Abstract
Erythropoietic and megakaryocytic programs are specified from multipotential progenitors by the transcription factor GATA1. FOG1, a GATA1-interaction partner, is critical for GATA1 function in several contexts by bringing multiple complexes into association with GATA1 to facilitate activation or repression of target genes. To further elucidate regulation of these associations by cellular and extracellular cues, we examined FOG1 for post-translational modifications. We found that FOG1 is SUMOylated and phosphorylated in erythroid cells in a differentiation-dependent manner. Removal of the SUMOylation sites in FOG1 does not impair nuclear localization, protein stability, or chromatin occupancy. However, SUMOylation of FOG1 modulates interactions with C-terminal binding protein family members, specifically promoting CTBP1 binding. Phosphorylation of FOG1 modulates SUMOylation and, therefore, indirectly regulates the CTBP interaction. Post-translational modification of FOG1 may contribute to control of co-occupancy by CTBP family members, the NuRD complex, and GATA1 at differentially regulated genes.
Keywords: Development, Erythropoiesis, Post-translational Modification, Protein Phosphorylation, Sumoylation, Tissue-specific Transcription Factors, Transcription Repressor, GATA Factor
Introduction
Erythropoietic and megakaryocytic programs are specified from multipotential progenitors by the transcription factor GATA1, a zinc finger transcription factor first identified by its ability to bind regulatory regions of globin gene enhancers (1, 2). The importance of this factor in the development of these lineages is underscored by the discovery of functionally important GATA sites in virtually all erythropoietic and megakaryocytic-specific genes (3, 4). In fact, GATA1 is essential for erythropoiesis (5, 6) and megakaryopoiesis (7, 8) in vivo as revealed by mouse knock-out studies. In addition to its essential role in normal development, mutations in GATA1 cause hematological disorders, including anemia and thrombocytopenia. Recently, aberrant function of GATA1 as a transcriptional regulator has also been implicated in the initiation of M7 leukemia in Down syndrome (9).
FOG13 (or Friend of GATA 1) is a direct interaction partner of GATA1 that is essential for GATA1 function in multiple contexts (10). In addition to GATA1, FOG1 has been shown to interact with two repression complexes: a CTBP-containing complex (11, 12) and the NuRD repression complex (13, 14).
Although the interaction of FOG1 with these complexes may account for its role in facilitating GATA-mediated gene repression, no clear mechanism exists to describe how FOG1 might contribute to GATA1-dependent gene activation. The function of many master regulators is modulated by post-translational modifications, including GATA1, which is SUMOylated (15), phosphorylated (16), and acetylated (17). To elucidate further regulation of FOG1 associations by cellular and extracellular cues, we examined it for post-translational modifications. We found that FOG1 is SUMOylated and phosphorylated in erythroid cells in a differentiation-dependent manner.
SUMO modification involves the reversible, covalent attachment of SUMO to a lysine residue, typically located within the consensus sequence ψKXE, where ψ represents a large hydrophobic amino acid (18). The enzymes involved in the reversible conjugation of SUMO are similar to those mediating ubiquitin conjugation but provide a reversible attachment of the SUMO peptide (19, 20). SUMO modification appears to play a role in diverse cellular processes, including protein-protein interaction, subcellular localization, protein stabilization, and transcriptional regulation (21). Interestingly, mounting evidence supports a major role for the regulation of transcription factors by the post-translational modification of SUMOylation (22). Within erythropoietic and megakaryopoietic development, transcriptional regulators demonstrated to be SUMOylated include GATA1 (15), erythroid Kruppel-like factor (EKLF) (23), basic Kruppel-like factor (BKLF) (24), nuclear factor erythroid-derived 2, 45kDa (p45/NF-E2) (25), SRY (sex-determining region Y)-box 6 (SOX6) (26), v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (MAFG) (27), and GATA2 (28).
In this report we show that FOG1 is SUMOylated in vivo and map the modified residues to two lysine residues between the fourth and fifth zinc finger. Mutation of these lysine residues abrogates SUMOylation of FOG1. Furthermore, we find that FOG1 is phosphorylated at a site proximal to the second SUMOylated lysine, allowing for an additional level of regulation. Both post-translational modifications occur in a differentiation-dependent manner. SUMOylation is not involved in the regulation of FOG1 stability, cellular localization, or chromatin occupancy. Instead, SUMOylation of FOG1 modulates the association with the CTBP complex, enhancing interaction with CTBP1 specifically.
