical domains. Likewise, the closed Cpn-Δlid, except for the apical domains, displays an increased average B-factor compared with the other two domains. In marked contrast to the closed Cpn structures, the open Cpn-Δlid structure shows a broad distribution of B-factors; the lowest values are found in the equatorial domain and increase through the intermediate domain rising to a maximum in the apical domain.
The group II chaperonins are a perfect example of large macromolecular complexes with dramatically different conformational states associated with function. The structural investigation of flexible and/or large macromolecular complexes is challenging because they frequently do not produce crystals that diffract to high resolution. However, topological information such as orientation of domains and medium to large conformational changes can be readily obtained from low resolution crystallographic maps. Additionally low resolution models can provide the basis for further experimentation (36). For these reasons, the model of open state Cpn-Δlid derived from low resolution crystallographic data can offer important insight into the macromolecular function of Cpn.
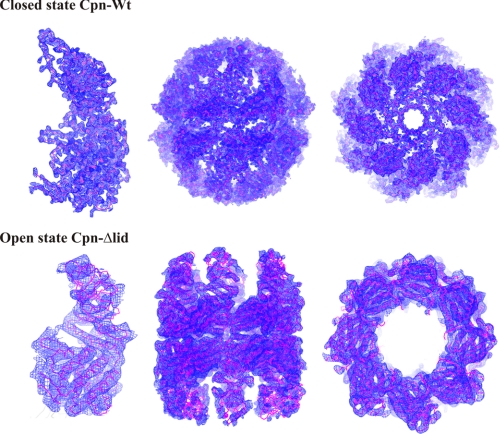
Crystals of the open state Cpn-Δlid diffracted to 6.0 Å of resolution and belonged to the orthorhombic space group I222, with four copies of the subunit per crystallographic asymmetric unit. Crystallographic 2-fold symmetry along two axes generates the two octameric rings observed in Cpn-Δlid. Although the open state of Cpn-Δlid has been solved at low resolution, the electron density map clearly shows the distinct arrangement of the open state Cpn-Δlid subunits and domains (Fig. 2). Extensive rigid body refinement of individual domains using a protocol with a large radius of convergence (39) was used to establish the positions of the equatorial, intermediate, and apical domains. The data collection and structure refinement statistics of closed Cpn-WT, closed Cpn-Δlid and open Cpn-Δlid structures are shown in Table 1.
FIGURE 2.
Representative cross-validated σA weighted electron density maps (2mFo − DFc Fourier synthesis) contoured at 1.0σ showing a single subunit, side and top view of a double octamer-ring complex of the closed Cpn-WT and the open state Cpn-Δlid.
TABLE 1.
Statistics for data collection and structure refinement of Cpn-Δlid
| Closed Cpn-WT | Closed Cpn-Δlid | Open Cpn-Δlid | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 1.000 | 0.978 | 1.000 |
| Resolution range (Å) | 50–3.30 (3.42–3.30) | 54.47–3.50 (3.69–3.50) | 60–6.0 (6.21–6.00) |
| Detector distance (mm) | 400 | 300 | 490 |
| Φ (degrees) | 180/1.0 | 160/1.0 | 180/1.0 |
| Exposure time (seconds) | 10 | 3 | 20 |
| Temperature of collect (Kelvin) | 100 | 100 | 100 |
| Data statistics | |||
| Space group | C2 | C2 | I222 |
| Unit cell parameters (Å) | a = 260.69, b = 162.22, and c = 184.73; α = γ = 90° and β = 135.05° | a = 261.45, b = 161.92, and c = 147.37; α = γ = 90° and β = 124.12° | a = 150.53, b = 209.52, and c = 266.86; α = β = γ = 90° |
| Unique reflections | 83,463 (5601) | 61,840 (8550) | 10,388 (721) |
| Multiplicity | 3.6 (2.4) | 2.7 (2.2) | 13.2 (5.3) |
| Data completeness (%) | 97.6 (82.1) | 96.5 (92.0) | 95.6 (68.4) |
| I/σ(I) | 5.8 (1.0) | 8.2 (1.3) | 17.1 (1.25) |
| Rsyma (lowest and highest shell) (%) | 17.9 (6.4, 75.2) | 14.9 (4.6, 71.4) | 11.9 (8.4, 86.9) |
| Structure refinement | |||
| Resolution range | 48.84–3.30 | 54.47–3.50 | 58.56–6.00 |
| R-factorb (%) | 20.5 | 23.1 | 24.0 |
| Rfreec (%) | 23.4 | 26.8 | 27.0 |
| RMSd from ideal geometry | |||
| Bond lengths (Å) | 0.010 | 0.010 | 0.035 |
| Bond angles | 1.201° | 1.273° | 1.470° |
| Protein residues per ASUe | 4344 | 4344 | 2172 |
| ATP analogue | 1 | 1 | |
| ADP | 1 | ||
| Ramachandran plot (%) | |||
| Favored region | 97.12 | 91.29 | 94.75 |
| Outliers region | 0.52 | 1.08 | 0.63 |
a Rsym = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Σhkl denotes the sum over all reflections, and Σi is the sum over all equivalent and symmetry-related reflections.
