Abstract
α-Galactosidases catalyze the hydrolysis of terminal α-1,6-galactosyl units from galacto-oligosaccharides and polymeric galactomannans. The crystal structures of tetrameric Saccharomyces cerevisiae α-galactosidase and its complexes with the substrates melibiose and raffinose have been determined to 1.95, 2.40, and 2.70 Å resolution. The monomer folds into a catalytic (α/β)8 barrel and a C-terminal β-sandwich domain with unassigned function. This pattern is conserved with other family 27 glycosidases, but this enzyme presents a unique 45-residue insertion in the β-sandwich domain that folds over the barrel protecting it from the solvent and likely explaining its high stability. The structure of the complexes and the mutational analysis show that oligomerization is a key factor in substrate binding, as the substrates are located in a deep cavity making direct interactions with the adjacent subunit. Furthermore, docking analysis suggests that the supplementary domain could be involved in binding sugar units distal from the scissile bond, therefore ascribing a role in fine-tuning substrate specificity to this domain. It may also have a role in promoting association with the polymeric substrate because of the ordered arrangement that the four domains present in one face of the tetramer. Our analysis extends to other family 27 glycosidases, where some traits regarding specificity and oligomerization can be formulated on the basis of their sequence and the structures available. These results improve our knowledge on the activity of this important family of enzymes and give a deeper insight into the structural features that rule modularity and protein-carbohydrate interactions.
Keywords: Carbohydrate-binding Protein, Carbohydrate Complex, Enzyme Structure, Protein Domains, Protein Structure, Protein-Carbohydrate Recognition, X-ray Crystallography
Introduction
Galactose is present in the oligosaccharides of many plant seeds and is also essential in structures as the hemicelluloses. These polymers build up the plant cell wall and represent a huge storage of carbon within the biosphere and might be an important source of renewable energy (1). In humans, mutations of the α-galactosidase gene cause incomplete degradation of glycolipids and glycoproteins, resulting in Fabry disease (2). Different strategies involving recombinant α-galactosidases are being developed for the treatment of this disease. Another interesting application is the conversion between the ABO blood groups, determined by differences in polysaccharide structures present in the surface of red blood cells. Some α-galactosidases are able to remove the α-linked terminal galactose that differs between O antigen (universal blood) and B antigen, and processes involving plant α-galactosidases are being developed to obtain O-type blood from B-type donors (3). Furthermore, the activity of α-galactosidase is of great interest in many biotechnological applications. It is used to improve the quality and yield of sucrose in the sugar beet industry by achieving an efficient raffinose and other galacto-oligosaccharide hydrolyses. In addition, the processing of soybean-related products and other legume-derived food with this enzyme reduces the content of nondigestible oligosaccharides. Moreover, molasses, the by-product of these industries, represent a potential environmental problem due to its high oligosaccharide content; α-galactosidase or organisms expressing it are being used for biomass and ethanol production coupled to molasses degradation (4). The enzyme is also used as a dietetic supplement for the treatment of gastric disorders. For animal feeding, α-galactosidase is added to maximize the energetic conversion of galacto-oligosaccharides by monogastric animals (4–7).
Glycosyl hydrolases are classified into 115 different families in the CAZy data base according to their amino acid similarities (8). They can also be grouped in two classes regarding their catalytic mechanism, retaining and inverting glycosidases. Although retaining glycosidases keeps the anomeric conformation of the substrate via a double displacement catalytic mechanism, inverting glycosidases induce its inversion in a one-step reaction. Most of them have two carboxylates (glutamate or aspartate) surrounding the glycosidic oxygen of the substrate, one of them acting as a proton donor and the other being the nucleophile. Saccharomyces cerevisiae α-galactosidase (ScAGal) is classified into family 27 of glycosyl hydrolases (GH27), for which several members are structurally known (CAZy). All enzymes within this family present a retaining mechanism, with two aspartic acids being the catalytic residues. It shares high sequence similarity with other α-galactosidases, α-N-acetylgalactosaminidases (EC 3.2.1.49), isomalto-dextranases (EC 3.2.1.94), and β-l-arabinopyranosidase (EC 3.2.1.88) included in the same family. Other α-galactosidases from bacteria are classified into family 36, and both families, together with family 31, are included in clan GH-D. There are other less related prokaryotic α-galactosidases in families 4, 57, and 110.
ScAGal is coded by the gene MEL1 that codifies a highly glycosylated 471-amino acid extracellular protein. The first 18 residues of the protein form a signal peptide that directs the protein to the secretion pathway, where post-translational modifications lead to a mature protein in which carbohydrates represent 30–40% of its final molecular weight. We report here the structure of the ScAGal at 1.95 Å and also the structure of the complexes with melibiose and raffinose at 2.4 and 2.7 Å resolution, respectively. Relevant residues at the active site have been mutated, and kinetic analysis of the mutants has been performed. Recently, the reaction mechanism of human α-galactosidase has been depicted by crystallographic analysis, and the reaction intermediate has been captured (9) showing new information regarding sugar ring deformation during the catalysis. However, little is known about the mechanisms of substrate specificity and recognition against natural substrates. We address here some structural features of ScAGal and other GH27 enzymes regarding substrate recognition and specificity, oligomeric state, and protein stability that remained unexplained. The results provide valuable information that might be most useful for further biotechnology experiments and protein engineering. Moreover, some features proposed here can be extended to other α-galactosidases, where modularity and oligomerization are key features in determining substrate recognition.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
The MEL1 gene (GB X03102) encoding an α-galactosidase from S. cerevisiae (ScAGal; Uniprot P04824) was amplified and cloned into YEpFLAG-1 vector (Eastman Kodak Co.) as described previously (10). Mutagenesis of ScAGal was done by PCR using the commercial kit QuikChange-XL (Stratagene). Oligonucleotide design and mutagenic procedures were performed following the manufacturers' recommendations. Mutants of ScAGal and their main characteristics are listed in Table 3.
TABLE 3.
Kinetic analysis of ScAGal mutants
kcat values were calculated assuming a protein molecular mass of 52 kDa. ± refers to standard errors based on the curve fitting using Sigmaplot. Mutants D149A and D209A showed no measurable activity. Mutants Y232R and N252A were highly unstable and kinetic data are not available. NC means not calculable.
| PNPG |
Melibiose |
Raffinose |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| mm | mm | mm | |||||||
| MEL1 | 4.5 ± 0.7 | 286 ± 21 | 63.5 | 11.2 ± 1.1 | 193 ± 5 | 17 | 54.1 ± 15.5 | 46.8 ± 7.02 | 0.9 |
| A41Y | 3.7 ± 1.2 | 90 ± 16 | 24 | 5.8 ± 0.5 | 131 ± 3 | 22 | 87.9 ± 66.1 | 91 ± 39 | 0.8 |
| Y232R | NC | NC | NC | NC | NC | NC | NC | NC | NC |
| Q251A | 1.4 ± 0.3 | 170 ± 11 | 121 | 254 ± 60 | 113 ± 16 | 0.45 | NC | NC | NC |
| Q251W | 0.24 ± 0.03 | 16 ± 0.35 | 66 | NC | NC | NC | NC | NC | NC |
| N252A | NC | NC | NC | NC | NC | NC | NC | NC | NC |
Expression and Purification
Both ScAGal and mutants were expressed in yeast BJ3505 (Kodak), deglycosylated with endoglycosidase H (New England Biolabs), and purified with anti-FLAG M2 affinity resin (Sigma) as described previously (10).
Crystallization and Data Collection
Crystallization of ScAGal (2.5 mg ml−1 in 0.05 m Tris-HCl, pH 7.4, 0.150 m NaCl, and 0.002 m DTT) and mutant D149A-ScAGal (1.8 mg ml−1 in 0.05 m Tris-HCl, pH 7.4, 0.150 m NaCl, and 0.002 m DTT) was performed on Cryschem (Hampton Research) sitting drop plates at 291 K as described previously (10). Crystals with polyhedrical shape grew from both samples in 18–20% (w/v) PEG 3350, 0.1 m Bistris propane,4 pH 8.5, 0.2 m KSCN within 2 weeks. For data collection, native crystals were transferred to cryoprotectant solutions consisting of mother liquor plus 20% (v/v) glycerol before being cooled to 100 K in liquid nitrogen. Complexes with the natural substrates melibiose (Sigma) and raffinose (Sigma) were obtained by the soaking method (11). To minimize crystal manipulation during the soaking with the substrates, drop solution was substituted with a stabilizing solution (20% PEG 3350, 0.1 m Bistris propane, pH 8.5, 0.2 m KSCN) containing the substrate at 50 mm concentration, incubated for 5 min, and then substituted again with cryoprotectant solution in which the PEG 3350 concentration was increased to 35%. In the case of the raffinose-soaked crystals, an additional 10% glycerol was needed to prevent ice formation. Total incubation time was about 10 min.
