Abstract
In trypanosomatids, all mRNAs are processed via trans-splicing, although cis-splicing also occurs. In trans-splicing, a common small exon, the spliced leader (SL), which is derived from a small SL RNA species, is added to all mRNAs. Sm and Lsm proteins are core proteins that bind to U snRNAs and are essential for both these splicing processes. In this study, SmD3- and Lsm3-associated complexes were purified to homogeneity from Leishmania tarentolae. The purified complexes were analyzed by mass spectrometry, and 54 and 39 proteins were purified from SmD3 and Lsm complexes, respectively. Interestingly, among the proteins purified from Lsm3, no mRNA degradation factors were detected, as in Lsm complexes from other eukaryotes. The U1A complex was purified and mass spectrometry analysis identified, in addition to U1 small nuclear ribonucleoprotein (snRNP) proteins, additional co-purified proteins, including the polyadenylation factor CPSF73. Defects observed in cells silenced for U1 snRNP proteins suggest that the U1 snRNP functions exclusively in cis-splicing, although U1A also participates in polyadenylation and affects trans-splicing. The study characterized several trypanosome-specific nuclear factors involved in snRNP biogenesis, whose function was elucidated in Trypanosoma brucei. Conserved factors, such as PRP19, which functions at the heart of every cis-spliceosome, also affect SL RNA modification; GEMIN2, a protein associated with SMN (survival of motor neurons) and implicated in selective association of U snRNA with core Sm proteins in trypanosomes, is a master regulator of snRNP assembly. This study demonstrates the existence of trypanosomatid-specific splicing factors but also that conserved snRNP proteins possess trypanosome-specific functions.
Keywords: RNA-binding Protein, RNA-Protein Interaction, RNA Splicing, Spliceosome, Trypanosome, Lsm, Sm, Small Nuclear RNPs, Splicing Factors, trans-Splicing
Introduction
Pre-mRNA processing is mediated by the spliceosome, which is assembled in a stepwise manner on pre-mRNA (1). Although pre-mRNA splicing is mediated by RNA, the splicing factors not only play key roles in the formation of the spliceosome but may even directly participate in catalysis (2).
The human spliceosome contains 45 distinct snRNP3-associated proteins, and up to 300 distinct proteins co-purify with the complex (3). However, only 170 of these proteins were identified as part of active spliceosomes (4).
In contrast to the mammalian and yeast systems, little is known about trypanosome snRNP proteins. In trypanosomes, all mRNAs are processed by trans-splicing. trans-Splicing is mediated by ligating a common spliced leader (SL) to all mRNAs from a small RNA donor, the SL RNA (5). Trypanosomes possess all the U snRNPs, but only U2, U4, U5, and U6 were suggested to function in trans-splicing (5, 6). U1 snRNP exists in trypanosomes, most probably for mediating the splicing of cis-spliced introns, but its role in trans-splicing has not been investigated (7, 8).
Over the past few years, progress has been made in describing a variety of trypanosome splicing factors, and their function was elucidated by down-regulation via RNAi. Among these factors are Sm proteins, Lsm proteins, and SSm proteins (9–13). SSm proteins are specific Sm proteins that bind either U2 or U4 snRNP and substitute for the canonical Sm proteins (12, 13). Splicing defects such as an increase in the level of SL RNA and changes in the level of the Y structure intermediate were observed upon the depletion of the splicing factors, PRP31 and PRP43 (14). More recently, U2AF35, U2AF65, and SF1, the factors that function to select the correct 3′ AG splice sites were identified, and their function in trans-splicing was elucidated (15). An ∼45-S complex that carries all the U snRNPs, including U1 snRNP and SL RNP and the splicing intermediates, was identified, suggesting the existence of a single spliceosome complex that can potentially conduct both trans- and cis-splicing reactions(14).
SL RNA, the donor of the trans-spliced exon, has a very unique biogenesis. As opposed to U snRNAs, which are transcribed in trypanosomes by polymerase III (16), the SL RNA is transcribed from a defined polymerase II promoter, and its transcription requires the tSNAP complex (17, 18). Transcription of SL RNA takes place in a distinct nuclear site, near the nucleolus (19). This compartment was termed the “SL RNP factory” because Sm assembly and modification, such as the pseudouridylation by spliced leader-associated (SLA1) RNA, take place in this subdomain (13, 20).
Two recent studies purified and analyzed Trypanosoma brucei snRNP proteins by mass spectrometry. The first study purified proteins associated with SmB (21) and led to the identification of 30 components of the trypanosome spliceosome. One protein, SMN (survival of motor neurons), was studied in detail. In metazoa, this protein, along with the proteins GEMIN2–8, serves as the scaffold for snRNP assembly in the cytoplasm (22). The trypanosome SMN homologue is smaller, localized in the nucleus, and specifically and stably interacts with the SmD3/B subcomplex. In vitro experiments demonstrated that SMN is sufficient to confer specificity to Sm core assembly and to discriminate against binding of the canonical Sm core to nonspecific RNA species and to snRNAs with noncanonical Sm sites, such as U2 and U4 snRNAs (21). A homologue of GEMIN2 was also identified, but its exact role during Sm core assembly was not established (21). Purification of SmD1 complexes from T. brucei was also recently published. This study identified 47 spliceosome proteins, as well as 21 novel proteins lacking a specific annotation (23).
Lsm proteins, unlike Sm proteins, are involved in nuclear processing and turnover of RNAs in eukaryotes. Lsm proteins form two distinct complexes, the Lsm2–8 complex, which binds U6 snRNA, and the Lsm1–7 complex, which governs mRNA degradation (24, 25). Initially, seven Sm-like (Lsm) proteins were identified in T. brucei (11). Functional studies on two of these proteins, Lsm3 and Lsm8, suggest that these proteins not only bind U6 but also affect mRNA stability (11). Two of these proteins were later identified as SSm proteins that bind to U2 and U4 snRNAs (13), and ultimately, the entire complex that binds the U6 snRNA was identified (26). Interestingly, localization studies demonstrated that the Lsm proteins localize near the nucleolus but cannot be detected in cytoplasmic bodies analogous to P-bodies in other eukaryotes (26).
In this study, the SmD3-, Lsm3-, and U1A-associated proteins were purified from Leishmania tarentolae and subjected to mass spectrometry. Interestingly, Lsm purification did not reveal any factors involved in mRNA degradation. The function of selected snRNP proteins that were identified by mass spectrometry in Leishmania was elucidated by RNAi silencing and in situ tagging in T. brucei. Silencing of U1A, as opposed to other U1 snRNP proteins, affected both cis- and trans-splicing as well as polyadenylation. The purification of U1A-associated proteins from L. tarentolae also identified, in addition to the U1 snRNP proteins, factors involved in splicing and polyadenylation. PRP19, a splicing factor that is associated with U5 snRNP in active cis-spliceosomes (27), affected the capping of SL RNA and is also found in the SL RNP factory, suggesting a unique function in trypanosomes. GEMIN2, the factor that associates with SMN was localized to the SL RNP factory, but also to other domains in the nucleus. Silencing of GEMIN2 affected the level of all sn and SL RNAs, suggesting a major role in snRNP biogenesis. Although U1, U5, and SL RNA assemble with the same core Sm proteins, U1 and U5 snRNP proteins are not found in the SL RNP factory, suggesting that the SL RNP factory is a distinct and exclusive site for SL RNP production. This study highlights the function of universal splicing factors that acquired unique functions in trypanosomes, as well as that of trypanosome-specific splicing factors.
EXPERIMENTAL PROCEDURES
Tagging of the SmD3, Lsm3, and U1A in L. tarentolae
For generation of the tagged constructs, genes were amplified with primers listed in supplemental S-1. The fragments were cloned into the pSAP1 vector (kindly provided to us by Dr. Larry Simpson, UCLA). The cloned vector (20 μg) was transfected into L. tarentolae and selected using neomycin resistance (28).
Purification of the Complexes Associated with SmD3, Lsm3, and U1A
Tandem affinity purification was performed from whole cell extracts. The cell pellet (∼2 × 1011 cells) was washed twice with PBS and once with buffer I (20 mm Tris-HCl (pH 7.7), 150 mm KCl, and 3 mm MgCl2). The cells were resuspended in 15 ml of buffer II (buffer I with 1 mm DTT and 10 μg/ml leupeptin), equilibrated in a nitrogen cavitation bomb (Parr Instruments Co.) with 750 psi N2 for 1 h at 4 °C, and disrupted by release from the bomb. After release of the pressure, protease inhibitor mixture (Roche Applied Science) was added, and the extract was treated with 0.5% Triton X-100. The extract was incubated at 4 °C for 15 min and cleared by centrifugation (15,000 × g), and the supernatant was incubated while rotating for 2 h with rabbit IgG-agarose beads (200 μl) (Sigma). The beads were washed five times with TEV buffer (buffer I with 0.5 mm DTT, 0.5 mm EDTA) and incubated overnight in 1.5 ml of TEV buffer with 200 units of tobacco etch virus protease (Promega). After centrifugation, the supernatant was incubated with 50 μl of Strep-T actin-Sepharose beads (IBA) for 1 h. The beads were washed with buffer III (TEV buffer with 2 mm CHAPS (GE Healthcare)), and the complexes were eluted with elution buffer (100 mm Tris-Cl (pH 8), 150 mm NaCl, 1 mm EDTA) containing 2.5 mm d-desthiobiotin (Sigma). The proteins were analyzed by mass spectrometry as follows. The samples for MS analysis were digested with trypsin, analyzed by LC-MS/MS on an LTQ-Orbitrap (Thermo), and identified by Sequest 3.31 software against the GeneDB Leishmania major specific data base.
T. brucei, Cell Lines, and Transformation
The silencing constructs using the T7 opposing and the stem-loop constructs were prepared using primers listed in supplemental S-1, as described previously (9, 29). To generate the YFP/CFP-tagged constructs, PCR fragments were amplified using the primers listed in supplemental S-1. The fragments were cloned into the p2828-YFP and p2709-CFP vectors as described previously (26, 30). To generate the PTP-tagged constructs that encode for a triple tag composed of the ProtC-binding site, tobacco etch virus protease recognition site, and protein A, the gene of interest was amplified with primers listed in supplemental S-1 and cloned into the PTP vector (8).
Northern and Primer Extension Analyses
Primer extension was performed as described previously (9). The extension products were analyzed on 6% acrylamide denaturing gels. Primers are listed in supplemental S-1. For Northern analysis, total RNA was extracted, separated on an agarose-formaldehyde gel, and analyzed using a DNA probe that was prepared by random labeling (9). Primers are listed in supplemental S-1. To determine changes in the level of RNA, the phosphorimages were subjected to densitometric analysis using ImageJ. The standard deviation is indicated for experiments that were repeated three times and more.
In Situ Hybridization Combined with Immunofluorescence
In situ hybridization with SL RNA was performed as described recently (20). The slides were incubated with 1:400 diluted primary anti-U1 70-kDa antibodies that were detected using IgG conjugated to FITC. Nuclei were stained using 4′-6′-diamidino-2-phenylindole (DAPI) or propidium iodide. The cells were visualized under a Zeiss LSM 510 META inverted microscope.
Generation of Antibodies against U1 70 kDa
The full-length sequence of U1 70 kDa (Tb927.8.4830) was cloned into the pHIS vector. Recombinant protein was purified using the Bugbuster reagent (Novagen, Inc.), and 400 μg of purified protein was used for multiple injections into rabbits, as described previously (26).
Poly(A) Tail Assay
Total RNA was subjected to splint labeling that labels only the poly(A) RNA. Ten to twenty μg of RNA was mixed with 150 pmol of an oligonucleotide that included poly(T) complementary to the poly(A), and runs of G (see supplemental S1 for the primer sequence). The mixture was heated for 2 min at 85 °C in 50 mm Tris-HCl (pH 7.8), 10 mm MgCl2, and 1 mm DTT. The annealing reaction was quenched on ice for 30 min; 50 μCi of [α-32P]dCTP (3000 Ci/mmol) and 5 units of T7 DNA polymerase (Sequenase version 2.0, United States Biochemical Corp.) were added, and the reaction was incubated for 1 h at 37 °C. The RNA was incubated with 2 μg of RNase A, 5 units of T1 (RNase/T1 mix, Fermentas), and 20 μg of yeast tRNA (Ambion) in 10 mm Tris-HCl, 2 mm MgCl2, 2 mm DTT, 0.2 mm ATP, 2% (v/v) DMSO, 300 mm NaCl, and 5 mm EDTA at 37 °C for 30 min. After phenol/chloroform extraction and ethanol precipitation, the tails were separated on 15% denaturing gel. Quantitation of intensity of the labeled tails was performed by analyzing the phosphorimages using ImageJ.