EXPERIMENTAL PROCEDURES
Cell Lines
MEL and 293T cells were grown as before in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (29). MEL cells were differentiated with 1.9% dimethyl sulfoxide (DMSO) for 4 days. L8057 cells were grown as before (30) and were differentiated with 12-O-tetradecanoylphorbol-13-acetate (TPA) for 3 days. GIE and GIER cells were grown as before (31). MEL cells expressing BirAV5his were a generous gift of A. Cantor. BirA-expressing MEL cells were then used for electroporation with plasmid constructs containing FLAG-biotin-tagged cDNAs. MEL cells expressing tagged cDNA were identified by Western blotting with anti-streptavidin-HRP of the total lysates or nuclear extracts.
Plasmid Construction
Plasmids for bacterial biotin ligase BirA and biotin tagging were as described (32). Plasmids for cDNA epitope tagging or expression of untagged wild-type or mutant FOG1 cDNA in 293T (pEF1aV5his series) were purchased from Invitrogen. Mutagenesis of FOG1 was performed using the QuikChange site-directed mutagenesis kit (Stratagene) as per the manufacturer's instructions, and all constructs were verified by DNA sequencing. In addition, vectors containing HA-tagged hSUMO3 (plasmid #17361) (33) and EGFP-SENP2 fusion (plasmid #13382) (34) were obtained from Addgene. All other vectors were gifts acknowledged in the footnotes section.
Total Lysate and Nuclear Extract Preparation and Western Blot Analyses
Total lysates and nuclear extracts were prepared as described (35), except N-ethylmaleimide (Sigma) was used to treat nuclear extracts in all SUMOylation experiments at 20 mm unless otherwise stated. For Western blot analysis, aliquots of total lysates or nuclear extracts (10∼20 mg) were fractionated on a SDS-polyacrylamide gel and electroblotted onto a PVDF membrane. Antibody incubation and chemiluminescence detection were performed according to the manufacturer's instructions (Amersham Biosciences). Antibodies used were all from Santa Cruz unless otherwise specified and included those against GFP (sc-8334), FOG1 (sc-9362), MTA-2 (sc-9447), GATA-1 (sc-265), CTBP1 (BD), CTBP2 (BD), anti-SUMO1 (sc-9060), and HA tag (sc-805), SENP1 (Abcam), SENP2 (Abcam), and FLAG tag (ECS, Bethyl).
Phosphatase Treatment of Nuclear Extracts
For dephosphorylation experiments, calf intestinal phosphatase or λ-phosphatase were used. Aliquots of extract were incubated with calf intestinal phosphatase (Roche Applied Science), or they were incubated with 400,000 units/ml λ-phosphatase (New England Biolabs) at 30 °C for 30 min in LS buffer containing 200 μg/ml bovine serum albumin and 0.2 mm MnCl2.
Affinity Capture and Immunoprecipitation
Transient co-transfection of 293T cells with plasmids expressing FOG1 cDNA, Tagged-FOG1, FOG1 mutants, empty vector, and GFP-SUMO1, HA-SUMO1, FLAG-SENP1, EGFP-SENP2, HA-SUMO3, FLAG-CTBP1, FLAG-CTBP2, and/or GATA1 constructs was performed according to manufacturer's protocol (Roche Applied Science FuGENE 6). Two days after transfection, total lysates or nuclear extracts were prepared and incubated with anti-FOG1 antibody or control Ig (sc-2028) and protein G-agarose overnight. On day 2, unbound material was washed away, and bound material was eluted by boiling in Laemmli buffer and subjected to Western blot analyses.
Co-immunoprecipitation of MEL cells stably transfected with various FLAG-biotin-tagged FOG1 versions was achieved using streptavidin-agarose, or anti-M2 FLAG-agarose was performed as described (30). In brief, nuclear extracts from MEL cells expressing BirA and biotin-tagged cDNA were incubated with streptavidin-agarose or anti-M2 FLAG-agarose in a buffer containing 20 mm Tris-HCl, 139 mm KCl, 12 mm NaCl, and 20% glycerol, and 0.5% Nonidet P40. Binding was performed at 4 °C for 1 h to overnight on a rocking platform followed by six washes in binding solution. Bound material was eluted by boiling for 5 min in Laemmli buffer.