X-ray diffraction data were collected at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). Diffraction images were processed with MOSFLM (12) and merged using the CCP4 package (13). A summary of data collection and data reduction statistics is shown in Table 1.
TABLE 1.
Crystallographic statistics
Values in parentheses are for the high resolution shell.
| ScAGal | ScAGal-melibiose | D149A ScAGal-raffinose | |
|---|---|---|---|
| Crystal data | |||
| Space group | P 4 21 2 | P 4 21 2 | P 21 21 21 |
| Unit cell parameters | |||
| a | 101.24 Å | 100.45 Å | 111.17 Å |
| b | 101.24 Å | 100.45 Å | 129.53 Å |
| c | 111.52 Å | 111.28 Å | 136.78 Å |
| Data collection | |||
| Beamline | ID23.1 (ESRF) | ID14.2 (ESRF) | ID23.1 (ESRF) |
| Temperature | 100 K | 100 K | 100 K |
| Wavelength | 0.979 Å | 0.933 Å | 1.072 Å |
| Resolution | 74.96 to 1.95 Å (2.06 to 1.95 Å) | 59.99 to 2.40 Å (2.53 to 2.40 Å) | 71.80 to 2.70 Å (2.85 to 2.70 Å) |
| Data processing | |||
| Total reflections | 455,518 (73,906) | 238,360 (32,904) | 327,060 (51,340) |
| Unique reflections | 42,919 (6140) | 22,940 (3262) | 54,965 (7932) |
| Multiplicity | 10.6 (12.0) | 10.4 (10.1) | 6 (6.5) |
| Completeness | 100.0% (100.0%) | 99.6% (99.2%) | 100.0% (100.0%) |
| I/σ (I) | 7.6 (1.7) | 6.9 (1.9) | 8.9 (1.8) |
| Mean I/σ (I) | 33.3 (7.1) | 19.4 (5.8) | 19.6 (4.9) |
| Rmergea | 6.7% (45.7%) | 11.1% (43.3%) | 8.1% (43.8%) |
| Rpimb | 2.2% (13.7%) | 3.5% (13.7%) | 3.8% (18.4%) |
| Molecules per ASU | 1 | 1 | 4 |
| Matthews coefficient (Å3 Da−1) | 2.75 | 2.67 | 2.43 |
| Solvent content | 55% | 54% | 49% |
| Refinement | |||
| Rwork/Rfreec (%) | 20.4/23.4% | 19.8/25.3% | 24.1/28.3% |
| No. of atoms | |||
| Protein | 3518 | 3518 | 10,545 |
| Carbohydrate | 130 | 135 | 430 |
| Other | 15 | 1 | 0 |
| Water molecules | 434 | 415 | 1022 |
| Ramachandran (Ref. 21) | |||
| Favored | 97.1% | 96.2% | 96.1% |
| Outliers | 0.00% | 0.00% | 0.00% |
| Root mean square deviations | |||
| Bonds | 0.01 Å | 0.012 Å | 0.008 Å |
| Angles | 1.372° | 1.543° | 1.057° |
| Protein Data Bank codes | 3LRK | 3LRL | 3LRM |
a Rmerge = Σhkl Σi|Ii(hkl) − (I(hkl))|/ΣhklΣi I(hkl), where Ii(hkl) is the ith measurement of reflection hkl, and (I(hkl)) is the weighted mean of all measurements.
b Rpim = Σhkl (1/(N − 1))1/2 Σi|Ii(hkl) − (I(hkl))|/Σhkl Σi Ii(hkl), where N is the multiplicity for the hkl reflection.
c Rwork/Rfree = Σhkl| Fo − Fc|/Σhkl|Fo|, where Fc is the calculated and Fo is the observed structure factor amplitude of reflection hkl for the working/free (5%) set, respectively.
Structure Solution and Refinement
The structure of ScAGal was solved by molecular replacement using the MOLREP program (14). The structure of Oryza sativa (rice) α-galactosidase (Protein Data Bank code 1UAS) (15) was used to prepare the search model using the program CHAINSAW (16), and the ScAGal sequence was aligned to that from rice α-galactosidase. A single solution containing one molecule in the asymmetric unit (ASU) was found using reflections within 74.96 to 2.12 Å resolution range and a Patterson radius of 40 Å, which after rigid body fitting led to an R factor of 0.50. Crystallographic refinement was performed using the program REFMAC5 (17) within the CCP4 suite with flat bulk-solvent correction, and using maximum likelihood target features. Several loops in the catalytic domain and two insertions in the C-terminal domain were excluded from the model during the first stages of the refinement because no electron density was observed for the polypeptide chain. After iterative refinement and rebuilding of these regions using the programs O (18) and COOT (19), the final 2Fo − Fc map showed continuous density for the whole molecule. At the latest stages, water molecules, several N-acetylgalactosamines (GalNAc), three glycerol molecules, and a sodium ion were included in the model, which, combined with more rounds of restrained refinement, led to a final R factor of 20.4 (Rfree = 23.4) for all data set up to 1.95 Å resolution. The structures of the complexes were solved by molecular replacement with the native model, and refinement was performed as described above. In the case of the raffinose-ScAGal complex, tight noncrystallographic symmetry restraints between the four molecules of the tetramer in the ASU was applied during refinement. Refinement parameters for the three structures are reported in Table 1.
Stereochemistry of the models was checked with PROCHECK (20) and MOLPROBITY (21), and the figures were generated with PyMOL (22). Analysis of the interfacial surfaces and the oligomer stability was done with the Protein Interfaces, Surfaces, and Assemblies service at the European Bioinformatics Institute (23).
Docking of Galactomannan into ScAGal Active Site
The galactomannan fragment (Gal4Man4) was modeled on the basis of the coordinates retrieved from the Protein Data Bank code 1OH4 (24) and manually docked into the ScAGal active site by superimposition of the Gal-Man moiety of galactomannan onto the melibiose (Gal-Glu) found in the ScAGal-melibiose and ScAGal-raffinose complexes. All hydrogen atoms were added to the ligand, and charges were assigned by the Gasteiger method using AUTODOCK TOOLS program (ADT) (25). With the exception of the Gal-Man link, all glycosidic linkages between mannose units, but not those of the sugar rings, were defined as rotatable bonds, and also all hydroxyl groups were set free. The protein model contained the coordinates of the tetrameric ScAGal structure presented here, after removing all nonpolypeptide atoms. Polar hydrogen atoms were then added using ADT. AUTODOCK 4.2 was executed 50 times with the hybrid genetic-local search algorithm (GA-LS) (26), a population size of 150, elitism set at 1, mutation rate at 0.02, and crossover rate of 0.8. Simulations were performed with a maximum of 2,500,000 energy evaluations and a maximum of 27,000 generations. Docking results were clustered using a cutoff of 2 Å root mean square deviations. The resulting three highest clusters contain 20, 6, and 13 conformations with mean binding energy of −10.5, −9.9, and −8.8 kcal/mol. The first and second clusters gave very similar conformations.
α-Galactosidase Activity and Kinetics
The enzymatic activity of α-galactosidase (EC 3.2.1.22) was followed using p-nitrophenyl-α-d-galactopyranoside (PNPG), melibiose, and raffinose as substrates. 1 μg of purified enzyme in 125 μl of 20 mm Tris, pH 7.4, was incubated for 5 min at 303 K. After incubation, the reaction was started by adding 200 μl of substrate in reaction buffer (61 mm citric acid and 77 mm Na2HPO4, pH 4) to the enzyme solution. Reaction was stopped at three different times by mixing 100-μl aliquots with 900 μl of 100 mm Tris, pH 9.5. Measurement of the released p-nitrophenol was performed by UV absorbance at 400 nm. When the natural substrates were used, the released glucose was measured chromatographically in an HPLC (Waters) using a Sugar Pack column (Waters). α-Galactosidase activity is expressed in enzyme units, with enzyme units indicating the amount of enzyme capable of liberating 1 mmol of product (p-nitrophenyl or glucose) per min under experimental conditions (mmol min−1 mg−1).