Live Cell Imaging of the YFP/CFP-tagged Proteins
T. brucei cells expressing the YFP-tagged proteins were stained with 1 μg/ml Hoechst 33342 (Molecular Probes) for 30 min, then washed in phosphate-buffered saline (PBS), and dropped on slides. Image acquisition was performed with a motorized Zeiss AxioImager Z1 wide field microscope equipped with an EC Plan-NeoFluar 100×/1.30 Oil Iris M27 objective and AxioCamHR CCD camera. The microscopic setup was controlled using the features of the AxioVision software (version 4.6.3.0, Carl Zeiss Imaging Solutions Gmbh). For all images, the exposure times were automatically adjusted to the desired maximum signal intensity, preventing overexposure. Three-dimensional images were acquired using the optimal sampling density derived from the optical setup as stacks of 30 images taken with a Z step size of 0.3 μm. Following image acquisition, the raw data were exported to the Huygens Essential software (version 3.4.0.1 for Windows x64, Scientific Volume Imaging B.V.), and digital de-convolution was performed using the “maximum likelihood estimation” algorithm (>100 iterations). The theoretical point spread function was used for the deconvolution process. The restored image data set was visualized and analyzed with the Imaris software package, featuring the “Surpass” module (version 6.1.5 for Windows x64, Bitplane AG).
RESULTS
Identification of Proteins Associated with SmD3 and Lsm3
The proteins associated with SmB and SmD1 were recently identified after purification of these TAP-PTP-tagged proteins in T. brucei. SmB binds to U1, U5, U4, and SL RNA (21), and SmD1 binds to U1, U2, U4, U5, and SL RNA (23). In this study, we chose to identify the proteins that co-purified with SmD3, because this protein binds only to U1, U5, and SL RNA, thus increasing the probability of identifying SL RNP-specific proteins. L. tarentolae was chosen as an experimental system, because it was used previously to purify RNP complexes (31) and was also utilized for functional analysis of SL RNA (32). Because the genome of L. tarentolae was not available, we expressed in L. tarentolae proteins derived from the closely related species L. major. The proteins were cloned into the pSAP1 vector, carrying a C-terminal tag composed of a protein A binding domain, the tobacco etch virus protease cleavage site, and the streptavidin-binding peptide (Fig. 1A). The results suggest that the tagged proteins expressed in the transgenic parasites were of the expected size (Fig. 1B). Purification of the complexes was carried out as described under “Experimental Procedures.” RNA was extracted from the final step of purification, separated on a denaturing gel alongside total RNA, and stained with silver. The results (Fig. 1C, panels a and b) demonstrate that during SmD3 purification three RNA species with sizes corresponding to SL RNA, U1, and U5 were selected (Fig. 1C, panel a). In addition, degradation products of SL RNA (Fig. 1C, marked with asterisks) were also observed. The identity of these fragments as SL RNA degradation products was verified by Northern analysis (results not shown). Staining of RNA purified from Lsm3 revealed a single RNA species of size corresponding to U6 (Fig. 1C, panel b). The identity of the RNAs enriched in the purification of both SmD3 and Lsm3 was verified by primer extension using U snRNA-specific probes for SmD3 purification (Fig. 1Dpanel a) and for Lsm3 purification (Fig. 1D, panel b). The results demonstrate that SmD3 purification enriched SL, U1, and U5 snRNAs, and the purification of Lsm enriched U6 snRNA. Proteins from the final step of purification were fractionated on a 12% SDS-polyacrylamide gel and stained with silver. The SmD3-associated proteins are presented in Fig. 1E, panel a, and the Lsm3-associated proteins in Fig. 1E, panel b. The proteins were then subjected to mass spectrometry.
FIGURE 1.
Purification of SmD3 and Lsm3 complexes from L. tarentolae. A, schematic presentation of the vector used to express the SmD3 and Lsm3 proteins. The tag complements, streptavidin-binding domain, tobacco etch virus (TEV) cleavage site, and protein A are indicated. B, Western analysis demonstrating the expression of SmD3- and Lsm3-tagged proteins; proteins were extracted from 107 cells, separated on a 12% SDS-polyacrylamide gel, and subjected to Western analysis by reaction with rabbit IgG (Sigma). C, silver stain of RNA extracted from the purified SmD3 (panel a) and Lsm3 (panel b) particles. Purification was carried out from ∼2 × 1011 cells as described under “Experimental Procedures.” Half of the final material was separated next to total RNA (10 μg) on a 6% denaturing acrylamide gel. The gel was stained with silver. The size markers and the identity of the RNAs are indicated. D, primer extension to identify the content of U snRNA in the purified complexes. SmD3, panel a; Lsm3, panel b. Half of the sample analyzed in C was subjected to primer extension using U snRNA-specific probes, listed in supplemental S-1. E, protein composition of the SmD3 (panel a), and Lsm3 (panel b) complexes. Purification was performed using ∼2 × 1011 cells, as described under “Experimental Procedures.” The purified proteins were separated on a 12% SDS-polyacrylamide gel and stained with silver. The molecular mass markers are indicated.
Proteins Associated with SmD3 Reveal snRNP Proteins and Novel Splicing or Splicing-associated Factors
The purification of SmD3 identified 54 proteins (Table 1), including the following: the entire repertoire of the seven Sm core proteins; Lsm2 and Lsm8; three of the U1 proteins, U1 70 kDa, U1A, and U1C; six U5 snRNP proteins; and proteins associated with the NineTeenComplex (33) such as CWC21, PRP19, SYF1, and PRP46. The other proteins with known functions that were selected include the following: importin α; a TFIIH factor implicated in SL RNA transcription (34); the helicase DHH1; poly(A)-binding proteins PAB1 and PAB2; ATPase; and Up13 ubiquitin protein ligase (Table 1). Of special interest are eight hypothetical proteins. The list in Table 1 was compared with the list described in published reports of the purification of SmB (21) and SmD1 (23). Of these, 30 of the proteins were already identified in the previous purification of the snRNP particles. In addition, 19 novel proteins were revealed for the first time in this study (see Table 1), and two additional proteins were detected here that were identified previously but were not annotated. One is the Brr2 homologue (Tb927.5.2290), which shares 26% identity and 46% similarity with the yeast protein, and the second one is DED1 (Tb927.10.14550), which shares 38% identity and 57% similarity with the yeast protein (Table 1). The purification revealed not only proteins stably associated with the snRNP particles but also stable complexes that are most probably released from the active spliceosome, such as PRP19 and its associated proteins. Surprisingly, although the purification identified proteins involved in SL RNA transcription (TFIIH factor) and biogenesis (SMN and GEMIN2), none of the enzymes involved in cap modification were identified, suggesting that the interaction of these enzymes during SL RNA biogenesis is transient and not very stable. Comparing the data in Table 1 with previous studies (21, 23) suggests that our purification revealed eight novel hypothetical proteins, three of which were also identified in Lsm3 purification (see below), but also including two proteins with distinct domains (numbers 39 and 40 in Table 1). As in the study of Palfi et al. (21), importin α and poly(A)-binding proteins were identified in the purified complex, but La protein, which was also purified by Luz Ambrósio et al. (23), was not detected in our purification. In addition, our studies as well as those of Luz Ambrósio et al. (23) failed to detect coatamer proteins that were identified in the purification of Palfi et al. (21). Although U2 is the most abundant U snRNP, it was absent from the purified sample, because SmD3 does not bind to U2 snRNA (12, 13). However, not all the co-purified proteins are necessarily genuine snRNP proteins, because the purification also included peptides from very abundant proteins such as the tubulins.
TABLE 1.
Mass spectrometric identification of SmD3 purification products
L. tarentolae proteins were identified by mass spectroscopy as described under “Experimental Procedures.” Each protein is described by the systematic GeneDB name, annotation, and molecular mass (in kilodaltons), T. brucei homologue, molecular mass of the T. brucei homologue, and reference. An asterisk after the annotation indicates that this protein was also identified by Lsm3 purification. Proteins were grouped into Sm proteins, Lsm proteins, U1 snRNP, U4/U6 snRNP, U5 snRNP, SMN/GEMIN proteins, Prp19/CDC5/CWC, proteins with known functions, proteins involved in RNA binding, snoRNA proteins, helicases/ATPases, and hypothetical proteins. The protein U5–200K/Brr2 (Tb927.5.2290) was identified in Luz Ambrósio et al. (23) but was not annotated as a U5 protein.
| Protein category and accession no | Annotation | Molecular mass | T. brucei homologue | Molecular mass | Source or Ref. | |
|---|---|---|---|---|---|---|
| kDa | kDa | |||||
| Sm proteins | ||||||
| 1 | LmjF27.1970 | SmB* | 11.8 | Tb927.2.4540 | 12.3 | 60 |
| 2 | LmjF22.1060 | SmD1* | 11.2 | Tb927.7.3120 | 11.7 | 60 |
| 3 | LmjF33.3190 | SmD2* | 11.7 | Tb927.2.5850 | 12.5 | 60 |
| 4 | LmjF34.3860 | SmD3* | 12.2 | Tb927.4.890 | 12.3 | 60 |
| 5 | LmjF30.1205 | SmE | 9.8 | Tb927.6.2700 | 9.6 | 60, 61 |
| 6 | LmjF35.4460 | SmF* | 8.5 | Tb09.211.1695 | 8.3 | 60 |
| 7 | LmjF32.1070 | SmG* | 8.7 | Tb11.01.5915 | 8.9 | 60 |
| Lsm proteins | ||||||
| 8 | LmjF16.1250 | Lsm2* | 15.8 | Tb927.8.5180 | 13.2 | 26 |
| 9 | LmjF25.1705 | Lsm8* | 13.8 | Tb927.3.1780 | 14 | 11 |
| U1 snRNP | ||||||
| 10 | LmjF16.1590 | U1–70K | 31.5 | Tb927.8.4830 | 31.7 | 8 |
| 11 | LmjF36.6985 | U1A | 17.9 | Tb927.10.8300 | 18 | 23 |
| Tb927.10.8280 | 16 | |||||
| 12 | LmjF21.0320 | U1C | 25.1 | Tb927.10.2120 | 21.7 | 8 |
| U4/U6 snRNP | ||||||
| 13 | LmjF01.0560 | PRP3 * | 67 | Tb09.160.2900 | 63.2 | 21, 23 |