b R-factor = Σ|Fobs − Fcalc |/ΣFobs.
c Rfree = R-factor for 5% of the data were not included during crystallographic refinement.
d RMS indicates root mean square.
e ASU indicates asymmetric unit.
General Architecture
The architecture of the closed states of Cpn-WT and Cpn-Δlid are similar to the previously described structures of group II chaperonins from T. acidophilum (32) and Thermococcus strain KS-1 (8). The double homo-octameric rings of closed Cpn-WT and Cpn-Δlid are stacked back-to-back, creating a spherically shaped complex, in contrast to the cylindrical form observed for the open state of Cpn-Δlid. The folding chamber is separated into two distinct cavities, one in each ring, by the crystallographically disordered segments composed of six N-terminal and 24 C-terminal amino-acids segments. In cryo-EM analysis of Cpn, these segments appear to divide the central folding chamber at the equatorial region (47, 48).
The equatorial domain is the largest subunit domain and is composed of amino acid residues present at both the N- and C-terminals (residues 1–141 and 400–543). A disulfide bond between residues Cys470 and Cys484 is observed in the equatorial domain of MmCpn; however, this interaction is not conserved in the structures of chaperonins from Thermoplasma and Thermococcus (32, 8). The equatorial domain is involved with inter- and intra-ring contacts in both the closed and open states, in contrast with the intermediate and apical domains, which only show intra-ring contacts in the closed conformation. The nucleotide binding site is located at the top of the equatorial domain and contains a conserved P-loop motif characterized by the sequence GDGTTT (49). The conserved hydrophobic residues Pro41, Phe476, and Val488 surround the nucleotide base moiety of ATP. Threonines Thr93, Thr94, and Thr95 in the P-loop provide hydrogen bonds to the α-, β-, and γ-phosphate groups. The γ-phosphate group is stabilized both by interactions with the threonine residues and by coordination to a magnesium ion. The proposed mechanism of ATP hydrolysis occurs by a water nucleophilic attack, in which H2O is held in place by hydrogen bonding to the side chains of catalytic residues Asp60 and Asp386 (32).
The intermediate domain is composed of two flexible hinges created by residues ranges 142–210 and 362–399. The lower region of the intermediate domain includes α-helices H7 and H14, which contact the equatorial domain, and H8. An intermediate domain residue present at the loop between H7/H8 interacts with the ATP binding site via the main chain of Gly160 with the α-phosphate group of ATP. The upper region of the intermediate domain consists of a β-sheet structure formed by S7, S8, and S20, opposite the β-sheet created by S9, S10, S18, and S19 in the apical domain.
The apical domain is composed of ∼150 residues (211–361). The overall folding consists of three α-helices (H10, H11, and H12) and 9 β-strands (S9–S17). The major difference between the apical domains of group II and group I chaperonins is a segment of 27 residues, which forms the built-in lid in the group II proteins. An extension to helix 10 is created by these extra residues at the tip of apical domain, isolating the folding chamber from the solvent. The built-in lid segment shows both hydrophobic and hydrophilic residues. The hydrophobic residues are involved with intra-ring contacts and the hydrophilic residues are pointed toward the center of the cavity in the protein folding state (closed). The apical domain is the most flexible region in the subunit and is involved with substrate binding via hydrophobic interactions (50–52).
Ring Rearrangement from the Open to a Closed State
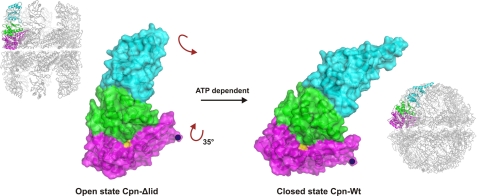
The superposition of closed Cpn-WT against the open Cpn-Δlid demonstrates that the equatorial domain undergoes a significant outward rotation of 35 ° from the open to a closed conformation, while the apical domain rotates inward (Fig. 3 and supplemental Movie 1), with the entire subunit moving as a rigid body around a pivot point near residue Pro41 at the ATP binding site. The complete rotation includes both this inward motion and an anticlockwise rotation of each subunit around the 8-fold symmetry axis of the octamer ring (Fig. 1, top view). Because the group II chaperonins depend exclusively on ATP hydrolysis to close the protein-folding chamber, a large conformational change with the ATP binding site as the pivot point is not unexpected for this class of proteins.