Kinetic characterization of ScAGal and mutants was performed assaying α-galactosidase activity of purified protein samples (described above) toward different substrate concentrations. Nonlinear fitting using least squares was performed to infer the apparent enzymatic kinetic parameters from Michaelis-Menten plots using SIGMAPLOT (Systat Software Inc.).
Analytical Ultracentrifugation
Sedimentation equilibrium experiments were performed in a Beckman Optima XL-A ultracentrifuge using a Ti50 rotor and six channel centerpieces of Epon charcoal (optical path length 12 mm). Samples of purified ScAGal in its native and denatured states in the concentration range 0.2–0.5 mg ml−1 were equilibrated against 2 mm Tris-HCl, pH 7.4, 15 mm NaCl, and 2 mm Tris-HCl, pH 7.4, 15 mm NaCl, 8 m urea, respectively. Samples were centrifuged at 9,000, 11,000, and 16,000 rpm at 20 K. Radial scans at 280 nm were taken at 12, 14, and 16 h. The three scans were identical (equilibrium conditions were reached). The weight average molecular weight was determined by using the program EQASSOC with the partial specific volume of ScAGal set to 0.72 at 293 K as calculated from its amino acid composition.
RESULTS AND DISCUSSION
Folding of ScAGal Monomer
As reported previously (10), we have purified and crystallized both the glycosylated and deglycosylated forms of ScAGal. The glycosylated enzyme always gave plate-shaped crystals that grew at low pH values (4, 5) and proved to be highly twinned. In contrast, treatment of the protein with endoglycosidase H reduced drastically the heterogeneity of the sample allowing us to obtain high quality crystals in a broad range of pH values, with the best crystals growing at pH higher than 8. It should be noted that the deglycosylated enzyme keeps almost 100% of the initial activity, and heterogeneity was reduced without affecting protein quaternary structure, as shown below (see Fig. 1). Details of crystallization conditions have been given before (10).
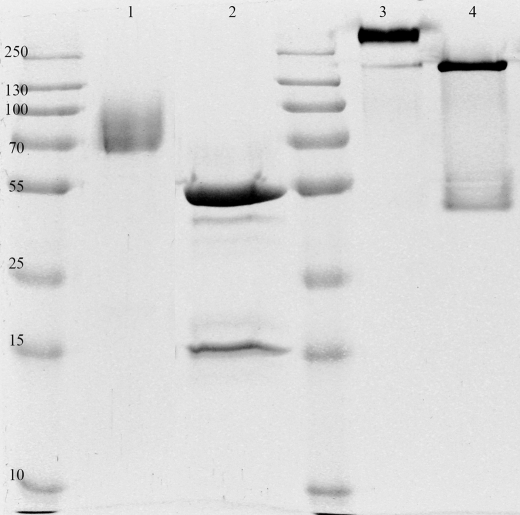
FIGURE 1.
SDS-PAGE analysis of the glycosylated (lanes 1 and 3) and deglycosylated (lanes 2 and 4) purified ScAGal. Glycosylated protein migrates as a broad band with an average molecular mass of 80 kDa (lane 1). Deglycosylated protein has a molecular mass of 52 kDa as predicted from protein sequence (lane 2). The second band in lane 2, which corresponds to endoglycosidase H, was removed in the last step of purification. In lanes 3 and 4, denaturing and reducing agents were avoided to perform a semi-native PAGE where oligomers are not disrupted.
The structure of ScAGal has been determined to 1.95 Å resolution (see Table 1 and “Experimental Procedures” for details). The final model contains the protein chain after cleavage of the signal peptide. Chain starts at residue 19 and extends to residue 470. The last 9 residues, which correspond to Ser and the eight amino acids from the purification FLAG tag, are missing and probably disordered. Two glycerol molecules can be clearly modeled in the catalytic pocket of the native ScAGal. Both molecules are mimicking the position of the natural substrates that are found in the complexes with melibiose and raffinose. Although Endo H treatment cleaves the oligosaccharide moieties and leaves, in general, single GalNAc residues, some poorly accessible glycosylation sites of ScAGal remained more glycosylated. From the electron density maps, GalNAc units were modeled at positions Asn-105, Asn-175, Asn-270, Asn-370, Asn-403, and Asn-422. Some additional density was found at the Asn-454 site, but its poor quality disallowed modeling of GalNAcs. It is interesting to note that this residue Asn-454 is predicted by the Protein Interfaces, Surfaces, and Assemblies service (23) to be involved in many crystal lattice contacts, and its heterogeneous glycosylation is probably the reason that prevented correct crystal growth of the glycosylated form.
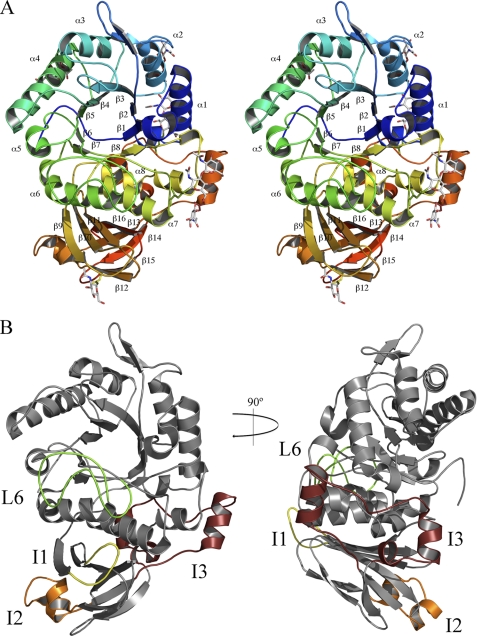
ScAGal is a globular protein that folds into two domains (Fig. 2A). The main domain is a (α/β)8 barrel that starts at residue 19 and extends to residue 324. It has eight parallel β-strands that form the central barrel connected by eight external α-helices. This motif is common to many glycosyl hydrolases, the loops at the C-terminal end of the β-strands, L1–L8, being the most variable regions and contouring the catalytic pocket that is located in the center of the barrel. There is a long insertion in one of these loops (L6, residues 211–238) that clearly emerges from the barrel (Fig. 2B), and it is involved in interdomain contacts, as will be explained below. Two disulfide bonds between residues 221–237 and 223–230 stabilize this L6 loop. The C-terminal domain presents a β-sandwich structure, which is less conserved within family GH27 and whose function has not been fully understood until now. It has eight antiparallel β-strands that fold into two β-sheets containing a Greek key motif that extends from residue 325 to 470. The first β-sheet is made up of six β-strands (β9, β10, β11, β13, β14, and β16), and the second is formed by two (β12 and β15). There is a 12-residue insertion in one of the loops within the large β-sheet, residues 331–342 (I1), which is interacting with L6 from the β-barrel and also participates in interdomain contacts, as stated above. A second insertion (I2) found in loop 378–386 from the same β-sheet is involved in oligomerization, as will be explained below. But the most outstanding feature of this β-sandwich domain is a 45-residue insertion (I3) in one of the loops (396–441) that remarkably emerges from the β-sandwich domain and packs over helix α8 from the barrel, in a very compact structure that protects this element from the solvent (Fig. 2B). Furthermore, as is seen in the figure, the segment 422–429 is located between the end of helices α1 and α8 in a kind of barrel “closure.” This is a remarkable trait taking into account that the short loops located at the opposite site of the active site cavity are generally considered to be essential in keeping the barrel stability; in fact, this side of the barrel has been termed the “stability face” (27). This insertion is probably increasing the overall stability of ScAGal, and it might be a reason that would explain the heat stability observed for this enzyme (data not shown) and why it remains folded when performing SDS-PAGE in mild conditions (Fig. 1). Moreover, as deduced from Blast analysis of the GH27 family sequences, this unique insertion is only present in yeast α-galactosidases from the genus Saccharomyces and in some Aspergillus species and clearly increases the association between both domains of the enzyme. Thus, the total buried surface area of this interface is 4220 Å2 in ScAGal, although reported value for other members of the GH27 family is between 2200 and 2800 Å2 (28, 29). This insertion seems to increase also the polar nature of the interface, as 72% of the total polar links within the interface are established by this region. Finally, the glycosylation chains of Asn-403 and Asn-422, both located in this segment, also contribute to make many polar interactions among residues from both domains, increasing even more the association between them.