Chromatin Immunoprecipitation and Antibodies
ChIP reactions and bioChIP reactions were performed as described previously with minor modifications (36, 37). For bioChIP reactions, streptavidin beads (Dynabeads MyOne Streptavidin T1) were used to precipitate chromatin, and 2% SDS was applied for one of the washing steps. At least three biological replicates were performed in each case. Primers for ChIP-PCR validation include Car2_d0.3 (5′ ACAATACCTATCGGGACCACAG, 3′-CCTTAACCTTTCTGCTACACACAA), Hba-a1_d0.2 (5′-AACTATGCTTTTCTGTGACCTCAAC, 3′-GCTTACATCAAAGTGAGGGAAGTAG), Sox2_3.7up (5′-GCAATGCTGAGAAATTCCAGTT, 3′-GTTCCCCTCCTCTCCTAATCTC), and Zfpm1_d1.1 (5′-CTAACAACGCTACACGAATGGAT, 3′-AGAAACTTCACTCGGGAACAAC).
Gene Expression in Erythroid Progenitors
Freshly isolated fetal liver cells (E14.5) were immunostained with phycoerythrin-conjugated anti-TER119 (BD Biosciences) and fluorescein isothiocyanate (FITC)-conjugated anti-CD71 (BD Biosciences) antibodies, as previously described (38). 7-Aminoactinomycin D (BD Biosciences) was added to exclude dead cells from analysis. For purification of RNA from fetal liver progenitors, populations of cells, called here Stage I-IV, corresponding to CD71loTer119− (committed erythroid progenitors, Stage I), CD71hiTer119− (proerythroblasts, Stage II), CD71hiTer119+ (basophilic erythroblasts, Stage III), and CD71loTer119+ (late erythroblasts, Stage IV), respectively, were sorted as before (38) using a BD FACS Aria sort machine (BD Biosciences).
RNA was prepared from the described populations using the TRIzol kit (Invitrogen), DNase I treated by RQ1 RNase-Free DNase (Promega, Madison, WI), and quantified. cDNA was synthesized using the 1 μg of RNA with the iScript cDNA synthesis kit (Bio-Rad). Typically, 1 μ of cDNA was then used as a template for quantitative PCR using the iQ SYBR Green Supermix (Bio-Rad) in an iCycler thermocycler (Bio-Rad). Quantitative PCR primers (found in the supplemental methods) were used to generate triplicate data sets, and results were always compared with β-actin reactions run in parallel.
RESULTS
FOG1 Is Modified by SUMO1 in Erythroid and Megakaryocytic Cells
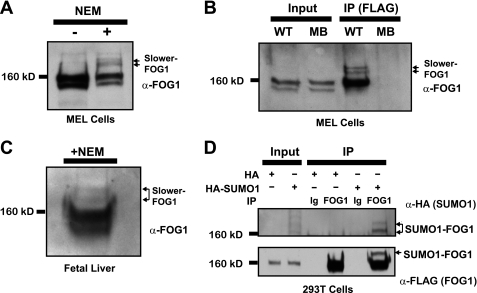
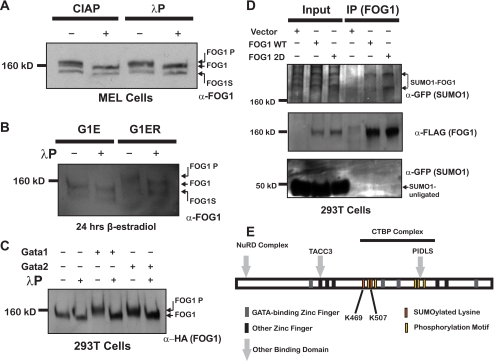
Nuclear extracts from MEL cells prepared in the presence of the SUMO-isopeptidase inhibitor N-ethylmaleimide (NEM) reveal two novel, slower migrating bands upon Western blotting with a FOG1-specific antibody (Fig. 1A). These two bands migrate with an apparent size of >160 kDa and are not detected in the absence of NEM. We have generated a MEL cell line that expresses the BirA biotin ligase alone (MB) or in combination with full-length FOG1 bearing an N-terminal FLAG tag as well as a biotin ligase substrate tag (WT). Immunoprecipitation of FOG1 with anti-FLAG M2-conjugated agarose beads reveals the same slower migrating bands by Western blot with an anti-FOG1 antibody selectively in the presence of NEM (Fig. 1B). These bands are not observed upon immunoprecipitation of FLAG-tagged FOG1 from nuclear extracts prepared in the absence of NEM. When we performed an immunoprecipitation experiment on MEL cell nuclear extracts using an antibody directed against FOG1 or normal Ig, we recovered endogenous FOG1 protein. SUMO1-positive bands corresponding in molecular weight to the slower migrating forms positive for FOG1 are selectively pulled down by the FOG1 antibody (supplemental Fig. S1A). To assess whether