FIGURE 3.
Conformational change for a single subunit between the open state of Cpn-Δlid and closed Cpn-WT. Each subunit of the complex rotates as a rigid body by ∼35 ° around a pivot point near residue Pro41 (colored in yellow) at the ATP-binding site. This single motion can be visually decomposed into a downward rotation (seen from the side of the subunit) and an anticlockwise rotation (see top of the subunit). As a result the apical domains tilt downward, closing the chamber. Seen from above, the conformation change has the appearance of an iris opening and closing.
The anticlockwise subunit rotation caused by ATP hydrolysis changes the ring dimension and shape (supplemental Movie 2). The closed states of Cpn-WT and Cpn-Δlid show heights of 162 and 148 Å, respectively, and a diameter of 161 Å. The open state of Cpn-Δlid shows a significant increase in height to 178 Å while maintaining a similar diameter of 159 Å. To visualize the orientation of the lid in an open conformation, a full-length model of the open state Cpn-WT was built using the open Cpn-Δlid structure as a template. The model of open state Cpn-WT shows a remarkable height of 204 Å. The change from the cylindrical form of the open state to the spherical shape of the closed state causes a decrease of ∼65% in the volume of the folding chamber. The chamber volume of each ring of the open Cpn-WT model was 350,000–370,000 Å3, whereas for the closed Cpn-WT state, the chamber volume was 130,000 Å3. The folding chamber in the open state is twice as large as that of the cis-cavity of the GroEL-GroES complex. The size of the central cavity observed for the open state readily explains the ability of group II chaperonins to fold much larger substrates than GroEL (53). A similar chamber volume for the closed state of Cpn-WT has been reported for the Thermosome structure (32).
Inter and Intra-ring Contacts
Open State
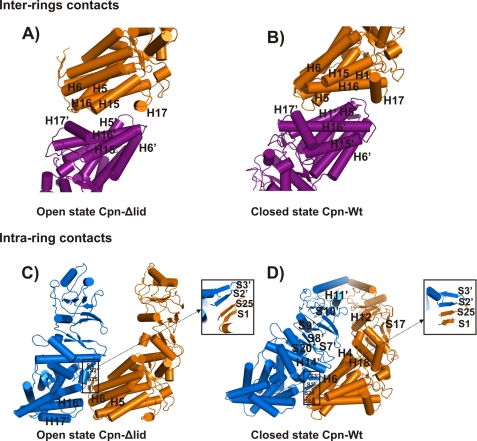
The inter-ring contacts of the open state of Cpn-Δlid occur between the subunits in the upper and lower rings as related by 2-fold symmetry. The inter-ring interactions consist principally of hydrophobic contacts between the residues on H17 with the residues of the loops located between helices H5/H6 and helices H15/H16 of the symmetry-related subunit (Fig. 4A). In contrast, in the closed state the helices H17 are distant from helices H15/H16 of the subunit across the ring (Fig. 4B). A similar change of ring-ring interface interactions between the open and closed states was observed in a recent MmCpn cryo-EM analysis (48). In addition to the hydrophobic contacts, the inter-ring interface shows a salt bridge involving residues Arg425 and Asp451. This electrostatic interaction is of particular interest because it is preserved between the open and closed states and may be essential to the double-ring formation found in group II chaperonins.
FIGURE 4.
A, inter-ring contacts for the open state Cpn-Δlid consist of hydrophobic contacts between the residues on H17 with the residues of the loops located between the helices H5/H6 and the helices H15/H16 of the symmetry-related subunit. B, inter-ring contacts for the closed state Cpn-WT are shown. The closed stage increases the contact area among the upper and lower rings compared with open conformation. A region of the loop H5/H6 of the upper ring hook into a cleft formed by helices H5, H16, and H17 of the lower ring. C, intra-ring contacts for the open state Cpn-Δlid are found at only at equatorial domain at two regions between neighboring subunits. The outer intra-ring contacts are made between loops H5/H6 and loop H16/H17 of the adjacent subunit and the inner intra-ring contact involving a β-sheet formation by the N- and C-terminal β-strands (S1 and S25) of one subunit and the β-strands (S2 and S3) of the adjacent subunit. D, intra-ring contacts for the closed state Cpn-WT are shown. The intra-ring interactions at the equatorial domain conserve the β-sheet formed by S1 and S25 of one subunit and S2 and S3 of an adjacent subunit. The intermediate domain contacts the upper region of the equatorial domain of the adjacent subunit via the loops S20/H14 and S7/S8. The intra-ring contacts at the apical domain occur mainly between the region of H11 and the loop S9/S10 with the loop region H12/S17.