FIGURE 2.
Structure of ScAGal monomer. A, schematic stereo representation of the ScAGal monomer showing a catalytic (α/β)8 domain and a β-sandwich domain. Secondary elements from both domains are labeled. Glycosylated Asn and GalNAc moieties are shown as sticks. B, two views of ScAGal monomer highlighting the important insertions in loops L6 (green), I1 (yellow), I2 (orange), and I3 (maroon).
ScAGal Is a Tetrameric Enzyme
The molecular weight and oligomeric state of ScAGal have been studied before, and a trimeric state of the protein was proposed (30–32); nonetheless, further analysis was necessary to determine its actual quaternary structure. Analytical ultracentrifugation of glycosylated ScAGal samples performed in native and denaturing conditions by us (see under “Experimental Procedures” for details) showed an average molecular mass of 80 and 320 kDa for the monomer and the oligomer, respectively, the glycosylation being 35% of the total molecular mass. These biochemical data are compatible with a tetrameric state of ScAGal, which was confirmed by the crystallographic analysis when the structure of ScAGal was solved.
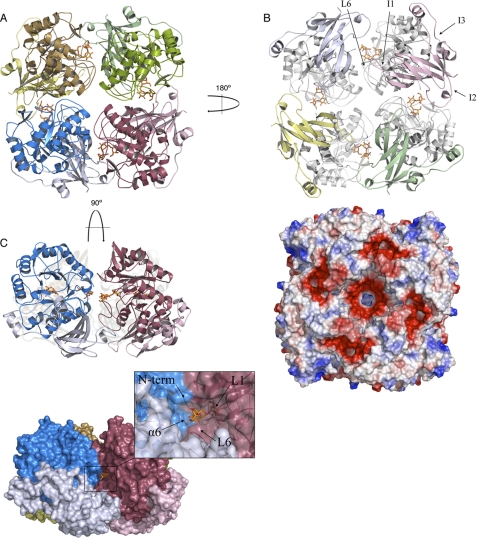
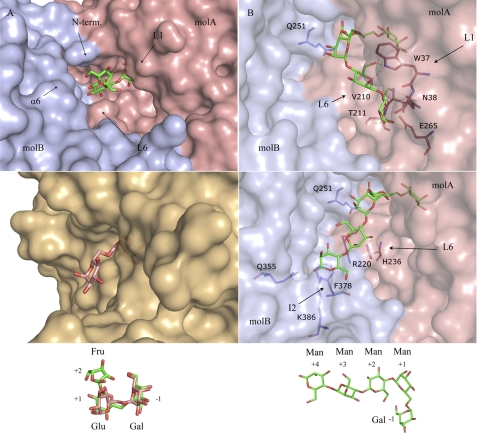
The tetramer is made up of four identical ScAGal subunits related by a crystallographic 4-fold axis in the free enzyme and the complex with melibiose. In the crystals from the mutant D149A-ScAGal complexed with raffinose, the asymmetric unit contains the whole tetramer. It is a flat square-shaped tetramer with dimensions 95 × 95 × 75 Å (Fig. 3A). Its molecular surface was 61,731 Å2, and the total surface area buried was 9292 Å2 (2323 Å2 within each interface). The interaction between subunits is mostly made by the catalytic domains through L3 to L7 loops, but the β-sandwich domains also make interactions through strand β9, and the insertions at loops β9–β10 (I1) and β12–β13 (I2). 11 hydrogen bonds and 1 salt bridge stabilize each monomer-monomer interface (Table 2). There is also an aromatic cluster made up of residues Phe-195, Phe-194, and Tyr-228 from a monomer and Tyr-232 and Phe-157 from the adjacent subunit that extends along the catalytic-catalytic domain interface, which may contribute to stabilize the oligomer. It is worth noting that the particular arrangement of the four units locates their supplementary β-sandwich domains in the same face of the tetramer (Fig. 3B), where five regions of highly negative/acidic patches are observed. These regions are defined by interaction between the long L6 and the insertions I1 and I2 of the β-sandwich domain. This ordered oligomerization pattern might suggest that the four supplementary β-sandwich domain of ScAGal play a concerted role as an ancillary module of the tetramer that may well promote the association of the enzyme with the substrate, but more analyses are necessary to confirm this hypothesis.
FIGURE 3.
Tetramer of ScAGal. The ScAGal tetramer is shown in schematic representation from three different views (A–C). A raffinose bound to the active site is represented as sticks. Important loops involved in oligomerization and substrate specificity are labeled in B. The electrostatic distribution in the tetramer surface calculated with PyMOL (22) (B) reveals the five acidic patches (red) that are generated upon oligomerization. The surface representation of the tetramer shows the active site cleft in C. The frame highlights the catalytic pocket with loops involved in substrate recognition.
TABLE 2.
Protein contacts in the interfaces between monomers
Contacts were predicted by Protein Interfaces, Surfaces. and Assemblies service (see Ref. 23).
| Hydrogen bonds | ||
|---|---|---|
| Residue (molecule A) | Distance | Residue (molecule B) |
| Å | ||
| Val-19(N) | 3.01 | Cys-121(O) |
| Ser-20(N) | 3.45 | Tyr-152(OH) |
| Tyr-228(OH) | 3.51 | Cys-223(SG) |
| Leu-382(N) | 3.00 | Asn-270(OD1) |
| Tyr-195(OH) | 3.51 | Ala-233(N) |
| Pro-248(O) | 3.08 | Tyr-232(OH) |
| Asn-252(OD1) | 2.77 | Tyr-232(OH) |
| Arg-326(O) | 2.65 | Ser-219(OG) |
| Val-327(O) | 2.92 | Arg-216(NH2) |
| Asp-379(OD1) | 3.00 | Arg-220(NH2) |
| Asp-379(OD2) | 2.70 | Arg-220(NH1) |
| Salt bridges | ||
|---|---|---|
| Residue (molecule A) | Distance (Å) | Residue (molecule B) |
| Asp-379(OD1) | 3.00 | Arg-220(NH2) |
| Asp-379(OD2) | 2.70 | Arg-220(NH1) |
The catalytic site is buried in a deep hole, almost 25 Å depth, placed in the thin faces of the square, being shaped by each pair of monomers within the tetramer (Fig. 3C). The catalytic residues are found at the bottom, in a narrow pocket shaped by loops L1–L3 and part of L6 from one molecule, and the catalytic domain of the other monomer through helix α6 and its N terminus. The cavity becomes wider as it opens to the surface, being surrounded at the entrance also by the β-sandwich domain of the second monomer, mainly through loops β10–β11 and β12–β13 (I2). It is therefore remarkable that oligomerization must affect the accessibility of the substrates to the active site, modulating according to the enzymatic specificity. It is also worth noting that the β-sandwich domain, with the less conserved sequence and up to now with unassigned function, is directly involved in shaping the active-site cavity and might participate in substrate recognition or binding.