| 14 | LmjF21.1120 | PRP4* | 63.8 | Tb927.10.960 | 65.4 | 21, 23 |
| 15 | LmjF15.1470 | SNU13* | 13.5 | Tb09.160.3670 | 13.5 | 62 |
| U5 snRNP | ||||||
| 16 | LmjF35.3950 | PRP8* | 277.4 | Tb09.211.2420 | 277 | 63 |
| 17 | LmjF32.2200 | U5–116K/Snu114* | 110.2 | Tb11.01.7080 | 105.4 | 21, 23 |
| 18 | Lmj17.0920 | U5–200K/Brr2* | 260.2 | Tb927.5.2290 | 249.2 | |
| 19 | LmjF32.2490 | U5–102K/PRP6* | 137.5 | Tb11.01.7330 | 111 | 23 |
| 20 | LmjF28.2960 | U5–40K* | 47.5 | Tb11.01.2940 | 35 | 21, 23 |
| 21 | LmjF23.0650 | U5–15K/Dib1* | 24.2 | Tb927.8.2560 | 17.7 | 23 |
| SMN/GEMIN proteins | ||||||
| 22 | LmjF32.1802 | SMN | 16.8 | Tb11.01.6640 | 17 | 21 |
| 23 | LmjF36.1300 | Gemin2 | 67.5 | Tb927.10.5640 | 55.4 | 21 |
| Prp19/CDC5/CWC | ||||||
| 24 | LmjF01.0190 | CWC21 | 21.2 | Tb09.160.2110 | 16.1 | 21, 23 |
| 25 | LmjF27.2480 | PRP19 | 53.1 | Tb927.2.5240 | 54.2 | 21, 23 |
| 26 | LmjF23.1550 | Splicing factor SYF1 | 88.8 | Tb927.5.1340 | 92.1 | 23 |
| 27 | LmjF36.4820 | PRP46* | 55.6 | Tb927.10.10170 | 48.7 | |
| Proteins with known functions | ||||||
| 28 | LmjF35.4130 | Poly(A)-binding protein 2/PABP2 | 65.3 | Tb09.211.2150 | 62.1 | 21, 23 |
| 29 | LmjF35.5040 | Poly(A)-binding protein 1/PABP1 | 62.6 | Tb09.211.0930 | 63 | |
| 30 | LmjF25.0080 | Poly(A)-binding protein | 60.9 | |||
| 31 | LmjF30.1120 | Importin α subunit | 58.1 | Tb927.6.2640 | 58 | 64 |
| 32 | LmjF28.2770 | HSP70* | 71.6 | Tb11.01.3110 | 75.3 | |
| 33 | LmjF16.1630 | SIK1P* | 26 | Tb927.8.4790 | 26 | 23 |
| 34 | LmjF33.0814 | β-Tubulin* | 49.7 | |||
| 35 | LmjF13.0380 | α-Tubulin* | 49.7 | |||
| 36 | LmjF17.0085 | Translation elongation factor TEF1* | 49.1 | Tb927.10.2100 | 49.1 | 21 |
| 37 | LmjF24.2280 | TFIIH | 91.7 | Tb927.8.5980 | 92.4 | 65 |
| 38 | LmjF07.0280 | Up13 ubiquitin protein ligase | 669.4 | Tb927.8.1590 | 470.7 | |
| Proteins involves in RNA binding | ||||||
| 39 | LmjF35.1510 | NIF domain (NLI interacting factor-like phosphatase) | 64.4 | Tb09.211.4670 | 59 | |
| 40 | LmjF18.0300 | RRM-containing protein | 38.4 | Tb927.10.13800 | 34.4 | |
| snoRNA proteins | ||||||
| 41 | LmjF15.1380 | NOP58 | 66 | Tb09.160.3820 | 55 | 62 |
| Helicases/ATPases | ||||||
| 42 | LmjF35.0370 | DHH1* | 46.4 | Tb927.10.3990 | 46.4 | 66 |
| 43 | LmjF07.0340 | RNA helicase* | 55 | Tb927.8.1510 | 62.4 | |
| 44 | LmjF21.0710 | ATPase | 73.9 | Tb927.10.1630 | 71 | |
| 45 | LmjF13.0500 | RNA helicase* | 225 | Tb11.02.1930 | 248.3 | |
| 46 | LmjF32.0400 | RNA helicase* | 66.8 | Tb927.10.14550 | 71.3 | |
| Hypothetical proteins | ||||||
| 47 | LmjF14.1430 | Hyp (WD40) | 192.7 | Tb927.7.3560 | 199.2 | |
| 48 | LmjF22.0650 | Hyp | 54.6 | Tb927.7.2570 | 52.5 | |
| 49 | LmjF21.0820 | Hyp | 149.7 | Tb927.10.1490 | 134.8 | |
| 50 | LmjF23.0290 | Hyp* | 37.2 | Tb927.8.2230 | 41.6 | |
| 51 | LmjF32.2360 | Hyp* | 87.4 | Tb11.01.7210 | 71 | |
| 52 | LmjF25.0540 | Hyp* | 32.3 | Tb11.03.0530 | 31.2 | |
| 53 | LmjF28.2330 | Hyp | 99 | Tb11.01.3480 | 64.5 | |
| 54 | LmjF02.0580 | Hyp | 399.7 | Tb927.2.2650 | 367.6 | |
Purification of Proteins Associated with Lsm3 Protein
The mass spectrometry analysis of the Lsm3-associated proteins revealed 39 proteins (Table 2). As expected, the additional Lsm proteins associated with U6 snRNA were identified, except Lsm6 and Lsm7. Interestingly, these proteins were also barely detected in the purification of the yeast Lsm complex, and it was suggested that these proteins might be only loosely associated with the complex (25). No other Lsm proteins were revealed, notably, including the absence of Lsm1; these results support the notion that trypanosomes may lack the Lsm1–7 complex, which in other eukaryotes is implicated in mRNA degradation (25). Indeed, as opposed to yeast, no proteins involved in mRNA degradation, such as XRN1 and PAT1, were identified in the purification (25). Six proteins that have homologues to U5-specific proteins were revealed. Among the proteins summarized in Table 2 are also U4-specific proteins, including PRP3, PRP4, and SNU13. In addition, the purification revealed five proteins (proteins 24–28) that might be contaminants. Of interest are those proteins that were enriched in both SmD3 and Lsm3 purifications (marked with asterisks in Table 2). Such proteins most probably are associated with U5 snRNP, because both purifications enriched this complex. The function of the helicases that are associated with the Lsm and Sm proteins is currently unknown. Further studies are needed to establish the role of these proteins in trypanosome splicing.
TABLE 2.
Mass spectrometric identification of Lsm3 purification products
L. tarentolae proteins were identified by mass spectroscopy as described under “Experimental Procedures.” Each protein is described by the systematic GeneDB name, annotation, molecular mass (kilodaltons), T. brucei homologue, molecular mass of the T. brucei homologue, and reference. An asterisk after the annotation indicates that this protein was also identified by SmD3 purification. Proteins were grouped into Lsm proteins, Sm proteins, U5 snRNP, U4/U6 snRNP, U5 snRNP, proteins with known functions, Prp19/CDC5/CWC, helicases/ATPases, and hypothetical proteins.
| Protein category and accession no | Annotation | Molecular mass | T. brucei homologies | Molecular mass | Source or Ref. | |
|---|---|---|---|---|---|---|
| kDa | kDa | |||||
| Lsm proteins | ||||||
| 1 | LmjF16.1250 | Lsm2* | 15.8 | Tb927.8.5180 | 13.2 | 26 |
| 2 | LmjF05.0540 | Lsm3 | 14.1 | Tb927.7.7380 | 10.1 | 11 |
| 3 | LmjF32.0715 | Lsm4 | 21.1 | Tb11.01.5535 | 14.1 | 11 |
| 4 | LmjF28.0970 | Lsm5 | 17.4 | tryp_XI-1036e06.p1k.embl | 13.9 | 26 |
| 5 | LmjF25.1705 | Lsm8* | 13.8 | Tb927.3.1780 | 14 | 11 |
| Sm proteins | ||||||
| 6 | LmjF27.1970 | SmB* | 11.8 | Tb927.2.4540 | 12.3 | 60 |
| 7 | LmjF22.1060 | SmD1* | 11.2 | Tb927.7.3120 | 11.7 | 60 |
| 8 | LmjF33.3190 | SmD2* | 11.7 | Tb927.2.5850 | 12.5 | 60 |
| 9 | LmjF34.3860 | SmD3* | 12.2 | Tb927.4.890 | 12.3 | 60 |
| 10 | LmjF35.4460 | SmF* | 8.5 | Tb09.211.1695 | 8.3 | 60 |
| 11 | LmjF32.1070 | SmG* | 8.7 | Tb11.01.5915 | 8.9 | 60 |
| U5 snRNP | ||||||
| 12 | LmjF35.3950 | PRP8* | 277.4 | Tb09.211.2420 | 277.4 | 63 |
| 13 | LmjF17.0920 | U5–200K/Brr2* | 260.2 | Tb927.5.2290 | 249.2 | |
| 14 | LmjF32.2200 | U5–116K/Snu114* | 110.2 | Tb11.01.7080 | 105.4 | 21, 23 |
| 15 | LmjF32.2490 | U5–102K/PRP6* | 137.5 | Tb11.01.7330 | 111 | 63 |
| 16 | LmjF28.2960 | U5–40K* | 47.5 | Tb11.01.2940 | 35 | 21, 23 |
| 17 | LmjF23.0650 | U5–15K/Dib1* | 24.2 | Tb927.8.2560 | 17.7 | 23 |
| U4/U6 snRNP | ||||||
| 18 | LmjF01.0560 | PRP3* | 67 | Tb09.160.2900 | 63.2 | 21, 23 |
| 19 | LmjF21.1120 | PRP4* | 63.8 | Tb927.10.960 | 65.4 | 21, 23 |
| 20 | LmjF15.1470 | SNU13* | 13.5 | Tb09.160.3670 | 13.5 | 62 |
| Proteins with known functions | ||||||
| 21 | LmjF28.2770 | HSP70* | 71.6 | Tb11.01.3110 | 75.3 | |
| 22 | LmjF26.1240 | HSP70 | 70.5 | Tb927.7.710 | 70.2 | |
| 23 | LmjF16.1630 | SIK1P* | 26 | Tb927.8.4790 | 26 | 23 |
| 24 | LmjF33.0814 | β-Tubulin* | 49.7 | |||
| 25 | LmjF13.0380 | α-Tubulin* | 49.7 | |||
| 26 | LmjF17.0085 | Translation elongation factor TEF1* | 49.1 | Tb927.10.2100 | 49.1 | 21 |
| 27 | LmjF25.0670 | Epsin | 65.9 | Tb11.50.0006 | 55.2 | |
| 28 | LmjF21.0885 | Mismatch repair protein | 148.3 | Tb927.10.1270 | 127.3 | |
| 29 | LmjF28.2750 | Activated protein kinase C receptor (LACK) | 34.3 | Tb11.01.3170 | 34.6 | |
| Prp19/CDC5/CWC | ||||||
| 30 | LmjF36.4820 | PRP46* | 55.6 | Tb927.10.10170 | 48.7 | |
| Helicases/ATPases | ||||||
| 31 | LmjF35.0370 | DHH1* | 46.4 | Tb927.10.3990 | 46.4 | 66 |
| 32 | LmjF07.0340 | RNA helicase* | 55 | Tb927.8.1510 | 62.4 | |
| 33 | LmjF35.3100 | RNA helicase | 100.2 | Tb09.211.3510 | 82.7 | |
| 34 | LmjF13.0500 | RNA helicase* | 225 | Tb11.02.1930 | 248.3 | |
| 35 | LmjF32.0400 | RNA helicase* | 66.8 | Tb927.10.14550 | 71.3 | |
| Hypothetical proteins | ||||||
| 36 | LmjF32.2360 | Hyp* | 87.4 | Tb11.01.7210 | 71 | |
| 37 | LmjF25.0540 | Hyp* | 32.3 | Tb11.03.0530 | 31.2 | |
| 38 | LmjF03.0790 | Hyp | 45.4 | Tb927.3.2700 | 42.6 | |
| 39 | LmjF23.0290 | Hyp* | 37.2 | Tb927.8.2230 | 41.6 | |
U1 snRNP Is Essential for cis- but Not trans-Splicing, although U1A, a Bona Fide U1 snRNP Protein, Is Essential for trans-Splicing as Well
Previously, we observed that U1 snRNP exists in the same ∼45 S complex as the SL RNP and all other snRNPs essential for trans-splicing (14). U1 snRNP was shown not to be essential for nematode trans-splicing (35). However, the role of U1 snRNP in trypanosomes is very puzzling because only three cis-spliced substrates have been identified so far (36). It was therefore of great interest to examine if U1 snRNP may have additional function(s) in trypanosomes, especially in trans-splicing. To this end, the function of U1 snRNP was examined by RNAi silencing of three of the U1 snRNP proteins, the U1 70-kDa, U1 24-kDa protein, and U1A. U1A was not detected in the initial purification of the U1 snRNP (8) nor in the purification of SmB complex (21), but it was first described in Luz Ambrósio et al. (23). In this study U1A was also detected (Table 1).
A stem-loop construct was prepared to silence the U1 70 kDa. The results (Fig. 2A) demonstrate that the U1 snRNP is essential for the parasite, because upon silencing growth was severely inhibited. Silencing was confirmed by Northern analysis and showed 95% reduction in the cognate mRNA (Fig. 2B). Silencing of the U1 70 kDa destabilized the U1 snRNP. The level of U1 snRNA was determined by primer extension. Because U1 snRNA is not only capped but also modified at the first nt by Mtr1, three extension products were revealed, reflecting the fully capped snRNA, and the partially modified intermediates (37). A reduction of 95 ± 3% in the level of U1 snRNA was observed, but no effect was observed on the level of other snRNAs (Fig. 2C). Such a specific reduction is expected, because this protein binds to the first stem-loop of the RNA (8). As expected, a change was observed for the cis-spliced substrate, poly(A) polymerase (PAP) (Tb927.3.3160). Silencing for 2 days resulted in 65% reduction in the mature PAP transcript and was accompanied by increased level of the precursor (Fig. 2D).
FIGURE 2.