The equatorial intra-ring contacts of the open Cpn-Δlid state are found at two regions between neighboring subunits. The outer intra-ring contacts are made between loops H5/H6 and loop H16/H17 of the adjacent subunit (Fig. 4C). The same loop H5/H6 region is involved in inter and intra-ring interaction and is potentially a key region for allosteric regulation. An inner intra-ring contact involving a β-sheet formation by the N- and C-terminal β-strands (S1 and S25) of one subunit and the β-strands (S2 and S3, also called the stem loop) of the adjacent subunit. This secondary structure is preserved in the open and closed conformations, despite the large rotation of the equatorial domain. The conformational change observed for β-sheet involving elements from neighboring subunits within one ring has been described as a “hand-shaking” movement, and it indicates a potential region for communication of cooperativity during the ATP hydrolysis cycle (48). The intermediate and apical domains did not show any intra-ring interactions, consistent with previous lower resolution cryo-EM experiments (47).
Closed State
The inter-ring interactions change from mostly hydrophobic contacts in the open state to several salt-bridge interactions in the closed state. The positively charged residues Arg18, Arg22, and Arg29 located at H1 of the upper ring interact, respectively, with the negatively charged residues Glu33 and Asp112 of H1 and H5 in the lower ring. The rotation observed from the open to a closed stage increases the contact area among the upper and lower rings. A region of the loop H5/H6 of the upper ring hook into a cleft formed by helices H5, H16, and H17 of the lower ring (Fig. 4B). A salt bridge between Arg425 and Asp451, as described for open state, completes the inter-ring electrostatic interactions. A similar interaction has been observed for the closed homologue structure of T. acidophilum involving residues Arg429 and Asp455 (32). Moreover, the structure of GroEL-GroES-ADP shows a salt bridge between Arg452 and Glu461 at the same location as Arg425 and Asp451 in the closed state Cpn-WT. A mutant form E461K of GroEL demonstrates that the inter-ring interaction Glu461 with Arg452 is involved directly with negative allosteric regulation (54). Therefore, the crystals structures of MmCpn suggest the possibility of a conserved allosteric interaction between the group I and II chaperonins.
The intra-ring interaction for the closed state occurs in all three domains. The contacts between the subunits are mediated principally by hydrophilic residues. Several of the residues involved with intra-ring interactions for the closed state are exposed highly to the solvent in the open state. The intra-ring interactions at the equatorial domain conserve the β-sheet formed by S1 and S25 of one subunit and S2 and S3 of an adjacent subunit. The intermediate domain contacts the upper region of the equatorial domain of the adjacent subunit via the loops S20/H14 and S7/S8. The intra-ring contacts at the apical domain occur mainly between the region of H11 and the loop S9/S10 with the loop region H12/S17 (Fig. 4D). The additional intra-ring interactions at the apical domain arise in the built-in lid region, creating a central β-barrel structure. A detailed description of the lid interactions is discussed below.
Built-in Lid and Substrate Binding
One of the major differences between the two classes of chaperonins is the presence of a built-in lid at the tip of the apical domains for group II, replacing the GroES-like cofactor used with group I chaperonins. The lid sequence of Cpn is composed of 27 highly conserved amino acid residues, which extend the long α-helix (helix 10) at the apical domain and provide a protective cap to the folding chamber. The presence of the built-in lid of Cpn is dispensable for both ATP hydrolysis and substrate binding, but deletion of the lid impairs the ability of MmCpn to fold substrate (4). In addition to providing a protected environment for folding substrate, the build-in lid limits the premature release of folded proteins from the central cavity. Therefore, the residues of the lid in group II chaperonins play an essential role in the protein folding mechanism.