Structure of Substrate-Enzyme Complexes
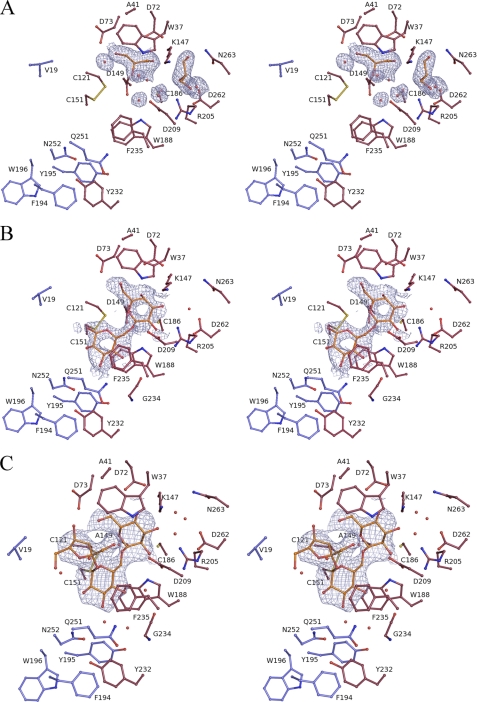
As native crystals were grown at pH 8.5, a value in which activity has been observed to be impaired, we attempted to get complexes by soaking these crystals into the natural substrates melibiose and raffinose. However, only melibiose soaking yields good diffracting crystals, and when the inactivated mutant became available, new soakings were carried out with raffinose. Although soaking with substrates seemed to have negative effects in the crystals, initial 2Fo − Fc and difference Fo − Fc maps showed clearly the presence of the substrates in the catalytic pocket (Fig. 4). Native and melibiose soaked crystals were isomorphous and belonged to the same space group (P4212) with little changes in unit cell parameters (see Table 1), whereas the raffinose complex presented a different space group (P212121). In this crystal, the asymmetric unit was a tetramer, and the substrate was detected in the four independent positions making the same interactions with the protein. Root mean square deviations between native ScAGal and both complexes for the backbone alignment (all molecule) were 0.2 Å.
FIGURE 4.
ScAGal-substrate complexes. Stereo view of the catalytic pocket of the native ScAGal (A), the complex with melibiose (B), and the complex with raffinose (C). Amino acid residues and substrates are shown in stick representation, and relevant water molecules are represented as red spheres. 2Fo − Fc maps contoured at 1σ (0.30, 0.30, and 0.25 eA−3, respectively) are also given for the molecules bound at the active site. Residues from one monomer are shown in red, and the residues from the adjacent molecule that are interacting with the substrate are in blue. Two glycerol molecules and ordered waters in the native structure (A) are found mimicking the position of the galactose at the subsite −1.
Previous works (15, 28, 33) have reported the complex of α-galactosidases with the reaction product galactose, and recently, the complexes of human α-galactosidase with an intermediate and the substrate melibiose have been reported (9). The main features observed at the galactose-binding pocket (subsite −1) in the two ScAGal-substrate complexes presented here are conserved with that described previously (for a description of subsites nomenclature within glycosyl hydrolases enzymes see Davies et al. (34)). The galactose unit of the substrates in ScAGal complexes is located between two aspartic acids (Asp-149 and Asp-209) that act as the catalytic residues. Asp-149, placed at the end of β4, was close to the anomeric carbon of the galactose in both complexes, and Asp-209 was at hydrogen bond distance from the galactose O1 and O2 atoms (Figs. 4 and 5). Moreover, when both residues were mutated to alanine, activity was reduced to undetectable levels (see Table 3).
FIGURE 5.
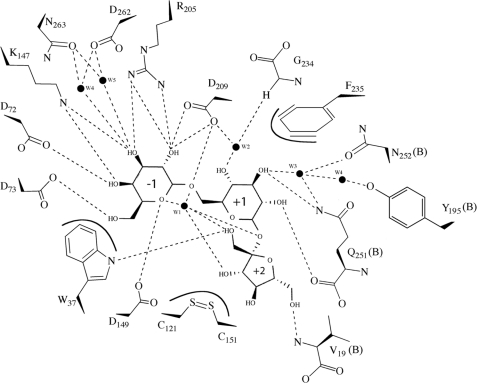
Protein-substrate interactions. Schematic representation of the protein-substrate interactions in melibiose and raffinose complexes. Dashed lines represent hydrogen bonds, and lines around Trp-37, Phe-235, and the disulfide bond Cys-121 to Cys-151 represent hydrophobic interactions. High ordered waters are represented here as black dots. (Schemes were drawn with ChemDraw (CambridgeSoft).)
The galactose ring is stabilized in the −1 subsite by stacking of its C4–C5–C6 moiety with Trp-37 side chain (Fig. 4). This feature is common for all GH27 galactosidases, and the Trp located in loop L1 is conserved. Nonetheless, both the loop L1 and Trp-37 are a bit farther from galactose in ScAGal than in the other enzymes with known structure. It is remarkable that a residue in this region, Ala-41, is substituted by more voluminous residues that stack against the tryptophan in the other enzymes, what might be pushing the tryptophan closer to the −1 subsite. This shift in the position of the Trp leaves more space that might allow the entrance of the substrates to the narrow catalytic center in the tetrameric structure. The effect of having bulkier residues at position 41 has been investigated by replacement of Ala-41 by a Tyr, as it is found in the rice α-galactosidase. As shown in Table 3, this replacement increases the affinity of ScAGal for short substrates (PNPGal and melibiose), although it is decreased against raffinose. However, this is counter-balanced by an opposite effect in the kcat value, which leads to mutant enzymes with a similar activity to the native form. It might be that this reduced affinity for long substrates is more important when polymeric or ramified substrates are considered.
On the other hand, the galactose moiety is further stabilized at the catalytic site through hydrogen bonds to Arg-205 (O2, O3), Lys-147 (O3, O4), Asp-72 (O4), and Asp-73 (O6). All these residues, located at strands β4 (Lys-147) and β6 (Arg-205) and loop L2 (Asp-72 and Asp-73), are conserved among the family. However, and despite this highly conserved −1 subsite, a remarkable feature of the ScAGal is Asn-263, substituted by a methionine in the other enzymes with known structure, which is located at a short distance of about 3.5 Å to the galactose O3 atom. In ScAGal, the shorter Asn-263 is linking the substrate O3 through two well ordered water molecules, but it leaves a pocket around O3 (Fig. 4). It is interesting to remark that in the native structure, one molecule of glycerol from the cryoprotectant and three ordered water molecules mimic the position of the galactose ring, but a second glycerol molecule is positioned in this pocket, being linked to both Asn-263 and the conserved Asp-265. It is tempting to suggest that this pocket could possibly allocate a putative substitution at the galactose O3, although the biological relevance of this feature remains to be explained.
In contrast to that reported for the other complexes of the family, where the glycosidic oxygen of galactose is at the protein surface pointing toward the solvent, the deep cavity that in ScAGal gives access to the catalytic pocket buries entirely the glucose and fructose moieties of raffinose (see Fig. 3C). In fact, the large number of interactions that the substrate makes with the enzyme at the +1 and +2 subsites implies some restrictions, which make the galactose ring to present a small shift with respect to the previously reported complexes. The orientation of the glucose ring at +1 subsite is also different from that reported for the human α-galactosidase-melibiose complex (Fig. 6A), and it should be highlighted that it is rotated by 180° around the glycosidic bond to avoid steric clashes with the long L6. This feature is consistent with the distinctive substrate specificity found in the mammal enzymes, which remove terminal α-galactose or α-GalNAc units from glycolipids and glycoproteins (28, 35). These substrates present a very different molecular structure to that found in mannan, what is reflected in the divergent geometry of the active site shaped by the different oligomerization pattern. On the other hand, the glucose moiety of melibiose and raffinose presents, essentially, the same interactions within the catalytic pocket that can be observed in Figs. 4 and 5. First of all, it is stabilized by stacking to the unconserved Phe-235, located in the L6 insertion unique to ScAGal. Moreover, several hydrogen bonds to the galactose O2 and O3 atoms are made by residues Gln-251 and Asn-252 from helix α6 of the adjacent monomer. Other residues from L6 (Gly-234) and the adjacent monomer (Tyr-195 and Val-19) are also close to the catalytic pocket and are stabilizing the ligand at this +1 subsite by making hydrogen bonds through well ordered water molecules to its O1, O3, and O4 atoms.
FIGURE 6.
ScAGal active-site specificity. A, comparison of the ScAGal-raffinose (top) and human α-galactosidase-melibiose (bottom, Protein Data Bank code 3HG3) complexes, showing the deep cavity that give access to the ScAGal catalytic pocket. The scheme shows the fixed conformation of raffinose, which is rotated by 180° around the −1/+1 glycosidic bond with respect to the melibiose found in the human enzyme. B, two putative positions of an α-1,6-galactose-substituted manno-tetrasaccharide (shown in the scheme), docked into the ScAGal active-site as explained in the text. The residues that may be involved in shaping subsites +1 to +4 are highlighted.