these slower migrating forms of FOG1, which are presumably SUMOylated forms, occur in primary cells in vivo, we prepared nuclear extracts from fetal livers of embryonic day 14.5 mice in the presence of NEM. At this stage, greater than 95% of cells in the fetal liver are committed to the erythroid lineage and express CD71, Ter119, or both (data not shown). A Western blot with FOG1-specific antibody permits detection of the two novel, slower migrating bands (Fig. 1C). After cotransfection of human epithelial 293T cell line with vectors expressing FLAG-tagged FOG1 and HA-tagged hSUMO1, we detected FOG1- and HA-positive bands by Western blot after immunoprecipitation with an antibody recognizing FOG1 (Fig. 1D). In addition, reblotting with an antibody against FLAG confirmed that these bands correspond to FOG1. To determine whether both types of SUMO, SUMO1 and SUMO2/3 (which share 95% identity (39), can be covalently attached to FOG1, we cotransfection 293T cells with vectors expressing FLAG-tagged FOG1 and HA-tagged hSUMO1 and HA-tagged hSUMO3 and performed immunoprecipitation with an antibody recognizing FOG1. We detect a slower form of FOG1, positive for both HA and FLAG, in those cells in which either HA-hSUMO1 or HA-hSUMO3 is expressed, demonstrating that FOG1 can be modified by both SUMO1 and SUMO2/3 (supplemental Fig. S1B).
FIGURE 1.
FOG1 is SUMOylated in vivo and in primary erythroid cells. A, two novel bands are illuminated by a FOG1 antibody in MEL cell nuclear extracts selectively in the presence of NEM. B, FLAG immunoprecipitation (IP) of a FLAG-tagged FOG1 stably expressed in MEL cells pulls down the two novel bands as detected by FOG1 antibody. C, novel bands are found in nuclear extracts of E14.5 fetal livers. D, FOG1 can be SUMOylated in vivo using a heterologous cell system. Constructs containing either a HA-hSUMO1 fusion protein or vector alone were co-expressed in 293T with a construct containing FLAG-Bio-tagged FOG1. After immunoprecipitation was performed on whole cell lysates with a FOG1 antibody, input and immunoprecipitates were run on a Western blot with antibodies against FLAG or HA.
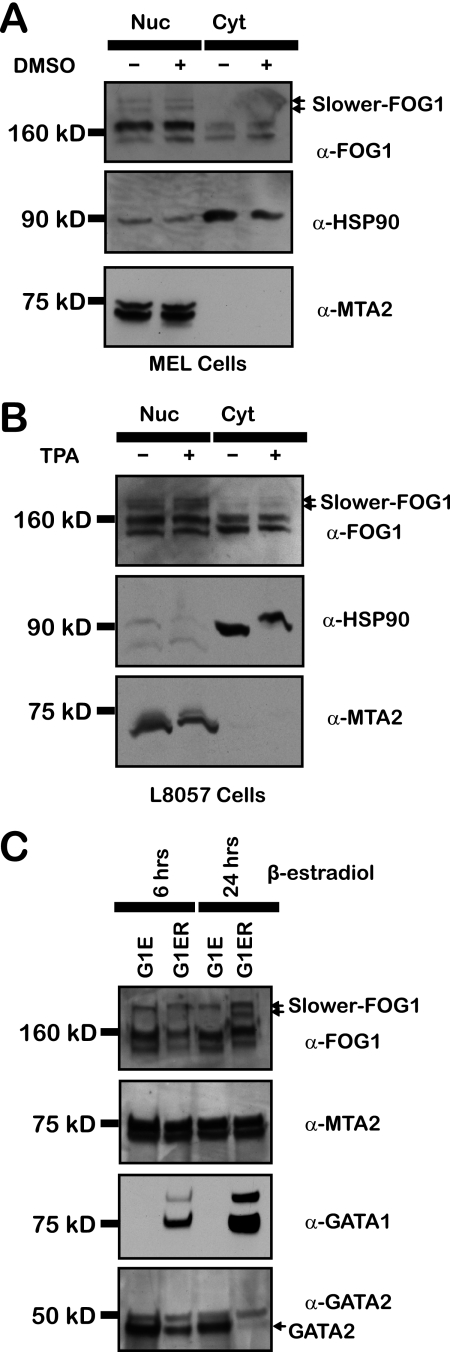
FOG1 is located in both the cytoplasm and nucleus in MEL cells (40). As SUMOylation of proteins is often involved in regulation of subcellular localization, we determined the distribution of SUMOylated versions of FOG1. MEL cells possess characteristics of partially differentiated erythroblasts and can be induced to resemble more mature erythroblasts with substantial induction of erythroid specific genes in the presence of DMSO (41). Thus, we prepared nuclear and cytoplasmic fractions of these cells with or without DMSO-induced differentiation followed by Western blotting with antibodies directed against MTA2 (nuclear) and HSP90 (cytoplasmic) to assess their purity. SUMOylated versions of FOG1, as detected with anti-FOG1 antibody, are found solely in the nuclear fraction (Fig. 2A). However, the majority of FOG1 is not SUMOylated, implying that SUMOylation of FOG1 is not required for nuclear localization of the protein. These results suggest that SUMOylation of FOG1 is important for the regulation of select FOG1-dependent gene loci, that SUMOylation of FOG1 is important in temporal regulation of FOG1 actions, or that SUMOylation of FOG1 does not play a substantive role in the function of this protein during late erythroid development.