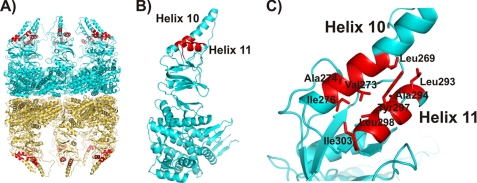
Hydrophobic interactions have been implicated in substrate binding in both group I and group II chaperonins (50–52, 55, 56). However, the previous closed state structures of group II chaperonins did not show any evident hydrophobic surface areas for substrate binding sites (32, 8). As described above, we constructed a full-length model of the open state Cpn-WT. The model shows two possible hydrophobic patches for substrate binding sites at the apical domain. The first observed hydrophobic area is formed by the residues Ile250, Ile252, Pro255, Leu258, Phe261, and Ile262. However, the Cpn-Δlid mutant (deletion of residues 241–267) retains the ability to bind substrate, thus excluding this hydrophobic region as the sole substrate binding site. These conserved hydrophobic residues in the lid region seem instead to be important in stabilizing the closed state of group II chaperonins because of the large number of van der Waals contacts in the lid-lid interface. The second hydrophobic patch, and more likely substrate binding site, is located at the interface between the apical helices H10 and H11. Residues Leu269, Val273, Ala274, and Ile276 of H10 and residues Leu293, Ala294, Tyr297, Leu298, and Ile303 of H11 create a hydrophobic pocket in the apical domain (Fig. 5). Sequence alignment showed that these residues are highly conserved among group II chaperonin members (data not shown). Residues Val273, Ala274, Leu293, and Tyr297 are exposed to solvent, and they can make direct contact with the substrate. Residues Leu269, Ile276, Ala294, Leu298, and Ile303 are important in creating a hydrophobic core and organizing the H10 and H11 helices. Similar to GroEL, the interface between H10 and H11 becomes less accessible in the closed conformation of the chaperonin. This is a consequence of the H10 and H11 interface making contacts with the loop consisting of residues 326 through 331 of a neighboring subunit. Superposition of the structure of the GroEL apical domain in complex with polypeptide substrate (19) with the open state Cpn-Δlid or Cpn-WT models showed that the substrate binding region observed between helices H and I for GroEL correspond to the hydrophobic core formed by the helices H10 and H11 in the apical domain of MmCpn. The charged residue Arg268, present at the N-terminal of helix I of GroEL implicated in direct hydrogen bonding with the substrate, is charge-conserved by Glu301 of H11 in the MmCpn sequence. Moreover, mutagenesis analysis of eukaryotic group II chaperonin TRiC/CTT suggests that the residues of H11 are directly involved in substrate interaction sites (57). Both the open state structure of MmCpn described here and previous TRiC/CTT biochemical studies indicate a similar site for substrate binding among the group I and group II chaperonins.
FIGURE 5.
Hydrophobic patch observed in apical domain of group II chaperonins (region shown in red). Hydrophobic interactions have been implicated in substrate binding in group II chaperonins and the residues Leu269, Val273, Ala274, and Ile276 of H10 and residues Leu293, Ala294, Tyr297, Leu298, and Ile303 of H11 create a potential substrate binding site at the apical domain. A full-length model of the open state Cpn-WT was built using the open Cpn-Δlid structure as a template. The upper and lower ring of open model Cpn-WT (A), the single subunit (B), and zoom view (C) of the potential substrate binding site involving the α-helix 10 and α-helix 11.
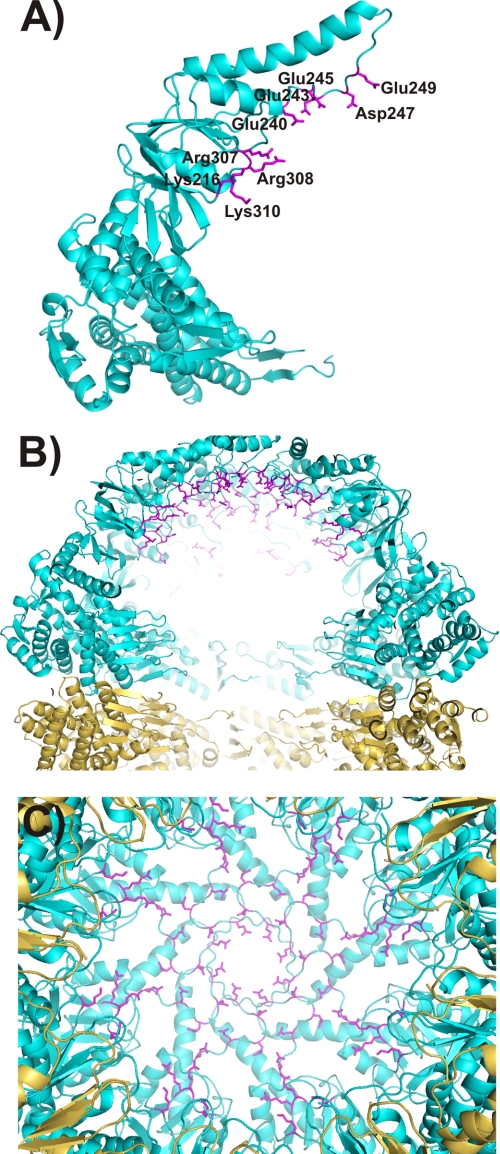
The crystal structure of the complex GroEL14-GroES7-ADP7 suggests that the hydrophilic nature of the interior of the cavity in the closed state is essential for protein folding (17). Similarly, the significant rotation observed in MmCpn in transitioning from an open state to a closed state creates a highly hydrophilic surface within the protein folding chamber. The negatively charged residues Glu240, Glu243, Glu245, Asp247, and Glu249 of the lid region and the positively charged residues Lys216, Arg307, Arg308, and Lys310 point to the inside of the folding chamber in the closed state. These residues create “rings” of extremely hydrophilic regions inside the folding chamber (Fig. 6). The deletion of hydrophilic residues present in the lid and/or the lost of the capping function provided by the lid, required to create a protected environment for protein folding, are potential reasons for the observed loss of folding activity in the Cpn-Δlid mutant (4).