Finally, the fructose moiety of raffinose, located at subsite +2, is recognized by direct hydrogen bond of its O1 atom to the Trp-37 main chain. The N-terminal segment of the contiguous molecule is also making direct interactions through hydrogen bond of Val-19 NH2 to the fructose O6, and this contribution of the N-terminal region in the active site could explain why the N-terminal tagged protein did not show activity against any substrate (data not shown). Remarkably, there is an intramolecular hydrogen link between the fructose O2 and the O6 from galactose, which keeps a very constrained conformation in the oligosaccharide. This conformation is further stabilized by the presence of a highly ordered water molecule that is hydrogen-bonded to the two glycoside linkages galactose-glucose and glucose-fructose and is also interacting to the fructose O1 and O3.
A last interesting feature to address is that, as a consequence of the tight interaction that takes place between ScAGal and the substrates, the overall structure of melibiose and raffinose bound to the enzyme greatly differs from what has been found in the crystalline state of both sugars (36, 37). The conformation around the α(1–6) linkage is of interest as it determines the overall molecular form, and in this sense, the largest difference is a change of the torsion angle around the C5–C6 bond (ω torsion) by ∼150°. This leads to an opposite orientation of the glucose a/b faces in both substrates and a different arrangement of the fructose unit in raffinose (supplemental Fig. S1). Therefore, binding to ScAGal promotes a great conformational change in the substrate glycosidic bond that is essential to allocate the subsequent sugar units into its active site.
Active Site of ScAGal
Following the above description, it is noticeable that the substrates are stabilized in the active site of ScAGal by many atomic interactions and that new binding pockets are created by the unique insertion found at L6 (Phe-235) and the proximity of the catalytic domain of the contiguous monomer found upon oligomerization (Gln-251, Asn-252, and the N-terminal amine group). All these residues should, accordingly, be essential for activity, a hypothesis that has been investigated by mutagenesis analysis. As deduced from Table 3, the mutant Q251A presents a loss of activity of about 98% against melibiose and is almost inactive against raffinose but, unexpectedly, seems to be more active against the synthetic substrate PNPG. As seen in Fig. 5, this glutamine is making interactions with the glucose moiety through both its main and side chain, consequently having an important role in substrate binding. However, the change of glutamine to alanine in the Q251A mutant is possibly making the catalytic pocket more accessible, which would explain the higher activity of the mutant against the substrate PNPG. Nevertheless, removal of the Gln-251 side chain prevents the formation of polar interactions that significantly decrease the affinity and are deleterious for the enzymatic activity when measured against raffinose. On the other hand, the change of glutamine by the bulkier tryptophan side chain in the Q251W mutant does not block the access to the active site, as the PNPG is hydrolyzed as efficiently as in the native form, but it seems deleterious to enzymatic activity against the other substrates. In the case of Asn-252, the N252A replacement yields a fully inactive enzyme, as is shown in Table 3. An inspection to the active site (Fig. 4) shows that the side chain of Asn-252 is located in a pocket surrounded by Tyr-232 from one monomer and Phe-194 and Tyr-195 from the adjacent monomer, and its removal could introduce structural changes that are detrimental for activity. It should be noted that all these residues participate in the aromatic cluster located at the dimer interface, which as stated above must be critical for the tetramer stability. This may also explain the absence of enzymatic activity in the mutant Y232R (Table 3) that probably introduces deleterious structural changes in the oligomer interface.
As described above, substrate specificity among α-galactosidases is maintained by a conserved configuration of the catalytic pocket that stabilizes the interactions with the substituents in the galactose ring. Differences in substrate specificity and affinity between different substrates (raffinose, stachiose, galactomannan, etc.) may be related to differences in the loops surrounding the catalytic pocket that interact and stabilize the substrates. Furthermore, it has been previously reported that ScAGal cleaves only terminal α-galactosyl units from galactomannan (38–40), and consequently, the inability to accommodate galactose linked to inner mannose should be attributed to the particular geometry of the deep cavity in which its active site pocket is found.
Therefore, in an attempt to have a picture of how ScAGal would recognize galactomannan, we have performed a molecular docking simulation. An α-1,6-galactose-substituted manno-tetrasaccharide, Gal4MAN4 (Fig. 6B), has been manually built into the ScAGal active site by superimposition of its Gal-Man portion to the Gal-Glu moiety of melibiose and raffinose found at the −1, +1 subsites in both ScAGal complexes. Subsequently, automatic molecular docking was performed using Autodock to allow for flexibility in the oligosaccharide chain at positions not observed experimentally, i.e. distal from the scissile bond. Most of the resulting conformations (40/50) can be clustered in two major positions shown in Fig. 6B. On the one hand, it can be assumed that an inner galactose would situate a mannose unit in a second hypothetical subsite +2 that would go deeply into the cavity and would crash with loop L6 and also with the contiguous subunit of the tetramer, which illustrates the inability of ScAGal in binding ramified galactomannans. On the other hand, the long insertion found at L6 may be involved in stabilization of the substrate in both putative conformations. However, although in one conformation the sugar units at positions +3 and +4 would be accommodated in subsites defined by L1 and L6 from one monomer, the second conformation extends to the other subunit and would be recognized by residues from its supplementary C-terminal domain, particularly from insertion I2, where Phe-378 seems to be properly positioned in the surface to stack against the mannose ring at subsite +4. Consequently, not only the adjacent subunit is essential in substrate binding through its catalytic domain, as has been shown above, but also the supplementary C-terminal domain might be involved in fine-tuning the specificity of ScAGal against long substrates through its unique insertion I2.
Finally, and apart from the inability of ScAGal to process nonterminal galactose residues, there is no apparent restriction in length or complexity of the galactomannans. In fact, analysis of different galactomannan degradation by the α-galactosidase from Umbelopsis vinacea (39) has shown that the tetrameric enzyme is able to hydrolyze the substrate even if the galactomannan presents inner ramified galactose (e.g. Gal3,4MAN4). This fact may be illustrated by our docking results by considering that the second extended conformation shows the mannose O6 atoms pointing to the solvent, and therefore, they seem able to allocate an attached galactose (Fig. 6B).
Determinants for Specificity and Oligomerization within GH27 Family
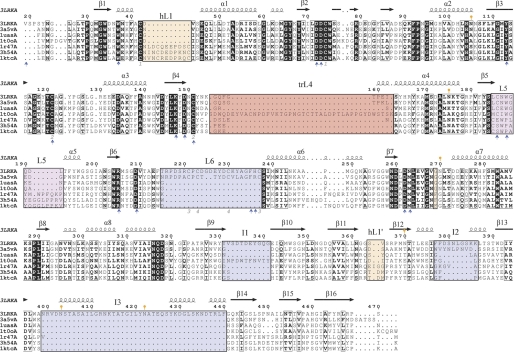
The overall structure of glycosyl hydrolases family 27 enzymes is very similar. It is well known that the (α/β)8 barrel domain acts as a scaffold for a wide variety of catalytic pockets (41) and that it allows changes in loop regions as long as they do not compromise the overall folding of the domain. As pointed out before, the function of the β-sandwich domain was not well known for this group of enzymes, with its sequence being the less conserved region among the family. Overall, ScAGal has similar sequence identity with rice α-galactosidase (38%), Trichoderma reesei α-galactosidase (34%), and the human enzyme (32%), although only 21% identity was observed with the Bacillus halodurans α-galactosidase. It also has strong identity with the α-N-acetylgalactosaminidases from Gallus gallus (32%) and Homo sapiens (34%). Interestingly, the recently reported U. vinacea α-galactosidase shares essential structural features with ScAGal, although their sequence identity (41%) was not significantly higher than that observed between ScAGal and the other enzymes discussed above.