U1 70-kDa silencing destabilizes the level of U1 snRNA and inhibits cis- but not trans-splicing. A, growth curves of T. brucei cells silenced for the U1 70 kDa. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 2 × 104 cells per ml. B, Northern blot analysis of the U1 70-kDa gene. RNA was prepared from uninduced cells (−Tet) and from cells after 2 days of induction (+Tet). Total RNA (20 μg) was subjected to Northern analysis with randomly labeled probes. The transcript of the U1 70 kDa and the dsRNA are shown, as well as 7SL RNA, which was used to control for equal loading. C, silencing of the U1 70 kDa specifically destabilized the level of U1 snRNA. Total RNA was extracted from uninduced cells or after 2 days of induction. The level of the U snRNAs was determined by primer extension, and the degree of reduction was determined by phosphorimaging. D, effect of U1 70-kDa silencing on cis-splicing. cDNA was prepared from the RNA extracted from uninduced cells (−Tet) or after 2 days of induction (+Tet). The cDNA was subjected to PCR with oligonucleotides specific to mature or pre-PAP transcript, as described under “Experimental Procedures.” The level of 7SL RNA was used to control for equal amounts of cDNA. E, panel a, reduction in the level of U1 70 kDa during silencing of gene expression. Proteins from cells (107) after 2–5 days of silencing were separated on a 10% SDS-polyacrylamide gel and subjected to Western analysis with anti-U1 70-kDa antibodies prepared as described under “Experimental Procedures.” An additional aliquot of tetracycline was added after 3 days to induce maximum silencing. Reactivity with PTB1 antibodies (59) was used as a control for equal loading. Panel b, trans-splicing during U1 70-kDa silencing. Total RNA (10 μg) from the same cells as described in panel a was subjected to primer extension with an oligonucleotide complementary to the intron region of SL RNA (supplemental S-1). Primer extension of U3 snoRNA was used to control for the level of the RNA in the samples. The products were separated on a 6% acrylamide denaturing gel. The results were subjected to phosphorimaging, and quantitation was performed using ImageJ. The levels of SL RNA and Y structure are given as fold change with respect to the amount present at day 0 and were normalized to the level of U3 snoRNA. Panel c, changes in the level of tubulin precursor during silencing of the U1 70 kDa. Total RNA (20 μg), from the cells described in panels a and b, was subjected to Northern analysis with antisense tubulin RNA probe. The blot was hybridized with random-labeled 7SL RNA probe to control for equal loading.
Next, the effect of U1 silencing on trans-splicing was examined. Cells were silenced for 5 days, and RNA and proteins were extracted from the cells from the 2nd day and analyzed. The level of the protein was examined by Western analysis using anti-U1 70-kDa antibodies. The results (Fig. 2E, panel a) show reduction of 85% after 2 days of silencing; the protein was completely absent after 4 days. The RNA was subjected to primer extension with antisense SL RNA primer, and the level of SL RNA and the Y structure intermediate were quantified by densitometry (Fig. 2E, panel b). The results suggest almost no change in SL RNA and the Y structure intermediate despite 85% reduction in the level of the protein, suggesting no effect on trans-splicing in the first days of silencing. Reduction in the level of Y structure and elevation of SL RNA were observed in the 4th and 5th days of silencing, suggesting that this might be a secondary effect due to a decrease in the level of PAP. Northern analysis demonstrated that tubulin precursors also accumulate after 4 days of silencing (Fig. 2E, panel c), suggesting that the depletion of U1 snRNP does not directly affect trans-splicing; the trans-splicing defect that appears at later stages is most probably the result of inhibiting polyadenylation. Because polyadenylation and trans-splicing are coupled (38, 39), inhibiting the polyadenylation process will compromise the trans-splicing process.
Next, the U1 24-kDa protein was silenced, using a stem-loop construct. The results (Fig. 3A) demonstrate that the U1 24-kDa protein is essential for the parasite, because upon silencing, growth was severely inhibited. The silencing was confirmed by Northern analysis and showed 93% reduction in the cognate mRNA (Fig. 3B). The silencing only slightly reduced the level of U1 snRNA (Fig. 3C). This is expected, because this protein interacts with U1 through a protein-protein interaction (8). Silencing did not have any effect on trans-splicing, because no change in the level of SL RNA or the Y structure intermediate were observed (Fig. 3D, panels a and b). However, as expected, the protein is essential for the function of cis-splicing, and silencing resulted in reduction in PAP (40%) and accumulation of its precursor (Fig. 3E).
FIGURE 3.
Effect of silencing the U1 24-kDa coding gene. A, growth curves of T. brucei cells silenced for the U1 24 kDa. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 1 × 105 cells per ml. B, Northern blot analysis of the U1 24-kDa protein. RNA was prepared from uninduced cells (−Tet) and cells after 2 days of induction (+Tet). Total RNA (20 μg) was subjected to Northern analysis with randomly labeled probes. Blot shows the transcript of the U1 24-kDa gene and the dsRNA, as well as 7SL RNA, which was used to control for equal loading. C, silencing of the U1 24 kDa slightly reduced the level of U1 snRNA. Total RNA (10 μg) was prepared from cells carrying the silencing construct uninduced (−Tet) or after 2 days of induction (+Tet) and was subjected to primer extension with U1-specific oligonucleotide, and the change in the level was determined by phosphorimaging. The level of U3 snoRNA was used to control for equal loading. D, effect of U1 24-kDa silencing on trans-splicing. Panel a, effect on the formation of the Y structure intermediate. Total RNA (10 μg) was prepared from cells carrying the silencing construct uninduced (−Tet) or after 2 days of induction (+Tet) and was subjected to primer extension with oligonucleotide complementary to the intron region of SL RNA (supplemental S-1). The products were separated on a 6% denaturing gel. The level of U3 snRNA was used to control for equal loading. Panel b, effect of U1 24-kDa silencing on SL RNA capping. RNA, as in panel a, was subjected to primer extension with oligonucleotide complementary to SL RNA (supplemental S-1). The RNA was separated on a 6% denaturing gel. The position of the capped nts is indicated. E, effect of U1 24-kDa silencing on cis-splicing. cDNA was prepared from RNA extracted from uninduced cells (−Tet) or after 2 days of induction (+Tet). The cDNA was subjected to PCR with oligonucleotide specific to mature or to pre-PAP transcript, as described under “Experimental Procedures.” The level of 7SL RNA was used to control for equal amounts of cDNA.
The recent identification of U1A was surprising given the fact that its canonical binding site on U1 snRNA is missing (8). The role of U1A in U1 snRNP function was therefore investigated as well by RNAi silencing. The results in Fig. 4A demonstrate that U1A is essential for parasite growth; silencing eliminated the U1A transcript by 95% (Fig. 4B). As expected, because U1A does not bind directly to the RNA, its silencing did not affect the steady-state level nor the modification of U1 snRNA (Fig. 4C). Silencing of U1A compromised cis-splicing, as expected; the level of PAP was reduced by 35%, and the precursor was increased (Fig. 4D).
FIGURE 4.
RNAi silencing of U1A suggests its function in both trans- and cis-splicing. A, growth curves of T. brucei cells silenced for U1A. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 5 × 104 cells per ml. B, Northern blot analysis. RNA was prepared from uninduced cells (−Tet) and cells after 2 days of induction (+Tet). Total RNA (20 μg) was subjected to Northern analysis with randomly labeled probes. The transcript of the U1A and the dsRNA as well as 7SL RNA that was used to control for equal loading are indicated. C, silencing of U1A has no effect on the level of U1 snRNA. Total RNA was extracted from uninduced cells or after 2 days of induction, and 10 μg was used to determine the level of the U1 snRNA by primer extension. The level of U3 was used to control for equal loading. D, cis-splicing of PAP in U1A silenced cells. RNA was prepared from uninduced cells or after 2 days of induction. cDNA was prepared and subjected to PCR amplification with oligonucleotide specific to mature or pre-PAP transcript, as described under “Experimental Procedures.” The level of 7SL RNA was used to control for equal amounts of cDNA. E, panel a, effect of U1 silencing on the level of U1A protein. Cells expressing PTP-U1A and the U1A silencing construct were silenced for the number of days indicated. Proteins from cells (107) after 2–5 days of silencing were separated on a 10% SDS-polyacrylamide gel and subjected to Western analysis as described under “Experimental Procedures.” Reactivity with PTB1 antibodies was used as a control for equal loading. Panel b, U1A silencing affects trans-splicing. Total RNA (10 μg) from the same cells as described in panel a was subjected to primer extension with an oligonucleotide complementary to the intron region of SL RNA (supplemental S-1). Primer extension of U3 was used to determine the RNA used. The products were separated on a 6% acrylamide denaturing gel. The results were subjected to phosphorimaging and quantitation using ImageJ. The levels of SL RNA and Y structure are given as fold change with respect to the amount present at day 0 and were normalized to the level of U3 snoRNA. Panel c, changes in the level of tubulin precursor during silencing of U1A. Total RNA (20 μg) from the cells described in panels a and b was subjected to Northern analysis with antisense tubulin RNA probe. The blot was hybridized with random-labeled 7SL RNA probe to control for equal loading. F, localization of U1A with respect to SL RNA. In situ hybridization combined with immunofluorescence was performed as described under “Experimental Procedures.” Panel a, in situ hybridization with SL RNA; panel b, immunofluorescence of PTP-tagged U1A; panel c, merge of panels a and b; panel d, DIC and DAPI-stained nuclei merge with panel c. Panel e, fluorescence emissions at 445–450 nm (blue, DAPI), 525–550 nm (green, the FITC bound to antibody), and 650–690 nm (red, SL RNA, Alexa fluor 647) were examined in the area surrounding the line (inset). The intensity of the different chromophores is plotted, demonstrating overlap between the SL RNA, U1A, and the nucleus signals.
Next, the effect of U1A on trans-splicing was examined. The U1A was PTP-tagged and introduced into the strain carrying the silencing construct. The results (Fig. 4E, panel a) demonstrate 70% reduction in U1A protein from the 2nd day of silencing. A clear trans-splicing defect was observed as early as the 2nd day of silencing, resulting in an increase of the SL RNA and reduction in the Y structure (Fig. 4E, panel b). Northern analysis indicated an increase in tubulin precursors, which started to accumulate on the 2nd day of silencing, suggesting a direct effect of U1A depletion on trans-splicing or polyadenylation (Fig. 4E, panel c). This finding is in contrast to a secondary effect on trans-splicing observed only after 4 days of silencing of the U1 70 kDa (Fig, 2E, panels b and c).
Next, the localization of U1A was examined with respect to the SL RNA. Immunofluorescence was used to detect the PTP-U1A fusion, and SL RNA was detected by in situ hybridization (Fig. 4F). The data suggest that U1A is not localized in the SL RNP factory because almost no overlap was found between the two chromophores (Fig. 4F, panel e).
To investigate the basis for the effect of U1A on trans-splicing, the U1A complex was tagged in L. tarentolae, and after purification, the proteins were subjected to mass spectrometry analysis. Proteins and RNA were extracted from the final step of purification. The eluted proteins were stained with silver (Fig. 5A). The level of the selected RNAs was determined by primer extension, and the results demonstrate enrichment of the U1 snRNA and minor selection of U2 and U5 snRNPs (Fig. 5B). The list of the proteins identified by mass spectrometry is given in Table 3. Surprisingly, in addition to the U1 snRNP proteins, other factors, including the polyadenylation factor CPSF73, were enriched. In a parallel experiment, the proteins associated in L. tarentolae with the polyadenylation factor CPSF73 were purified and identified by mass spectrometry (results not shown). Out of the 45 proteins associated with U1A, 25 were found in both complexes, suggesting that U1A might be involved in polyadenylation. Interestingly, among the selected proteins, factors involved in RNA metabolism and protein modification such as ADP-ribosylation, polyubiquitinylation, and proteasome functions were also identified (see under “Discussion”).
FIGURE 5.