FIGURE 6.
The hydrophilic nature of the cavity in the closed state is essential for protein folding in group I chaperonin (17). The closed state Cpn-WT shows a hydrophilic character. The negatively charged residues Glu240, Glu243, Glu245, Asp247, and Glu249 of the lid region and the positively charged residues Lys216, Arg307, Arg308, and Lys310 create a hydrophilic character inside the folding chamber. A, a single subunit of the closed state Cpn-WT. B, side view of the chamber. C, an inside view of the chamber looking from the equatorial domain to the apical domain. The upper ring is shown in green, the lower ring is shown in gold, and charged residues are shown in magenta.
Mechanism of Closing
Previous studies on MmCpn and TriC suggest that at intermediate ATP concentrations, hydrolysis of ATP in the trans-ring is dependent on dissociation of ADP from the cis-ring (4); this negative allostery is overcome at higher ATP concentrations, where both rings can close simultaneously. Interestingly, for the “lidless” mutant, no such cooperativity is observed, indicating that the lid region is important in communication between the rings, perhaps by stabilizing the closed form. Recent cryo-EM studies on GroEL have suggested the negative allostery between the two rings is mediated by a “relay helix” involving the D helices, which interact at the ring-ring interface (58).
The ring-ring allosteric mechanism in MmCpn is as yet unclear but would likely involve some of the same residues. The β-sheet intra-ring interactions appear unchanged in the closed and the open form, suggesting that these residues maintain stability of the ring during rotation of the subunits. Similarly, the inter-ring salt bridge between Asp451 and Arg425 remains undisturbed between the open and closed forms, suggesting that this might be a hinge point between the trans- and cis-rings.
In the thermosome, nucleotide binding induces a major structural change in the H4/H5 loop (32) at the top of the equatorial domain. It is possible that in MmCpn, a similar structural change upon ATP hydrolysis at the nucleotide binding site leads to rearrangement of the H5 helix. If the nucleotide binding site is at the pivot of the rotation, then a small movement at the binding site is translated into a larger movement at the H5/H6 loop at the bottom of the equatorial domain where the ring-ring contacts are made. The rotation of the equatorial domain upon closing the structure brings the H5/H6 loops into greater proximity to adjacent subunits, which help stabilize the closed form.
Conclusion
Cellular accumulation of unfolded proteins is linked with several diseases and the group II chaperonins, which are found in the eukaryotic cytosol, are crucial in the folding process. This paper describes the peptide acceptor (open) state and a protein folding (closed) state in a group II chaperonin, which we observe to be very different from the well characterized group I chaperonins. The open and closed states of MmCpn show a large rearrangement of all subunits, each moving as a rigid body, and consequently, a significant change in the overall dimension and shape between the two conformations. This result suggests that the considerable structural difference between the group II chaperonins, which contain a built-in lid, and the group I chaperonins, which require a ring-shaped GroES-like cofactor, translate to two radically different chamber-closing mechanisms. Despite these different mechanisms, similar substrate binding sites and mechanisms of allosteric regulation appear to be conserved between the two groups (57). These similar characteristics support the idea of a common ancestor for the chaperonins. Future experiments that create a single ring of MmCpn by disrupting inter-ring salt bridges or by forming electrostatic repulsion between the lower and upper rings will contribute to a more detailed understanding of the protein folding mechanism and allosteric regulation used in group II chaperonins.
Acknowledgments
We are grateful to the staff of the Berkeley Center for Structural Biology at the Advanced Light Source of Lawrence Berkeley National Laboratory and, in particular, Peter Zwart for help with data processing.
This work was performed as part of the Center for Protein Folding Machinery and National Institutes of Health Roadmap-supported Nanomedicine Development Center (Grant 2PN2EY016525). The Berkeley Center for Structural Biology is supported in part by NIGMS, National Institutes of Health, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U. S. Department of Energy under Contract DE-AC02-05CH11231.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3KFB, 3KFE, and 3KFK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies 1 and 2.
- WT
- wild type
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate
- MmCpn
- group II chaperonin from Methanococcus maripaludis.