Fig. 7 displays the structural alignment of the GH27 known enzymes from eukaryotes, which reveals interesting details. First of all, the residues that build up the catalytic pocket are well conserved through evolution, as they are essential for substrate recognition and catalysis. Only differences in the sequence of the “2-position recognition loop” (L5) are related to changes in the specificity α-galactosidase/α-N-acetylgalactosaminidase among the family. Thus, a short insertion rearranges the position of this loop that relocates farther from the catalytic pocket, making up the cavity for the N-acetyl substituent (28). On the other hand and as explained above, it has been previously reported that yeasts α-galactosidase releases only terminal galactose residues from galactomannan substrates (40), whereas the α-galactosidase from fungus such as Aspergillus niger and Penicillium purpurogenum (39, 42) hydrolyze only inner galactose. Interestingly, other enzymes such as rice α-galactosidase are active against both the side chain and the terminal α-galactosyl residue (43). These differences in specificity toward long substrates must be, in principle, related to the long insertions found in loops surrounding the catalytic center, although oligomerization must also be taken into account.
FIGURE 7.
Structural alignment of GH27 members. The structural alignment of ScAGal (3LRKA), the α-galactosidases from U. vinacea (3a5vA), rice (1uasA), T. reesei (1t0oA), H. sapiens (1r47A), and the α-N-acetyl-galactosaminidases from H. sapiens (3h54A) and G. gallus (1ktcA) was generated with the DALI server (45) and ESPript (46). ScAGal secondary structure is shown above the sequence alignment. The black squares indicate sequence similarity. The insertions in ScAGal loops (L6, I1, I2, and I3) are highlighted with a blue box. Those insertions involved in dimerization in human and chicken enzymes are highlighted with orange boxes, and the 2-position recognition loop, at L5, is in the magenta square. The insertion in T. reesei L4 is highlighted with a red box. Gray numbers refer to disulfide bonds in the ScAGal structure. Orange marks refer to glycosylated asparagines. Blue arrows highlight the residues involved in substrate recognition. B. halodurans α-galactosidase is more distant from the eukaryotic enzymes, and some motifs are unconserved. A full structural alignment containing also the prokaryotic enzyme is given in (supplemental Fig. S2).
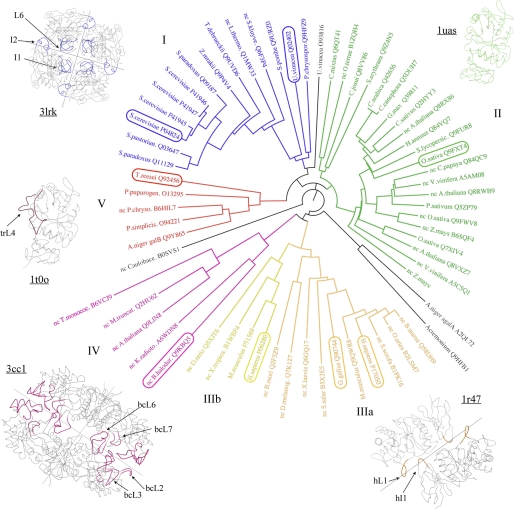
As observed in Fig. 7, a 10-residue insertion is found in loop L1 of all mammalian enzymes, i.e. chicken and human α-galactosidases and human α-N-acetylgalactosaminidase. All these enzymes have been reported to be dimeric, with the L1 insertion being directly involved in dimerization. These dimers are very similar and share the topology of the interface, which is very different from the interfaces found in the tetramer of ScAGal and U. vinacea α-galactosidase I. There is also a small insertion (359–361) at a loop in the β-sandwich domain that is important in the dimer interface by making interaction with Phe-273, a residue that is a Gly in the other enzymes. On the other hand, a long 32-residue insertion is found in loop L4 in the α-galactosidase from T. reesei, which has been reported to be a monomer. However, a clear specificity has not been ascribed to this fungal enzyme that could be correlated with this pattern. On the contrary and in the case of ScAGal, the long insertion found at loop L6 has been shown to be directly involved in creating new binding sites, as described above, and it is the structural determinant of the restricted accessibility of inner α-galactosyl residues to the catalytic pocket. Moreover, L6 together with insertions I1 and I2 are involved in the monomer-monomer interface (see Fig. 3B). Interestingly, the U. vinacea α-galactosidase, reported to also be a tetramer, presents the L6 and I2 insertions but not I1 and I3, and consequently, the last two segments must not be essential for oligomerization, although they could be responsible for the high stability observed in ScAGal, as commented before. Finally, monomeric rice α-galactosidase that presents broad specificity does not contain any of the described insertions. In conclusion and from the comparison of the reported structures, some trends in the oligomeric state and substrate specificity within GH27 enzymes may be envisaged.
To extend the above observations to other GH27 enzymes, we have performed a phylogenetic analysis of the family (Fig. 8). The analysis has been carried out by using the conserved regions of the protein sequences and excluding the variable regions and the insertions in an attempt to keep the results of the alignment independent from the presence/absence of any loop or insertion. A more exhaustive phylogenetic analysis of the family carried out by Naumoff (44) supports our results and is consistent with our analysis. Family GH27 members can be clustered within five groups. Interestingly, all protein sequences annotated in GH27 that present a long insertion in loop L6 and the insertion I2 are grouped in one branch together with the ScAGal (see Fig. 8), which therefore are predicted to be tetramers and to present similar specificity against terminal galactosyl residues. This group I includes enzymes from yeast as U. vinacea, and Phanerochaete chrysosporium. On the other hand, group II harboring plant enzymes, such as rice α-galactosidase but also bacterial enzymes such as those from Cellvibrio mixtus and Clostridium josui, lack any insertion involved in oligomerization and are also predicted to have broad specificity recognizing both the terminal and inner galactose. Group III includes α-N-acetylgalactosaminidases (IIIa) and α-galactosidases (IIIb) from mammals and other superior eukaryotes whose enzymatic activities are determined by different configurations at the “2-position loop” as discussed above. All the enzymes from this cluster present the insertions in L1 and in the β-sandwich domain that are involved in dimerization in the known structures from human and chicken. As found in group II, group IV harbors plant enzymes, such as those from Triticum monoccocum and Arabidopsis thaliana, and enzymes from prokaryotes, such as the B. halodurans α-galactosidase. Sequence similarity within this group is also high and all have insertions in the loops L2, L3, L6, and L7 when compared with other members of the family. Moreover, these loops are involved in the dimerization of the B. halodurans enzyme, which forms a tetramer through interactions between the β-sandwich domains. These bacterial enzymes are different from all the prokaryotic enzymes classified within family GH36, and as proposed by Naumoff (44), they possibly have eukaryotic origin, which is in accordance with the clustering of these proteins with plant enzymes in groups II and IV. Finally, proteins grouped in cluster V, along with A. niger, P. purpurogenum, and T. reesei α-galactosidases, have a long insertion in loop L4 that is likely determining the specificity of these enzymes to recognize only inner galactosyl residues. All these proteins lack the insertions involved in dimerization and tetramerization. Therefore, and on the basis of sequence homology, the oligomeric state and some details of substrate specificity could be predicted for family GH27 members.
FIGURE 8.
Phylogenetic analysis of GH27 family. The enzymes are clustered in five groups colored differently. Those members with known structure are highlighted in a box, and their foldings are given with the same color code as the groups to which they belong. Relevant loops common to all the enzymes in a cluster are represented and labeled. Protein sequences from enzymes classified in CAZy family GH27 as α-galactosidases (EC 3.2.1.22) or α-N-acetyl-galactosaminidases (EC 3.2.1.49) and also some noncharacterized protein sequences were retrieved from the UNIPROT data base and labeled with the name of the organism and the UNIPROT reference code. Not characterized protein sequences are labeled with “nc” before the name of the protein. Sequence alignment was performed with ClustalW2 (47), and then converted into a phylogenetic tree using the program Phylip Drawgram at the Pasteur Institute server (48).
To conclude, we determined the crystal structure of ScAGal and its complexes with the natural substrates melibiose and raffinose, which give further insights into the substrate recognition and specificity of this biotechnologically relevant enzyme. The structure of the complexes and the mutational analysis of ScAGal show that oligomerization is key in determining substrate binding. Furthermore, the structure presented here is a new example of the role that supplementary domains with, in principle, unknown function may play in fine-tuning polysaccharide recognition, as this domain could be putatively involved in binding sugar units distal from the catalytic site as suggested by docking. On the other hand, additional concerted role in promoting association with the substrate could be envisaged by the ordered arrangement that the supplementary domains of the different subunits display in the ScAGal tetramer. This domain may also be a key stability factor through its unique insertions that fold over the catalytic domain and seems to protect it from the solvent. Our analysis extends to other members of GH27 family, where some traits regarding oligomerization and substrate specificity can be formulated on the basis of their sequence and the structures available. It is outstanding how evolution may tailor enzymes highly specific to a particular substrate by only a few insertions that determine key structural features. Unraveling the molecular determinants of this versatility is of great theoretical interest for a better understanding of the protein-carbohydrate interaction. Furthermore, it will also allow the design of new enzymes with improved activity for biotechnological purposes.