Purification of U1A-associated proteins. A, protein composition of the U1A complex. Purification was performed using ∼2 × 1011 cells, as described under “Experimental Procedures.” The purified proteins were separated on a 12% SDS-polyacrylamide gel and stained with silver. The molecular mass markers are indicated. B, specificity of snRNA selection using the U1A-tagged protein. Extract was prepared from cells as described (13). RNA was extracted from IgG-agarose beads and subjected to primer extension with the different probes (supplemental S-1). RNA was prepared from an aliquot of cells before selection (Total), and RNA from the beads (Final). Only an aliquot (2%) of the sample was analyzed from the total RNA, whereas the entire sample from the beads was analyzed. C, U1A functions in polyadenylation. Panel a, changes in poly(A) tail during U1A silencing. Cell expressing silencing constructs for either PAP,4 U1 70 kDa, and U1A were silenced. RNA was prepared from uninduced cells (−) or after 2 days of silencing (+). RNA was splint-labeled, digested with RNase A and T1, and the labeled tails were separated on a 15% denaturing gel next to a pBR322 DNA-MspI digest. The level of U3 snoRNA was used to determine the amount of RNA in each sample. Panel b, quantitation of the poly(A) tail length. The phosphorimages were scanned, and the intensity of the signals was measured and plotted. The black and gray histograms represent the tail length distributions of uninduced and induced cells, respectively. D, fractionation of U1 snRNP-associated proteins. Whole cell extracts were prepared from 5 × 108 cells and layered on continuous 10–30% (w/v) sucrose gradient in buffer A containing 150 mm KCl. Gradients were centrifuged at 4 °C for 3 h at 35,000 rpm in a Beckman SW41 rotor. S values were determined using the following standards: 30 S, 50 S, and 70 S E. coli ribosomes and the enzyme, catalase (10 S). Following centrifugation, 24 fractions were collected, and RNA and protein samples were prepared after ethanol precipitation. The proteins were subjected to Western analysis with anti-U1 70-kDa antibodies that recognize the PTP-tagged U1A and the U1 70 kDa. U1 snRNA level was determined by primer extension.
TABLE 3.
Mass spectrometric identification of U1A purification products
L. tarentolae proteins were identified by mass spectroscopy as described under “Experimental Procedures.” Each protein is described by the systematic GeneDB name, annotation, molecular mass (kilodaltons), T. brucei homologue, molecular mass of the T. brucei homologue, and reference. An asterisk after the annotation indicates that this protein was also identified by CPSF73 purification. Proteins were grouped U1 snRNP, RNA processing factors, helicases/ATPases, chaperons, translation factors, post-translation factors, and other proteins.
| Protein category and accession no. | Annotation | Molecular mass | T. brucei homologue | Molecular mass | Source or Ref. | |
|---|---|---|---|---|---|---|
| kDa | kDa | |||||
| U1snRNP | ||||||
| 1 | LmjF16.1590 | U1–70K | 31.5 | Tb927.8.4830 | 31.7 | 8 |
| 2 | LmjF36.6985 | U1A | 17.9 | Tb927.10.8300 | 18 | 23 |
| Tb927.10.8280 | 16 | |||||
| 3 | LmjF21.0320 | U1C | 25.1 | Tb927.10.2120 | 21.7 | 8 |
| 4 | LmjF27.1970 | SmB | 11.8 | Tb927.2.4540 | 12.3 | 60 |
| 5 | LmjF22.1060 | SmD1 | 11.2 | Tb927.7.3120 | 11.7 | 60 |
| 6 | LmjF33.3190 | SmD2* | 11.7 | Tb927.2.5850 | 12.5 | 60 |
| 7 | LmjF34.3860 | SmD3 | 12.2 | Tb927.4.890 | 12.3 | 60 |
| 8 | LmjF30.1205 | SmE | 9.8 | Tb927.6.2700 | 9.6 | 60, 61 |
| 9 | LmjF35.4460 | SmF | 8.5 | Tb09.211.1695 | 8.3 | 60 |
| RNA processing factors | ||||||
| 10 | LmjF28.2960 | U5–40K | 47.5 | Tb11.01.2940 | 35 | 21, 23 |
| 11 | LmjF27.2480 | PRP19 | 53.1 | Tb927.2.5240 | 54.2 | 21, 23 |
| 12 | LmjF17.0710 | CEF1 | 89 | Tb927.5.2060 | 80.1 | |
| 13 | LmjF35.5040 | Poly(A)-binding protein 1/PABP1 (Cytoplasma) | 62.6 | Tb09.211.0930 | 63 | |
| 15 | LmjF33.1150 | Pumilio/PUF RNA-binding protein 6* | 93 | Tb927.10.11760 | 93 | |
| 16 | LmjF36.5100 | Pumillio/PUF RNA-binding protein 11 | 105.3 | Tb11.01.2580 | 99.3 | |
| 17 | LmjF34.3430 | CPSF73* | 84.1 | Tb927.4.1340, | 85.3 | |
| 18 | LmjF35.2330 | Importin β2 subunit | 99.4 | Tb09.211.4360 | 103.4 | |
| Helicases/ATPases | ||||||
| 19 | LmjF32.0400 | RNA helicase* | 66.8 | Tb927.10.14550 | 71.3 | |
| 20 | LmjF35.0370 | DHH1* | 46.4 | Tb927.10.3990 | 46.4 | 66 |
| 21 | LmjF07.0340 | RNA helicase* | 55 | Tb927.8.1510 | 62.4 | |
| 22 | LmjF35.3100 | RNA helicase | 100.2 | Tb09.211.3510 | 82.7 | |
| 23 | LmjF21.1552 | RNA helicase* | 49.4 | Tb927.10.540 | 49.2 | |
| 24 | LmjF05.0510 | ATPase alpha subunit* | 62.5 | Tb927.7.7420 | 63.5 | |
| Tb927.7.7430 | ||||||
| Chaperons | ||||||
| 25 | LmjF23.1220 | T-complex protein 1, γ subunit* | 60.3 | Tb927.8.3150 | 60.8 | |
| 26 | LmjF36.6910 | Chaperonin, putative, T-complex protein 1 (θ subunit)* | 58.1 | Tb927.10.8190 | 58 | |
| Translation factors | ||||||
| 27 | LmjF17.0085 | Translation elongation factor TEF1* | 49.1 | Tb927.10.2100 | 49.1 | 21 |
| 28 | LmjF09.1070 | Eukaryotic translation initiation factor 2 subunit* | 52.6 | Tb11.01.4830 | 52 | |
| 29 | LmjF33.2740 | Translation initiation factor IF-2* | 92.4 | Tb927.2.3780 | 94.4 | |
| Post-translation factors | ||||||
| 30 | LmjF36.3530 | Polyubiquitin* | 94 | Tb11.01.1680 | 76.6 | |
| 31 | LmjF31.2280 | ADP-ribosylation factor* | 20.5 | |||
| 32 | LmjF04.0480 | ADP-ribosylation factor, putative | 21 | Tb09.160.5600 | 21.2 | |
| 33 | LmjF29.0880 | ADP ribosylation factor 3, putative* | 20.1 | Tb927.3.3450 | 19.8 | |
| 34 | LmjF05.0030 | Ras-like small GTPases* | 21.7 | Tb927.5.4500 | 21.7 | |
| 35 | LmjF29.0120 | Proteasome regulatory non-ATPase subunit* | 99.3 | Tb927.3.5520 | 99.9 | |
| 36 | LmjF28.1730 | Proteasome regulatory non-ATPase subunit 2* | 106 | Tb11.01.0960 | 106.5 | |
| 37 | LmjF22.0570 | Proteasome regulatory ATPase subunit 1* | 49 | Tb927.7.2500 | 48.4 | |
| 38 | LmjF13.1360 | Hyp* (alkaline phosphatase) | 90.9 | Tb11.02.0980 | 87.8 | |
| Other proteins | ||||||
| 39 | LmjF25.1420 | GTP-binding protein* | 24.2 | Tb927.3.1120 | 24.4 | |
| 40 | LmjF05.0280 | Protein-tyrosine phosphatase* | 25.1 | Tb927.10.10610 | 25.4 | |
| 41 | LmjF23.0290 | HUS1-LIKE/Hyp* | 37.2 | Tb927.8.2230 | 41.6 | |
| 42 | LmjF21.0820 | Hyp | 149.7 | Tb927.10.1490 | 134.8 | |
| 43 | LmjF32.3210 | Hyp* | 24.2 | Tb11.01.8590 | 24.4 | |
| 44 | LmjF33.0814 | β-Tubulin* | 49.7 | |||
| 45 | LmjF13.0380 | α-Tubulin* | 49.7 | |||
To examine the intriguing possibility that U1A participates in polyadenylation, the size distribution of the poly(A) tails was examined under U1A depletion. To this end, the poly(A) tails were labeled at the 3′ end, and the mRNA was digested with RNases A and T1, leaving intact the poly(A) tails. The poly(A) tails were compared in uninduced cells and after 2 days of silencing. The same amount of RNA was used in each assay as judged by primer extension analysis using U3 snoRNA (Fig. 5C, panel a). The poly(A) tails were measured in cells silenced for PAP, U1A, or the U1 70 kDa. The results indicate significant shortening of the tails in cells silenced for PAP or U1A but not in the U1 70-kDa-silenced cells (Fig. 5C, panels a and b). This can be clearly observed by comparing the abundance of each poly(A) tail length from the uninduced cells (shown in black) to that of the silenced cells (gray) (Fig. 5C, panel b). These results suggest that silencing of U1A affects the poly(A) tail length much like silencing of poly(A) polymerase, clearly suggesting a direct role of U1A in polyadenylation. However, at the early stages of silencing, the U1 70 kDa does not affect polyadenylation. As discussed above, the effects of U1 70-kDa silencing on trans-splicing and polyadenylation are most probably secondary. Next, the presence of U1A in a complex outside the U1 snRNP was examined. To this end, whole cell extracts were prepared and fractionated on a 10–30% sucrose gradient. The fractions from the gradient were subjected to Western analysis (Fig. 5D, panels a and b). The U1A and 70-kDa proteins were found mostly at the top of the gradient in the 10 S U1 snRNP complex, which was identified by primer extension of U1 snRNA (Fig. 5D, panel c). As opposed to the 70 kDa, the U1A was found in an additional complex of ∼20 S. The data presented in Fig. 5 suggest that U1A participates in polyadenylation but most probably not as part of U1 snRNP. Because polyadenylation is linked to trans-splicing (38, 39), we cannot rule out the possibility that the effect on trans-splicing is not direct but stems only from the effect of U1A on polyadenylation.
PRP19 Is Essential for the First Step of trans-Splicing, Functions Early in SL RNA Biogenesis, and Is Localized to the SL RNP Factory
The role of U1A in processes other than cis-splicing demonstrated that in trypanosomes conserved splicing factors might have evolved to carry out additional unique and special functions. The PRP19 complex was shown to be part of a salt-resistant complex that is released from active mammalian spliceosomes (40). This complex is known as the PRP19-CDC5 complex in mammals and NTC in yeast (33, 40). The PRP19 complex is required for stable association of U5 and U6 with the pre-mRNA after U1 and U4 have dissociated during the splicing reaction (33, 40). It was interesting to find that SmD3 purification revealed the U5-associated NTC complex. Five proteins of the NTC were identified in this study, PRP19, HSP70, SYF1, CWC21, and PRP46. Other components of this complex were also identified in the purification of the T. brucei snRNP complex, such as CWC15 (21, 23). This suggests that the purification of Sm proteins is not only enriched for mono-particles and small oligomeric snRNP complexes but also for a complex that is most probably released from the active spliceosome. The trypanosome PRP19 shares 27% identity and 51% similarity with the human protein, and contains the following three domains: an N-terminal U box (amino acids 2–59), a predicted coiled-coil domain (amino acids 67–121), and a WD repeat domain at its C terminus (amino acids 215–504) (see supplemental S-2).
To investigate the role of PRP19 in trans-splicing, the gene was silenced by RNAi. The results in Fig. 6A demonstrate that this factor is essential for growth. Silencing reduced the level of its mRNA by 98% (Fig. 6B). The silencing of PRP19 also inhibited cis-splicing, and a 50% reduction in PAP mRNA production was observed, together with an increase in the level of its precursor (Fig. 6C).
FIGURE 6.
PRP19 is essential for trans-splicing and is localized in the SL RNP factory. A, growth curves of T. brucei cells silenced for PRP19 homologue. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 5 × 105 cells per ml. B, Northern blot analysis of the prp19 gene. RNA was prepared from uninduced cells (−Tet) and cells after 2 days of induction (+Tet). Total RNA (20 μg) was subjected to Northern analysis with randomly labeled probes. The transcript of PRP19, dsRNA, as well as 7SL RNA that was used to control for equal loading are indicated. C, silencing of PRP19 affects cis-splicing. RNA was prepared from uninduced cells or after 2 days of induction. cDNA was prepared and subjected to PCR amplification with oligonucleotide specific to mature or pre-PAP transcript, as described under “Experimental Procedures.” The level of 7SL RNA was used to control for equal amounts of cDNA. D, silencing of PRP19 affects trans-splicing and SL RNA capping. Panel a, total RNA was extracted from uninduced cells or after 2 days of induction and was subjected to primer extension with oligonucleotide complementary to the intron region of SL RNA (supplemental S-1). The products were separated on a 6% acrylamide denaturing gel. The level of U3 snoRNA was used as a control for equal loading. Panel b, capping of SL RNA is perturbed in PRP19 silenced cells. RNA as in panel a was subjected to primer extension with oligonucleotide complementary to SL RNA (supplemental S-1), and extension products were separated next to the DNA sequence of the SL RNA gene. The positions of the cap nts are indicated. E, localization of PRP19. Transgenic cells for tSNAP42-CFP and YFP-PRP19 were stained with Hoechst. Panels b and c depict an enlargement of the cell located in the left side, and panels d and e show an enlargement of the cell on the right side. The cells were visualized as described under “Experimental Procedures.” Fluorescence image stacks were deconvoluted and rendered as an isosurface model showing overlapping signals tSNAP42 and PRP19. The Hoechst stain is shown in red.