FIGURE 2.
FOG1 is SUMOylated in multiple hematopoietic lineages in a differentiation-dependent manner and are predominantly nuclear. Nuclear and cytoplasmic extracts from MEL cells (A) and L8057 cells (B) were prepared. Equal amounts of protein were run on SDS-PAGE gels for Western blot analysis using anti-FOG1 antibody, and fraction purity was demonstrated using antibodies against MTA2 (nuclear (Nuc)) and Hsp90 (cytoplasmic (Cyt)). Nuclear extracts from G1E and G1ER cells were prepared (C). Equal amounts of protein were run on SDS-PAGE gels for Western blot analysis using antibodies directed toward FOG1, GATA1, GATA2, and MTA2.
We performed similar experiments using the megakaryocytic cell line L8057, which most closely resembles progenitor cells initially committed to this lineage. These cells can be induced to differentiate into cells possessing characteristics of mature megakaryocyte cells using TPA (30). We made nuclear and cytoplasmic extracts of these cells with or without TPA-induced differentiation that yielded results similar to that observed with MEL cells regarding nuclear localization and relative amount of SUMOylated FOG1 during differentiation (Fig. 2B), indicating that FOG1 SUMOylation occurs in different hematopoietic lineages in which FOG1 plays an important role.
The G1E cell line represents an ideal model to examine earlier stages in erythroid differentiation. Derived from GATA1-deficient embryonic stem cells (31), these cells are blocked in differentiation at an earlier stage of erythroid differentiation than MEL cells and express high levels of GATA2. G1ER cells are derived from G1E and express an exogenous version of GATA1 fused to the estrogen receptor, allowing for β-estradiol-dependent differentiation. G1ER cells exposed to β-estradiol for 24 h possessed a pattern of SUMOylation similar to MEL cells (Fig. 2, A and C). In contrast, we observed both a striking decrease in FOG1 SUMOylation and a highly distinctive pattern of SUMOylation in G1E cells expressing only GATA2. These results indicate that FOG1 SUMOylation occurs in a differentiation stage-dependent manner. A similar configuration was observed in G1ER cells after 6 h of β-estradiol treatment. G1ER cells at this time point still express moderate levels of both GATA2 and GATA1. This result suggests that the changes in FOG1 SUMOylation are not directly due to the addition of GATA1.
FOG1 SUMOylation on Lysines K469R and K507R Is Regulated by Multiple SENP Family Members
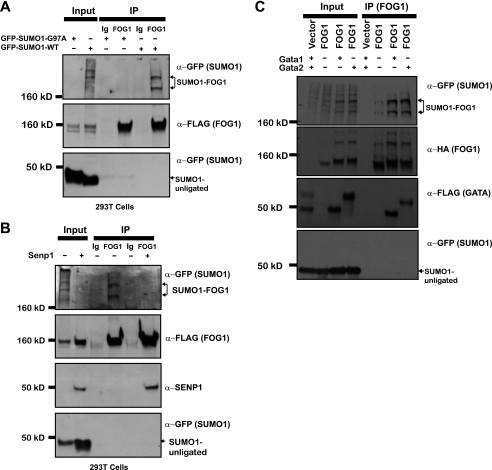
We co-expressed FLAG-tagged FOG1 in 293T cells in combination with GFP-tagged mouse SUMO1 or GFP-tagged mouse SUMO1-G79A, a mutant that cannot be conjugated to target proteins. Immunoprecipitation with FOG1 antibody followed by Western blotting with anti-GFP antibody reveals two slower migrating bands that are also recognized by FLAG specific antibody (Fig. 3A). We previously identified two translational isoforms of FOG1 (40), FOG1L and FOG1S. When we co-expressed constructs with either FLAG-tagged FOG1L or FLAG-tagged FOG1S with GFP-tagged SUMO1, we found that immunoprecipitation by a FOG1 antibody pulled down the two slower migrating bands that were positive for GFP in either case (supplemental Fig. S1C), indicating that both isoforms are SUMOylated. FOG1S does not contain the NuRD binding domain, indicating that binding to the NuRD complex is not necessary for FOG1 SUMOylation.