Supplementary Material
Acknowledgments
We thank the staff of the European Synchrotron Radiation Facility for providing beam time and for technical assistance at beamlines ID14.2 and ID23.1.
Research performed at the Universidade da Coruña was supported by Grant 07TAL010103PR from Xunta de Galicia co-financed by FEDER (CEE). General support during 2008–2010 was provided by Xunta de Galicia. Research performed at the Consejo Superior de Investigaciones Científicas was supported by Grant BIO2007-67708-C04-04 from the Dirección General de Investigación. This work is a product of the project “Factoría de Cristalización” from the program Consolider-Ingenio 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3LRK, 3LRL, and 3LRM) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- PNPG
- p-nitrophenyl-α-d-galactopyranoside
- ASU
- asymmetric unit.
REFERENCES
- 1.Centeno M. S., Guerreiro C. I., Dias F. M., Morland C., Tailford L. E., Goyal A., Prates J. A., Ferreira L. M., Caldeira R. M., Mongodin E. F., Nelson K. E., Gilbert H. J., Fontes C. M. (2006) FEMS Microbiol. Lett. 261, 123–132 [DOI] [PubMed] [Google Scholar]
- 2.Sakuraba H., Chiba Y., Kotani M., Kawashima I., Ohsawa M., Tajima Y., Takaoka Y., Jigami Y., Takahashi H., Hirai Y., Shimada T., Hashimoto Y., Ishii K., Kobayashi T., Watabe K., Fukushige T., Kanzaki T. (2006) J. Hum. Genet. 51, 341–352 [DOI] [PubMed] [Google Scholar]
- 3.Wang J., Zhang S., Tan H., Zhao Z. K. (2007) J. Microbiol. Methods 71, 225–230 [DOI] [PubMed] [Google Scholar]
- 4.Murphy R., Power R. (2002) J. Microbiol. Biotechnol. 28, 97–102 [DOI] [PubMed] [Google Scholar]
- 5.Di Stefano M., Miceli E., Gotti S., Missanelli A., Mazzocchi S., Corazza G. R. (2007) Dig. Dis. Sci. 52, 78–83 [DOI] [PubMed] [Google Scholar]
- 6.Rezessy-Szabó J. M., Nguyen Q. D., Hoschke A., Braet C., Hajós G., Claeyssens M. (2007) Biochim. Biophys. Acta 1770, 55–62 [DOI] [PubMed] [Google Scholar]
- 7.Viana P. A., de Rezende S. T., Marques V. M., Trevizano L. M., Passos F. M., Oliveira M. G., Bemquerer M. P., Oliveira J. S., Guimarães V. M. (2006) J. Agric. Food Chem. 54, 2385–2391 [DOI] [PubMed] [Google Scholar]
- 8.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guce A. I., Clark N. E., Salgado E. N., Ivanen D. R., Kulminskaya A. A., Brumer H., 3rd., Garman S. C. (2010) J. Biol. Chem. 285, 3625–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Leiro R., Pereira-Rodríguez A., Cerdan M. E., Becerra M., Sanz-Aparicio J. (2010) Acta Crystallogr. F Struct. Biol. Crystallogr. Commun. 66, 44–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassell A. M., An G., Bledsoe R. K., Bynum J. M., Carter H. L., 3rd., Deng S. J., Gampe R. T., Grisard T. E., Madauss K. P., Nolte R. T., Rocque W. J., Wang L., Weaver K. L., Williams S. P., Wisely G. B., Xu R., Shewchuk L. M. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie A. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26, 27–33 [Google Scholar]
- 13.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 14.Vagin A. A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 15.Fujimoto Z., Kaneko S., Momma M., Kobayashi H., Mizuno H. (2003) J. Biol. Chem. 278, 20313–20318 [DOI] [PubMed] [Google Scholar]
- 16.Stein N. (2008) J. Appl. Crystallogr. 41, 641–643 [Google Scholar]
- 17.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 18.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 19.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20.Laskowski R., MacArthur M., Moss D. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 21.Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 23.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 24.Boraston A. B., Revett T. J., Boraston C. M., Nurizzo D., Davies G. J. (2003) Structure 11, 665–675 [DOI] [PubMed] [Google Scholar]
- 25.Sanner M. F. (1999) J. Mol. Graph. Model. 17, 57–61 [PubMed] [Google Scholar]
- 26.Morris G., Goodsell D., Halliday R., Huey R. (1998) J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 27.Sterner R., Höcker B. (2005) Chem. Rev. 105, 4038–4055 [DOI] [PubMed] [Google Scholar]
- 28.Garman S. C., Garboczi D. N. (2004) J. Mol. Biol. 337, 319–335 [DOI] [PubMed] [Google Scholar]
- 29.Garman S. C., Hannick L., Zhu A., Garboczi D. N. (2002) Structure 10, 425–434 [DOI] [PubMed] [Google Scholar]
- 30.Lazo P. S., Ochoa A. G., Gascón S. (1977) Eur. J. Biochem. 77, 375–382 [DOI] [PubMed] [Google Scholar]
- 31.Lazo P. S., Ochoa A. G., Gascón S. (1978) Arch. Biochem. Biophys. 191, 316–324 [DOI] [PubMed] [Google Scholar]
- 32.Sumner-Smith M., Bozzato R. P., Skipper N., Davies R. W., Hopper J. E. (1985) Gene 36, 333–340 [DOI] [PubMed] [Google Scholar]
- 33.Golubev A. M., Nagem R. A., Brandão Neto J. R., Neustroev K. N., Eneyskaya E. V., Kulminskaya A. A., Shabalin K. A., Savel'ev A. N., Polikarpov I. (2004) J. Mol. Biol. 339, 413–422 [DOI] [PubMed] [Google Scholar]
- 34.Davies G. J., Wilson K. S., Henrissat B. (1997) Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark N. E., Garman S. C. (2009) J. Mol. Biol. 393, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeffrey G. A., Huang D. B. (1990) Carbohydr. Res. 206, 173–182 [DOI] [PubMed] [Google Scholar]
- 37.Hirotsu K., Higuchi T. (1976) Bull. Chem. Soc. Jpn. 49, 1240–1245 [Google Scholar]
- 38.Kim W. D., Kobayashi O., Kaneko S., Sakakibara Y., Park G. G., Kusakabe I., Tanaka H., Kobayashi H. (2002) Phytochemistry 61, 621–630 [DOI] [PubMed] [Google Scholar]
- 39.Kaneko R., Kusakabe I., Ida E., Murakami K. (1991) Agric. Biol. Chem. 55, 109–115 [PubMed] [Google Scholar]
- 40.Yoshida S., Tan C. H., Shimokawa T., Turakainen H., Kusakabe I. (1997) Biosci. Biotechnol. Biochem. 61, 359–361 [DOI] [PubMed] [Google Scholar]
- 41.Rigden D. J., Jedrzejas M. J., de Mello L. V. (2003) FEBS Lett. 544, 103–111 [DOI] [PubMed] [Google Scholar]
- 42.Shibuya H., Kobayashi H., Park G. G., Komatsu Y., Sato T., Kaneko R., Nagasaki H., Yoshida S., Kasamo K., Kusakabe I. (1995) Biosci. Biotechnol. Biochem. 59, 2333–2335 [DOI] [PubMed] [Google Scholar]
- 43.Dey P. M., Pridham J. B. (1972) Adv. Enzymol. Relat. Areas Mol. Biol. 36, 91–130 [DOI] [PubMed] [Google Scholar]
- 44.Naumoff D. G. (2004) Mol. Biol. 38, 388–399 [Google Scholar]
- 45.Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 46.Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 48.Felsenstein J. (1995) PHYLIP, Version 3.57c, University of Washington, Seattle [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.