Next, the effect of PRP19 silencing on trans-splicing was examined. The silencing resulted in classical splicing defects, resulting in increased levels of SL RNA and reduction of the Y structure intermediate (Fig. 6D, panel a). Surprisingly, silencing also induced changes in SL RNA capping, as can be seen by the accumulation of +2 cap nt, suggesting that PRP19 may be essential for the activity of MT57, the SL RNA specific cap-4 methylating enzyme, which carries out the modifications on the +3 and +4 nt of the SL RNA (Fig. 6D, panel b) (see “Discussion”) (41).
Because PRP19 seems to affect SL RNA capping, we investigated its localization in the SL RNP factory, the subnuclear domain where SL RNA transcription, modification, and Sm assembly take place (13, 20). To investigate the presence of PRP19 in the SL RNP factory, the co-localization of PRP19 with the SL RNA transcription factor, tSNAP42, was investigated. PRP19 was tagged in situ by YFP, and tSNAP42 was similarly tagged with CFP. The three-dimensional distribution of the two proteins was analyzed by fluorescence microscopy. As shown in Fig. 6E, the three-dimensional isosurface model, generated from the deconvoluted image stacks, demonstrates that PRP19 co-localized with tSNAP42, showing that this factor is intertwined with tSNAP42 and suggesting that PRP19 interacts with the SL RNA early in its biogenesis. As expected, because PRP19 is present in the active spliceosome, it is also found in “speckles” in other nuclear domains.
GEMIN2 Binds to snRNAs and Has a General Role in snRNA Biogenesis
Recently, the SMN homologue was identified in T. brucei (21). It was demonstrated that this factor functions to avoid illegitimate assembly of the canonical Sm proteins on RNAs that do not contain a bona fide canonical Sm site. The discriminatory activity of SMN in vitro requires preincubation of the SMN SmD3/B complex. Although GEMIN2 was shown to interact with SMN, its role in selective binding of the Sm core in trypanosomes is currently not known.
To investigate the role of GEMIN2 in snRNP biogenesis, the expression of the gene was silenced by RNAi, using a stem-loop construct (29). The results in Fig. 7A, panel a suggest that this factor is essential for growth. Silencing was confirmed by Northern analysis and showed 94% reduction in the cognate mRNA (Fig. 7A, panel b). The effect of silencing on the level of snRNAs was examined by primer extension. The results demonstrate a decrease in the level of U1 (43 ± 11%) and U5 (30 ± 5%) (Fig. 7B, lanes 1 and 4) and accumulation of defective SL RNA lacking modification at the +4 cap nt (35 ± 5%) (Fig. 7B, lane 6). This phenotype resembles the phenotype of Sm depletion, leading to a decrease in the U snRNAs but an increase in defective SL RNA (9). Of special interest is the phenotype observed for U2 and U4 snRNAs. Silencing increased the level of U2 snRNA (10 ± 4%). In the absence of Sm core assembly for U1, U5, and especially SL RNA, the pool of remaining Sm proteins becomes available for U2 snRNP. This may explain why the level of U2 increases during GEMIN2 silencing. However, the U4 snRNA level was decreased (10 ± 2%) (Fig. 7B, lane 3). U4 snRNA requires SmB for its proper assembly. In the absence of GEMIN2, which binds and may stabilize the steady-state level of SmD3/B, the level of SmB might be reduced, thus affecting the assembly of U4 snRNP. GEMIN2 is also required for cis-splicing, because in the silenced cells the level of PAP was decreased by 50%, whereas the precursor was increased (Fig. 7C).
FIGURE 7.
GEMIN2 exists in the SL RNP factory and directly and indirectly controls the level of snRNAs. A, RNAi silencing of GEMIN2. Panel a, growth curves of T. brucei cells silenced for the GEMIN2 homologue. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 5 × 104 cells/ml. Panel b, Northern blot analysis of the GEMIN2 gene. RNA was prepared from uninduced cells (−Tet) and cells after 2 days of induction (+Tet). Total RNA (20 μg) was subjected to Northern analysis with randomly labeled probes. The transcripts of GEMIN2, dsRNA, as well as 7SL RNA that was used to control for equal loading are indicated. B, level of U snRNAs upon GEMIN2 silencing. Total RNA was extracted from uninduced cells or after 2 days of induction. The level of the U snRNAs was determined by primer extension using specific oligonucleotide, as listed in supplemental S-1. U3 level was used as a control for equal loading. C, silencing of GEMIN2 affects cis-splicing. RNA was prepared from uninduced cells or after 2 days of induction. cDNA was prepared and subjected to PCR amplification with oligonucleotide specific to mature or pre-PAP transcript, as described under “Experimental Procedures.” The level of 7SL RNA was used to control for equal amounts of cDNA. D, localization of GEMIN2 with respect to SL RNA. In situ hybridization combined with immunofluorescence was performed as described under “Experimental Procedures.” Panel a, in situ hybridization with SL RNA; panel b, immunofluorescence of PTP-tagged GEMIN2; panel c, merge of panels a and b; panel d, DIC and DAPI-stained nuclei merge with panel c. Panel e, fluorescence emissions at 445–450 nm (blue, DAPI), 525–550 nm (green, the FITC bound to antibody), and 650–690 nm (red, SL RNA, Alexa Fluor 647) were examined in the area surrounding the line (inset). The intensity of the different chromophores is plotted, demonstrating overlap between the SL RNA, GEMIN2, and the nucleus signals.
Because SL RNA assembles in the SL RNP factory, it was of interest to examine if GEMIN2 is localized to this site. The results in Fig. 7D, using immunofluorescence with PTP-tagged protein and in situ with SL RNA, suggest co-localization. To quantitate the degree of co-localization, the fluorescence of both chromophores was measured; the results indicate the presence of a distinct overlapping peak between the SL RNA and GEMIN2 (Fig. 7D, panel e). However, GEMIN2 exists in other domains in addition to the SL RNP factory, as expected from a factor that also participates in the biogenesis of other snRNPs.
Identification of Trypanosome-specific Splicing Factors
The localization of GEMIN2 outside the SL RNP factory, observed in Fig. 7D, led us to examine where U1 and U5 assembly may takes place in the nucleus, and especially to explore whether these RNPs assemble in the SL RNP factory.
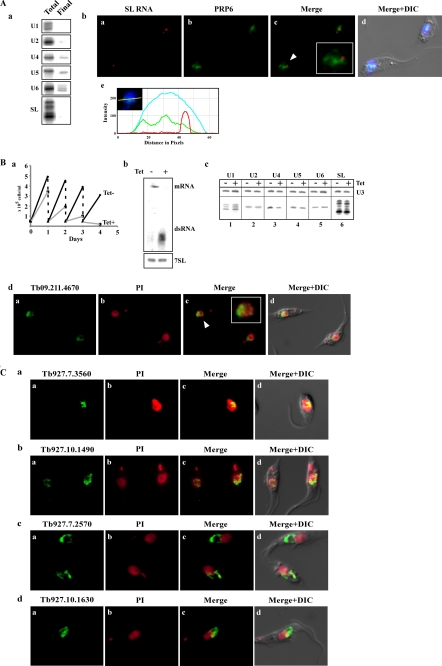
PRP6 is a splicing factor that binds the 20 S free U5 snRNP, but in mammals it also functions to mediate the interaction with U4/U6 to form the tri-snRNP complex (42). PRP6 was tagged by PTP, and the association of this factor with the different snRNAs was examined by quantitating the amount of snRNAs co-precipitated by the tagged protein. The results in Fig. 8A, panel a, clearly show that PRP6 binds efficiently to U5 but also binds to U4 and U6, albeit at a lower level, suggesting that the factor binds not only to U5 snRNP but also to the tri-snRNP complex. The localization of PRP6 was examined with respect to the SL RNA, and the results suggest that this factor does not co-localize with SL RNA in the SL RNP factory (Fig. 8A, panel b); this result suggests that the SL RNP factory is exclusively dedicated to SL RNP production.
FIGURE 8.
Localization of splicing factors co-purifying with SmD3 complex. A, PRP6. Panel a, PRP6 is associated with U5 but also with U4 and U6 snRNPs. Extract was prepared from cells as described (13). RNA was extracted from IgG-agarose beads and subjected to primer extension with the different probes (supplemental S-1). RNA was prepared from an aliquot of cells before selection (Total), and RNA from the beads (Final). Only an aliquot (2%) of the total RNA sample was analyzed, whereas the entire sample from the beads was analyzed. Panel b, localization of PRP6 with respect to SL RNA. In situ hybridization combined with immunofluorescence was performed as described under “Experimental Procedures.” Panel a, in situ hybridization with SL RNA; panel b, immunofluorescence of PTP-tagged PRP6; panel c, merge of panels a and b; panel d, DIC and DAPI-stained nuclei merge with panel c; panel e, intensity plot of the three chromophores under the line (inset) as described in Fig. 7D, panel e, demonstrating no significant overlap between the SL RNA and PRP6. B, gene coding for Tb09.211.4670 is essential and its silencing affects the level of U4 and U5 snRNAs. Panel a, growth curves of T. brucei cells silenced for the factor. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 5 × 104 cells per ml. Panel b, Northern blot analysis demonstrating the silencing of Tb09.211.4670. RNA was prepared from uninduced cells (−Tet) and cells after 2 days of induction (+Tet). Total RNA (20 μg) was subjected to Northern analysis with randomly labeled probes. The transcripts of the gene, dsRNA as well as 7SL RNA, that were used to control for equal loading are indicated. Panel c, level of U snRNAs upon silencing. Total RNA as was extracted from uninduced cells or after 2 days of induction. The level of the U snRNAs was determined by primer extension using specific oligonucleotides as listed in supplemental S-1. U3 snoRNA level was used as a control for equal loading. Panel d, localization of the protein encoded by Tb09.211.4670. The factor was PTP-tagged, and the expression was detected by immunofluorescence. Panel a, immunofluorescence of PTP-tagged factor; panel b, propidium iodide-stained nuclei; panel c, merging of panels a and b; panel d, DIC merged with panel c. C, localization of the factors associated with SmD3. The details are the same as described in B, panel d, identity of each gene is indicated.
The purification of SmD3 particles identified several hypothetical proteins. To examine if these factors are bona fide splicing factors, we have begun to analyze their function, searching for SL RNP-specific proteins. One of the criteria for a protein that may function in SL RNP biogenesis is its concentration in the SL RNP factory. Several factors that were consistently purified with the SmD3 complex were further analyzed. One such factor is the protein encoded by Tb09.211.4670. This factor is found only in trypanosomatid species. It contains two distinct domains as follows: a NIF domain known to function in interaction with DNA-binding proteins, and a zinc finger domain, which is absent in T. brucei but exists in other trypanosomatid species. ClustalW multiple sequence alignment of the factor in the different trypanosomatid species was performed (supplemental S-3). To explore its function, the gene was silenced using a T7-opposing silencing construct (29). The results in Fig. 8B, panel a, suggest that this factor is essential for growth. Silencing was confirmed by Northern analysis and showed 95% reduction in the cognate mRNA (Fig. 8B, panel b). The effect of silencing on the level of snRNAs was determined by primer extension. The results demonstrate reduction in the level of U4 (45 ± 5%), U5 (12 ± 2%), and U6 (10 ± 1%) snRNAs (Fig. 8B, panel c). The localization of this factor was determined using a PTP-tagged protein, and the results indicate that it is a nuclear factor, present in speckles (Fig. 8B, panel d). Altogether, these results suggest that Tb09.211.4670 encodes for a trypanosomatid-specific splicing factor that is involved in tri-snRNP complex biogenesis.