REFERENCES
- 1.Frydman J. (2001) Annu. Rev. Biochem. 70, 603–647 [DOI] [PubMed] [Google Scholar]
- 2.Taubes G. (1996) Science 271, 1493–1495 [DOI] [PubMed] [Google Scholar]
- 3.Dobson C. M. (2004) Semin. Cell Dev. Biol. 15, 3–16 [DOI] [PubMed] [Google Scholar]
- 4.Reissmann S., Parnot C., Booth C. R., Chiu W., Frydman J. (2007) Nat. Struct. Mol. Biol. 14, 432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth C. R., Meyer A. S., Cong Y., Topf M., Sali A., Ludtke S. J., Chiu W., Frydman J. (2008) Nat. Struct. Mol. Biol. 15, 746–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwich A. L., Farr G. W., Fenton W. A. (2006) Chem. Rev. 106, 1917–1930 [DOI] [PubMed] [Google Scholar]
- 7.Frydman J., Nimmesgern E., Erdjument-Bromage H., Wall J. S., Tempst P., Hartl F. U. (1992) EMBO J. 11, 4767–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shomura Y., Yoshida T., Iizuka R., Maruyama T., Yohda M., Miki K. (2004) J. Mol. Biol. 335, 1265–1278 [DOI] [PubMed] [Google Scholar]
- 9.Chen S., Roseman A. M., Hunter A. S., Wood S. P., Burston S. G., Ranson N. A., Clarke A. R., Saibil H. R. (1994) Nature 371, 261–264 [DOI] [PubMed] [Google Scholar]
- 10.Roseman A. M., Chen S., White H., Braig K., Saibil H. R. (1996) Cell 87, 241–251 [DOI] [PubMed] [Google Scholar]
- 11.Clare D. K., Bakkes P. J., van Heerikhuizen H., van der Vies S. M., Saibil H. R. (2009) Nature 457, 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braig K., Otwinowski Z., Hegde R., Boisvert D. C., Joachimiak A., Horwich A. L., Sigler P. B. (1994) Nature 371, 578–586 [DOI] [PubMed] [Google Scholar]
- 13.Chaudhry C., Horwich A. L., Brunger A. T., Adams P. D. (2004) J. Mol. Biol. 342, 229–245 [DOI] [PubMed] [Google Scholar]
- 14.Hunt J. F., Weaver A. J., Landry S. J., Gierasch L., Deisenhofer J. (1996) Nature 379, 37–45 [DOI] [PubMed] [Google Scholar]
- 15.Mande S. C., Mehra V., Bloom B. R., Hol W. G. (1996) Science 271, 203–207 [DOI] [PubMed] [Google Scholar]
- 16.Braig K., Adams P. D., Brünger A. T. (1995) Nature Struct. Biol. 2, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 17.Xu Z., Horwich A. L., Sigler P. B. (1997) Nature 388, 741–750 [DOI] [PubMed] [Google Scholar]
- 18.Lin Z., Schwartz F. P., Eisenstein E. (1995) J. Biol. Chem. 270, 1011–1014 [DOI] [PubMed] [Google Scholar]
- 19.Chen L., Sigler P. B. (1999) Cell 99, 757–768 [DOI] [PubMed] [Google Scholar]
- 20.Weissman J. S., Hohl C. M., Kovalenko O., Kashi Y., Chen S., Braig K., Saibil H. R., Fenton W. A., Horwich A. L. (1995) Cell 83, 577–587 [DOI] [PubMed] [Google Scholar]
- 21.Mayhew M., da Silva A. C., Martin J., Erdjument-Bromage H., Tempst P., Hartl F. U. (1996) Nature 379, 420–426 [DOI] [PubMed] [Google Scholar]
- 22.Roseman A. M., Ranson N. A., Gowen B., Fuller S. D., Saibil H. R. (2001) J. Struct. Biol. 135, 115–125 [DOI] [PubMed] [Google Scholar]
- 23.Weissman J. S., Kashi Y., Fenton W. A., Horwich A. L. (1994) Cell 78, 693–702 [DOI] [PubMed] [Google Scholar]
- 24.Ranson N. A., Burston S. G., Clarke A. R. (1997) J. Mol. Biol. 266, 656–664 [DOI] [PubMed] [Google Scholar]
- 25.Horovitz A., Fridmann Y., Kafri G., Yifrach O. (2001) J. Struct. Biol. 135, 104–114 [DOI] [PubMed] [Google Scholar]
- 26.Saibil H. R., Horwich A. L., Fenton W. A. (2001) Adv. Protein Chem. 59, 45–72 [DOI] [PubMed] [Google Scholar]
- 27.Swain J. F., Gierasch L. M. (2006) Curr. Opin. Struct. Biol. 16, 102–108 [DOI] [PubMed] [Google Scholar]
- 28.Kafri G., Horovitz A. (2003) J. Mol. Biol. 326, 981–987 [DOI] [PubMed] [Google Scholar]
- 29.Yifrach O., Horovitz A. (1998) Biochemistry 37, 7083–7088 [DOI] [PubMed] [Google Scholar]
- 30.Yifrach O., Horovitz A. (1995) Biochemistry 34, 5303–5308 [DOI] [PubMed] [Google Scholar]