Finally, we examined the localization of additional proteins that consistently co-purified with the SmD3 complex. The intracellular localization of Tb927.7.3560, Tb927.10.1490, Tb927.7.2570, and Tb927.10.1630 was determined by immunofluorescence of PTP-tagged proteins. The results presented in Fig. 8C suggest that Tb.927.7.3560, possessing a WD domain, is localized in nuclear speckles, although in situ hybridization failed to detect this factor in the SL RNP factory.4 Likewise, Tb927.10.1490 also localized in nuclear speckles but was not found within the SL RNP factory. Of additional interest are two factors, Tb927.10.1630 and Tb927.7.2570, that have para-nuclear staining. This might indicate that snRNP biogenesis (not SL RNP) may also have a cytoplasmic phase (see under “Discussion”).
DISCUSSION
In this study, the Sm, Lsm, and U1A complexes were purified from Leishmania. This is the first purification of Lsm complex from trypanosomatids, leading to the identification of 39 snRNP U6-associated proteins. Interestingly, the purification did not reveal factors associated with mRNA stability, suggesting that Lsm proteins are either not involved in this process or that these factors are not tightly bound to the Lsm complex, as in other eukaryotes (25). The purification of SmD3-associated proteins revealed 30 snRNP proteins already identified in T. brucei. However, 20 additional proteins were also discovered that were either not previously identified or were not annotated before. The function of proteins identified by mass spectrometry in Leishmania was studied in T. brucei, including that of U1- and U5-binding proteins, as well as PRP19 and GEMIN2, and additional proteins that have no homologues in other eukaryotes. The results suggest that U1 snRNP is not essential for trans-splicing, but one of the U1 snRNP proteins U1A is. To investigate the function of U1A in mRNA processing, the U1A complex was purified, and 45 additional proteins were identified, including the polyadenylation factor, CPSF73. U1A was found to be required for polyadenylation, because 3′ end formation is linked to trans-splicing (38, 39), explaining how its depletion affects the trans-splicing process. This study also revealed conventional splicing factors that acquired trypanosome-specific functions. One such factor is PRP19, which is not only essential for trans-splicing but also plays a role in SL RNA biogenesis and is localized in the SL RNP factory. Another factor belonging to this family is the SMN complex protein, GEMIN2, which was found to be a master regulator of snRNP assembly. The factor is localized to the SL RNP factory, but is also found in other nuclear locations and most probably functions to deliver the SmD3/B proteins to SMN, and initiates the assembly of Sm core proteins on U1, U5, and SL RNAs. The SL RNP factory does not contain mature U1 and U5 snRNPs, suggesting that this site is exclusively devoted to SL RNP production. Several proteins that were co-purified with SmD3 are trypanosome-specific, are localized in nuclear speckles, and affect snRNP biogenesis, suggesting that in addition to canonical splicing factors, trypanosomes express specific splicing factors.
U1 Functions in cis-Splicing but U1A Affects trans-Splicing via Its Direct Role in Polyadenylation
The function of U1 snRNP in trypanosome processing was always very puzzling, given that only three cis-splicing substrates have been found in the genome (36). The finding that U1 snRNP seems to be essential only for cis-splicing but not for trans-splicing (Figs. 2 and 3) is in agreement with results obtained in vitro in a nematode trans-splicing system (35). However, our analysis of U1A silencing demonstrates that U1A is essential for both cis- and trans-splicing (Fig. 4), suggesting that this factor might have additional functions in the cell in addition to its cis-splicing role. U1 snRNP, and especially U1A, were shown in other eukaryotes to regulate 3′ end processing of its own mRNA (43). The effect of U1A depletion on trans-splicing may stem from its role in polyadenylation, due to the linkage between these processes (38, 39). Indeed, U1A was shown to associate with mammalian poly(A) polymerase (44, 45). To examine if the effect of U1A depletion on trans-splicing stems from its direct role in polyadenylation, two different approaches were taken; these included purification of protein complexes carrying U1A and examining the direct role of the factor in polyadenylation. Because poly(A) polymerase is itself a substrate of cis-splicing, one can argue that affecting the level of U1 snRNP affects the production of PAP. However, this does not appear to be the case, as can be deduced from the different phenotypes observed for the U1A and U1- 70-kDa silencing. Whereas silencing of the U1 70 kDa was already efficient after 2 days of silencing, no trans-splicing defect was observed. However, U1A silencing resulted in a clear defect in trans-splicing after 2 days of silencing, and the effect became stronger as a result of more complete depletion of the protein (Fig. 4E, panels b and c). More convincing support for the direct involvement of U1A in polyadenylation comes from the effect of silencing on the length of the poly(A) tail. The effect of U1A silencing on the poly(A) tails was similar to that in cells silenced for the poly(A) polymerase itself. No effect on the length of poly(A) was observed in the first days of U1 70-kDa silencing. Additional, strong evidence for the involvement of U1A in polyadenylation comes from the mass spectrometry analysis of the U1A-associated proteins. First, the polyadenylation factor CPSF73 was detected in association with U1A. In addition, out of 45 proteins identified, 25 were found to be associated with complexes purified in L. tarentolae with CPSF73. Note that these common proteins are unique to these complexes, because the majority of these proteins did not appear in the purification of SmD3 or Lsm3 nor in the purification of splicing factors such as U2AF35, SF1, TSR1, and TSRIP.4
Interestingly, a recently purified mammalian complex active in 3′ end formation was shown to contain, in addition to known polyadenylation factors, factors involved in splicing, transcription, signaling, translation, and DNA-damage response (46). The purification of U1A revealed the expected U1 snRNP proteins but also other factors (Table 3), including splicing factors, helicases, and proteins involved in DNA damage and repair. One such protein is encoding for Tb927.8.2230, related to HUS-1 protein implicated in DNA damage response, and factors involved in ADP-ribosylation. ADP-ribosylation controls various processes, including DNA-repair pathways (47). The purification also revealed factors involved in ubiquitinylation and the proteasome. Indeed it was demonstrated in mammalian cells that small ubiquitin-like modifier regulates PAP and CPSF73 activity (48, 49). In addition, PAP was shown to be regulated by ubiquitinylation in yeast (50). Interestingly, the same ADP-ribosylation and ubiquitinylation factors co-purified with U1A were shown to associate with L. tarentolae CPSF73,4 suggesting that these factors are important to execute or regulate 3′ end formation.
PRP19 Is an Essential Splicing Factor That Is Also Involved in Regulating cap-4 Formation
The purification of PRP19 with its associated factors using Sm proteins as bait suggests that this purification approach is able not only to capture the U5 mono-particles and the tri-snRNP complex but also the U5 complex that exists in the active spliceosome, known as the NTC complex (27). Interestingly, the trypanosome NTC-like complex with its associated proteins was detected only in the purification of SmD3 but not in the purification of the Lsm complex. It was shown that Lsm proteins are released in the active spliceosome, whereas Sm proteins remain associated with the active spliceosomal complex (27, 40). This explains why the NTC complex was not detected among the proteins co-purified with the Lsm complex.
PRP19 is a very essential component of the NTC complex. The trypanosome PRP19, like its homologues, contains three domains (indicated in supplemental S-2). The three domains include an N-terminal U box, a predicted coiled-coil domain, and a WD repeat domain at its C terminus (51). The U-box is structurally similar to RING finger domains. Like many proteins containing RING finger domains, PRP19 proteins exhibit E3 ubiquitin ligase activity in vitro (51). Recently, it was demonstrated that ubiquitinylation/de-ubiquitinylation takes place during spliceosome activation (52). The trypanosome PRP19 might also serve as a ubiquitin ligase during splicing activation, as in cis-splicing. Of special interest are the following: 1) the localization of PRP19 in the SL RNP factory, which was established based on its distinct co-localization with tSNAP42 (Fig. 6), and 2) the effect of PRP19 on capping of the SL RNA. Silencing led to the accumulation of capped SL RNA at the +2 position. The effect on capping is surprising. It will be interesting to examine whether MT57, which carries out the +2 modification (41), is controlled by PRP19. Further research is necessary to elucidate the exact role of PRP19 during SL RNA capping and biogenesis. Because of the close association of PRP19 with the SL RNP factory and because this factor is retained throughout the splicing reaction, it might serve as the factor that helps incorporate the SL RNP into the spliceosome.
Lsm Complex Purification Revealed the Entire Subset of U4, U5, and U6 Proteins but No Factors Involved in mRNA Stability
The purification of the Lsm complex led to the identification of the following: 1) five out of the seven Lsm proteins; 2) six of the Sm proteins (all except SmE); 3) six U5 snRNP proteins; and 4) three U4/U6 proteins, suggesting that the U4/U6 and the tri-snRNP complex are sufficiently stable to be purified via an Lsm protein. Of special interest are the five helicases revealed in this purification, one of which was identified as DED1 (Tb927.10.14550). Among the helicases was also DHH1, which was shown to exist in P-bodies in other eukaryotes and in stress granules in trypanosomes (53). The other helicases are of unknown function. These helicases are not homologues of known helicases that function in cis-splicing such as SUB2, PRP2, PRP16, PRP22, and PRP434 and might be specific to trans-splicing. Note that U5 snRNA interacts by base-pairing with SL RNA in both intron and exon domains, and therefore helicase(s) may be needed to unwind this interaction (54).
The purification of Lsm3 also did not reveal factors involved in mRNA decay. However, our previous study of Lsm8 silencing indicated changes in mRNA stability as a result of depleting this factor, suggesting the involvement of Lsm proteins in mRNA degradation (11). Previously, we also provided evidence that Lsm proteins could not be detected in P-bodies or stress granules. However, these bodies in trypanosomes may function in the storage of mRNA but not their degradation (26). It is still possible that Lsm2–8 function in mRNA decay, but these factors are not stored in P-bodies, and their association with the decay machinery is weak and was disrupted during the purification performed in this study.
GEMIN2 Both Directly and Indirectly Affects the Biogenesis of Trypanosome snRNP
As stated above, SMN was shown to be essential for directing the Sm core to U1, U5, and SL RNA, avoiding the binding of the Sm core to RNAs carrying noncanonical Sm sites, such as U2 and U4 snRNAs. In vitro, this selective binding can be achieved solely by SMN. However, to achieve this selective interaction, preincubation of SMN with SmD3/B was required (21). Because SMN alone can mediate the selective association of snRNA with the canonical Sm-binding site, the function of GEMIN2 found in the same complex remains unknown (21). The phenotype of GEMIN2 silencing may hint at its function. Its depletion reduced the level of U1 and U5 as expected, because in the absence of a functional SMN complex, the assembly of Sm core proteins on these RNAs is perturbed. GEMIN2 might be responsible for selectively recruiting the SmD3/B to associate with SMN, delivering the dimeric complex to SMN and thereby specifying and initiating the assembly of the canonical Sm core. The association of GEMIN2 with snRNA directs the assembly of the canonical core on snRNAs. However, the signals that specify the association of specific snRNAs carrying a canonical Sm site with SMN are currently unknown. In mammalian cells, the specificity of recognition is mediated by GEMIN5 (55). However, no other GEMINS were identified in our purification. Interestingly, a possibility exists that GEMIN2, which is larger in trypanosomes, may manifest this binding ability. It is possible that the primordial SMN complex is composed of a single type of GEMIN.
SL RNP Factory Functions Exclusively in SL RNP Production
The SL RNP factory was first noted as a distinct site near the nucleolus that contains the SL RNA and polymerase II (19). tSNAP42 was shown to reside at this site (56), and later it was noticed that this domain also contains core Sm proteins but not the SSm2 proteins that specifically bind to U2 snRNP (13). The finding that Sm core assembly involves the SMN complex raised the possibility that this domain might include the assembly site of all U snRNAs binding to the canonical Sm core. To investigate this possibility, we followed the localization of U1- and U5-specific proteins with respect to SL RNA, and no evidence was found for the localization of these proteins in the SL RNP factory.
Microscopically, the SL RNP factory appears either in a single spot or sometimes as two spots. This pattern of imaging was obtained either by in situ hybridization with SL RNA probe (9, 19, 57) or by localization of Sm proteins and tSNAP42 protein (13, 56). The SL RNP factory might assemble around the transcriptionally active SL repeat units, much like the assembly of the nucleolus around the actively transcribed rRNA repeats. This may explain why U1 and U5 snRNAs are not found in these sites. All the data from our laboratory support the notion that SL RNA is transcribed and modified within the nucleus (13, 20, 57). The enzymes that carry out cap-4 modification also reside in the nucleus (37, 41, 58), as well as SNIP, a nuclease that is involved in processing the SL RNA tail (10). In addition, cap-4 formation was shown to be coupled to nascent transcription of the SL RNA (7). However, it is not currently known whether snRNA assembly takes place exclusively in the nucleus, as this process may also have a cytoplasmic phase. It is therefore interesting to note that among the factors that associate with SmD3 are also cytoplasmic factors with distinct para-nuclear localization. It has yet to be determined if these factors are involved in regulating the formation of the snRNPs or in re-cycling its different components.
Despite the detailed description of the U1, U5, U6, and tri-snRNP complexes, this study as well as previous studies (21, 23) failed thus far to identify SL RNP-specific proteins. Such proteins might be either known splicing factors that also stably bind the SL RNA, such as PRP19, which associates with SL RNP early in its biogenesis (Fig. 6E), or they may be found among the hypothetical proteins identified in these studies. A few such proteins were already analyzed in this study (Fig. 8, B and C). The next challenge will be to identify additional key trypanosome-specific factor(s), possibly among the hypothetical proteins identified here, and to further classify the splicing factor(s) with which the SL RNP-specific proteins interact to mediate the recruitment of the SL RNP to the spliceosome.
Supplementary Material
Acknowledgment
We thank Asher Pivko for the preparation of anti-U1 70-kDa antibodies.
This work was supported the Deutsche Forcshungsgemeinschaft and by an International Research Scholar grant from the Howard Hughes Medical Institute (to S. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental S-1–S-3 and additional references.
I. D. Tkacz, S. K. Gupta, V. Volkov, M. Romano, T. Haham, P. Tulinski, I. Lebenthal, and S. Michaeli, unpublished results.
- snRNP
- small nuclear ribonucleoprotein
- SL
- spliced leader
- CFP
- cyan fluorescent protein
- nt
- nucleotide
- PTP
- ProtC-TEV protease site-ProtA
- snoRNA
- small nucleolar RNA
- PAP
- poly(A) polymerase
- DIC
- differential interference contrast.
REFERENCES
- 1.Brow D. A. (2002) Annu. Rev. Genet. 36, 333–360 [DOI] [PubMed] [Google Scholar]
- 2.Abelson J. (2008) Nat. Struct. Mol. Biol. 15, 1235–1237 [DOI] [PubMed] [Google Scholar]
- 3.Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002) Genome Res. 12, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. (2002) Science 298, 2205–2208 [DOI] [PubMed] [Google Scholar]
- 5.Liang X. H., Haritan A., Uliel S., Michaeli S. (2003) Eukaryot. Cell 2, 830–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tschudi C., Ullu E. (1990) Cell 61, 459–466 [DOI] [PubMed] [Google Scholar]
- 7.Mair G., Shi H., Li H., Djikeng A., Aviles H. O., Bishop J. R., Falcone F. H., Gavrilescu C., Montgomery J. L., Santori M. I., Stern L. S., Wang Z., Ullu E., Tschudi C. (2000) RNA 6, 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palfi Z., Schimanski B., Günzl A., Lücke S., Bindereif A. (2005) Nucleic Acids Res. 33, 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandelboim M., Barth S., Biton M., Liang X. H., Michaeli S. (2003) J. Biol. Chem. 278, 51469–51478 [DOI] [PubMed] [Google Scholar]
- 10.Zeiner G. M., Hitchcock R. A., Sturm N. R., Campbell D. A. (2004) Mol. Cell. Biol. 24, 10390–10396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q., Liang X. H., Uliel S., Belahcen M., Unger R., Michaeli S. (2004) J. Biol. Chem. 279, 18210–18219 [DOI] [PubMed] [Google Scholar]
- 12.Wang P., Palfi Z., Preusser C., Lücke S., Lane W. S., Kambach C., Bindereif A. (2006) EMBO J. 25, 4513–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tkacz I. D., Lustig Y., Stern M. Z., Biton M., Salmon-Divon M., Das A., Bellofatto V., Michaeli S. (2007) RNA 13, 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang X. H., Liu Q., Liu L., Tschudi C., Michaeli S. (2006) Mol. Biochem. Parasitol. 145, 29–39 [DOI] [PubMed] [Google Scholar]
- 15.Vazquez M. P., Mualem D., Bercovich N., Stern M. Z., Nyambega B., Barda O., Nasiga D., Gupta S. K., Michaeli S., Levin M. J. (2009) Mol. Biochem. Parasitol. 164, 137–146 [DOI] [PubMed] [Google Scholar]
- 16.Nakaar V., Dare A. O., Hong D., Ullu E., Tschudi C. (1994) Mol. Cell. Biol. 14, 6736–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilinger G., Bellofatto V. (2001) Nucleic Acids Res. 29, 1556–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das A., Zhang Q., Palenchar J. B., Chatterjee B., Cross G. A., Bellofatto V. (2005) Mol. Cell. Biol. 25, 7314–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dossin Fde M., Schenkman S. (2005) Eukaryot. Cell 4, 960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hury A., Goldshmidt H., Tkacz I. D., Michaeli S. (2009) Eukaryot. Cell 8, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palfi Z., Jaé N., Preusser C., Kaminska K. H., Bujnicki J. M., Lee J. H., Günzl A., Kambach C., Urlaub H., Bindereif A. (2009) Genes Dev. 23, 1650–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yong J., Wan L., Dreyfuss G. (2004) Trends Cell Biol. 14, 226–232 [DOI] [PubMed] [Google Scholar]
- 23.Luz Ambrósio D., Lee J. H., Panigrahi A. K., Nguyen T. N., Cicarelli R. M., Günzl A. (2009) Eukaryot. Cell 8, 990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salgado-Garrido J., Bragado-Nilsson E., Kandels-Lewis S., Séraphin B. (1999) EMBO J. 18, 3451–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouveret E., Rigaut G., Shevchenko A., Wilm M., Séraphin B. (2000) EMBO J. 19, 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tkacz I. D., Cohen S., Salmon-Divon M., Michaeli S. (2008) Mol. Biochem. Parasitol. 160, 22–31 [DOI] [PubMed] [Google Scholar]
- 27.Wahl M. C., Will C. L., Lührmann R. (2009) Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 28.Aphasizhev R., Aphasizheva I., Nelson R. E., Gao G., Simpson A. M., Kang X., Falick A. M., Sbicego S., Simpson L. (2003) EMBO J. 22, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Morris J. C., Drew M. E., Englund P. T. (2000) J. Biol. Chem. 275, 40174–40179 [DOI] [PubMed] [Google Scholar]
- 30.Kelly S., Reed J., Kramer S., Ellis L., Webb H., Sunter J., Salje J., Marinsek N., Gull K., Wickstead B., Carrington M. (2007) Mol. Biochem. Parasitol. 154, 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li F., Ge P., Hui W. H., Atanasov I., Rogers K., Guo Q., Osato D., Falick A. M., Zhou Z. H., Simpson L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12306–12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturm N. R., Campbell D. A. (1999) J. Biol. Chem. 274, 19361–19367 [DOI] [PubMed] [Google Scholar]
- 33.Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. (2003) Science 302, 279–282 [DOI] [PubMed] [Google Scholar]
- 34.Lee J. H., Jung H. S., Günzl A. (2009) Nucleic Acids Res. 37, 3811–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denker J. A., Maroney P. A., Yu Y. T., Kanost R. A., Nilsen T. W. (1996) RNA 2, 746–755 [PMC free article] [PubMed] [Google Scholar]
- 36.Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S. M., Bianchettin G., Borzym K., Bothe G., Bruschi C. V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R. M., Cronin A., Cruz A. K., Davies R. M., De Gaudenzi J., Dobson D. E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A. C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J. C., Müller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O'neil S., Pentony M., Pohl T. M., Price C., Purnelle B., Quail M. A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J. C., Rutter S., Saunders D., Schäfer M., Schein J., Schwartz D. C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D. F., Blackwell J. M., Stuart K. D., Barrell B., Myler P. J. (2005) Science 309, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamudio J. R., Mittra B., Foldynová-Trantírková S., Zeiner G. M., Lukes J., Bujnicki J. M., Sturm N. R., Campbell D. A. (2007) Mol. Cell. Biol. 27, 6084–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews K. R., Tschudi C., Ullu E. (1994) Genes Dev. 8, 491–501 [DOI] [PubMed] [Google Scholar]
- 39.LeBowitz J. H., Smith H. Q., Rusche L., Beverley S. M. (1993) Genes Dev. 7, 996–1007 [DOI] [PubMed] [Google Scholar]
- 40.Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. (2008) Nature 452, 846–850 [DOI] [PubMed] [Google Scholar]
- 41.Arhin G. K., Li H., Ullu E., Tschudi C. (2006) RNA 12, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarov E. M., Makarova O. V., Achsel T., Lührmann R. (2000) J. Mol. Biol. 298, 567–575 [DOI] [PubMed] [Google Scholar]
- 43.Boelens W. C., Jansen E. J., van Venrooij W. J., Stripecke R., Mattaj I. W., Gunderson S. I. (1993) Cell 72, 881–892 [DOI] [PubMed] [Google Scholar]
- 44.Gunderson S. I., Beyer K., Martin G., Keller W., Boelens W. C., Mattaj L. W. (1994) Cell 76, 531–541 [DOI] [PubMed] [Google Scholar]
- 45.Guan F., Caratozzolo R. M., Goraczniak R., Ho E. S., Gunderson S. I. (2007) RNA 13, 2129–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y., Di Giammartino D. C., Taylor D., Sarkeshik A., Rice W. J., Yates J. R., 3rd, Frank J., Manley J. L. (2009) Mol. Cell 33, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hassa P. O., Haenni S. S., Elser M., Hottiger M. O. (2006) Microbiol. Mol. Biol. Rev. 70, 789–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vethantham V., Rao N., Manley J. L. (2007) Mol. Cell. Biol. 27, 8848–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vethantham V., Rao N., Manley J. L. (2008) Genes Dev. 22, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.del Olmo M., Mizrahi N., Gross S., Moore C. L. (1997) Mol. Gen. Genet. 255, 209–218 [DOI] [PubMed] [Google Scholar]
- 51.Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Ren L., Hirsch J. P., Chazin W. J., Walz T., Gould K. L. (2005) Mol. Cell. Biol. 25, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellare P., Small E. C., Huang X., Wohlschlegel J. A., Staley J. P., Sontheimer E. J. (2008) Nat. Struct. Mol. Biol. 15, 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kramer S., Queiroz R., Ellis L., Webb H., Hoheisel J. D., Clayton C., Carrington M. (2008) J. Cell Sci. 121, 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y., Liu L., Michaeli S. (2000) J. Biol. Chem. 275, 27883–27892 [DOI] [PubMed] [Google Scholar]
- 55.Battle D. J., Lau C. K., Wan L., Deng H., Lotti F., Dreyfuss G. (2006) Mol. Cell 23, 273–279 [DOI] [PubMed] [Google Scholar]
- 56.Lustig Y., Sheiner L., Vagima Y., Goldshmidt H., Das A., Bellofatto V., Michaeli S. (2007) EMBO Rep. 8, 408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biton M., Mandelboim M., Arvatz G., Michaeli S. (2006) Mol. Biochem. Parasitol. 150, 132–143 [DOI] [PubMed] [Google Scholar]
- 58.Zamudio J. R., Mittra B., Zeiner G. M., Feder M., Bujnicki J. M., Sturm N. R., Campbell D. A. (2006) Eukaryot. Cell 5, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stern M. Z., Gupta S. K., Salmon-Divon M., Haham T., Barda O., Levi S., Wachtel C., Nilsen T. W., Michaeli S. (2009) RNA 15, 648–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palfi Z., Lücke S., Lahm H. W., Lane W. S., Kruft V., Bragado-Nilsson E., Séraphin B., Bindereif A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8967–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goncharov I., Palfi Z., Bindereif A., Michaeli S. (1999) J. Biol. Chem. 274, 12217–12221 [DOI] [PubMed] [Google Scholar]
- 62.Barth S., Shalem B., Hury A., Tkacz I. D., Liang X. H., Uliel S., Myslyuk I., Doniger T., Salmon-Divon M., Unger R., Michaeli S. (2008) Eukaryot. Cell 7, 86–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lücke S., Klöckner T., Palfi Z., Boshart M., Bindereif A. (1997) EMBO J. 16, 4433–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H., Tschudi C. (2005) Mol. Cell. Biol. 25, 2216–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J. H., Nguyen T. N., Schimanski B., Günzl A. (2007) Eukaryot. Cell 6, 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holetz F. B., Correa A., Avila A. R., Nakamura C. V., Krieger M. A., Goldenberg S. (2007) Biochem. Biophys. Res. Commun. 356, 1062–1067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.