- 31.Kafri G., Willison K. R., Horovitz A. (2001) Protein Sci. 10, 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ditzel L., Löwe J., Stock D., Stetter K. O., Huber H., Huber R., Steinbacher S. (1998) Cell 93, 125–138 [DOI] [PubMed] [Google Scholar]
- 33.Jancarik J., Kim S. H. (1991) J. Appl. Cryst. 24, 409–411 [Google Scholar]
- 34.Otwinowski Z., Minor W. (1997) in Methods Enzymology: Macromolecular Crystallography (Carter C. W., Sweet R. M. eds) Vol. 276, pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 35.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLaBarre B., Brunger A. T. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 923–932 [DOI] [PubMed] [Google Scholar]
- 37.Ling H., Boodhoo A., Hazes B., Cummings M. D., Armstrong G. D., Brunton J. L., Read R. J. (1998) Biochemistry 37, 1777–1788 [DOI] [PubMed] [Google Scholar]
- 38.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 39.Afonine P. V., Grosse-Kunstleve R. W., Urzhumtsev A., Adams P. D. (2009) J. Appl. Crystallogr. 42, 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Soeyink J., Richardson J. C., Richardson D. C. (2007) Nucleic Acids Res. 35, 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llorca O., Smyth M. G., Carrascosa J. L., Willison K. R., Radermacher M., Steinbacher S., Valpuesta J. M. (1999) Nat. Struct. Biol. 6, 639–642 [DOI] [PubMed] [Google Scholar]
- 43.Nitsch M., Walz J., Typke D., Klumpp M., Essen L. O., Baumeister W. (1998) Nat. Struct. Biol. 5, 855–857 [DOI] [PubMed] [Google Scholar]
- 44.Gutsche I., Holzinger J., Rössle M., Heumann H., Baumeister W., May R. P. (2000) Curr. Biol. 10, 405–408 [DOI] [PubMed] [Google Scholar]
- 45.Gutsche I., Holzinger J., Rauh N., Baumeister W., May R. P. (2001) J. Struct. Biol. 135, 139–146 [DOI] [PubMed] [Google Scholar]
- 46.Iizuka R., Yoshida T., Shomura Y., Miki K., Maruyama T., Odaka M., Yohda M. (2003) J. Biol. Chem. 278, 44959–44965 [DOI] [PubMed] [Google Scholar]
- 47.Clare D. K., Stagg S., Quispe J., Farr G. W., Horwich A. L., Saibil H. R. (2008) Structure 16, 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., Baker M. L., Schröder G. F., Douglas N. R., Reissmann S., Jakana J., Dougherty M., Fu C. J., Levitt M., Ludtke S. J., Frydman J., Chiu W. (2010) Nature 463, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saraste M., Sibbald P. R., Wittinghofer A. (1990) Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 50.Rommelaere H., De Neve M., Melki R., Vandekerckhove J., Ampe C. (1999) Biochemistry 38, 3246–3257 [DOI] [PubMed] [Google Scholar]
- 51.Feldman D. E., Spiess C., Howard D. E., Frydman J. (2003) Mol. Cell 12, 1213–1224 [DOI] [PubMed] [Google Scholar]
- 52.Kubota S., Kubota H., Nagata K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8360–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spiess C., Meyer A. S., Reissmann S., Frydman J. (2004) Trends Cell Biol. 14, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sewell B. T., Best R. B., Chen S., Roseman A. M., Farr G. W., Horwich A. L., Saibil H. R. (2004) Nat. Struct. Mol. Biol. 11, 1128–1133 [DOI] [PubMed] [Google Scholar]
- 55.Guagliardi A., Cerchia L., Bartolucci S., Rossi M. (1994) Protein Sci. 3, 1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobrzynski J. K., Sternlicht M. L., Farr G. W., Sternlicht H. (1996) Biochemistry 35, 15870–15882 [DOI] [PubMed] [Google Scholar]
- 57.Spiess C., Miller E. J., McClellan A. J., Frydman J. (2006) Mol. Cell 24, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ranson N. A., Clare D. K., Farr G. W., Houldershaw D., Horwich A. L., Saibil H. R. (2006) Nat. Struct. Mol. Biol. 13, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]