FIGURE 3.
FOG1 SUMOylation is regulated by SENP1 and GATA factors. Constructs containing either a GFP-SUMO1 fusion protein or a GFP-SUMO1 fusion in which a G97A mutation (to abrogate SUMO1 attachment) was engineered were co-expressed in 293T cells with a construct containing FLAG-Bio-tagged FOG1. After immunoprecipitation (IP) was performed on whole cell lysates with a FOG1 antibody, input and immunoprecipitates were run for Western blot with anti-FLAG or anti-GFP (A). A construct containing a GFP-SUMO1 fusion protein was co-expressed with a construct containing FLAG-Bio-tagged FOG1 in the presence or absence of the FLAG-tagged SUMO-isopeptidase SENP1. After immunoprecipitation was performed on whole cell lysates with a FOG1 antibody, input and immunoprecipitates were run for Western blot with anti-FLAG and anti-GFP (B). A construct containing a GFP-SUMO1 fusion protein was co-expressed with a construct containing HA-tagged FOG1 alone or with FLAG-tagged GATA1 or FLAG-tagged GATA2. After immunoprecipitation was performed on whole cell lysates with a FOG1 antibody, input and immunoprecipitates were run for Western blot with anti-HA, anti-FLAG, and anti-GFP (C).
SUMO moieties are removed from SUMOylated proteins by the action of one of seven related SUMO-isopeptidases (SENP1-SENP7) with various tissue distributions and specificities for the three SUMO peptides (39). Of potential relevance to our studies in erythroid cells, loss of SENP1, a SUMO-isopeptidase with specificity for SUMO1, leads to in utero death in mice with notable defects in erythroid development (42). We co-expressed FLAG-tagged FOG1 with GFP-SUMO1 alone or with SENP1 and immunoprecipitated FOG1 to assess its level of SUMOylation. Co-expression of SENP1 completely abrogated FOG1 SUMOylation (Fig. 3B). Additionally, blotting with antibodies to SENP1 revealed that this protein is co-immunoprecipitated by FOG1, demonstrating an interaction between FOG1 and this SUMO-isopeptidase. Our results indicate that SENP1 is able to efficiently remove SUMO1 moieties from FOG1 and that the interaction of SENP1 with FOG1 is sufficiently stable for their co-immunoprecipitation. Furthermore, our findings suggest that a critical function of SENP1 in vivo may be to remove SUMO1 from FOG1 and perhaps other transcription factors during erythroid development.
To determine whether FOG1 SUMOylation is selectively regulated by SENP1, we also examined the ability of SENP2 to deSUMOylate FOG1. We co-expressed FLAG-tagged FOG1 with GFP-SUMO1 alone or with EGFP-SENP2 and immunoprecipitated FOG1 to assess its level of SUMOylation. Co-expression of SENP2 completely abrogated FOG1 SUMOylation (supplemental Fig. S1D). Additionally, blotting with antibodies to SENP2 revealed that this protein is co-immunoprecipitated by FOG1, demonstrating an interaction between FOG1 and this SUMO-isopeptidase. Our results indicate that SENP2 is also able to efficiently remove SUMO1 moieties from FOG1 and that the interaction of SENP2 with FOG1 is sufficiently stable for their co-immunoprecipitation.
The G1E experiments above (Fig. 2C) left open the possibility of direct effects of GATA1 or GATA2 on FOG1 SUMOylation. To examine this directly, we co-expressed HA-tagged FOG1 with GFP-SUMO1 alone and with FLAG-GATA1 or FLAG-GATA2 and immunoprecipitated FOG1 to assess its level of SUMOylation. Co-expression of either GATA1 or GATA2 resulted in increases in FOG1 SUMOylation (Fig. 3C). As expected, blotting with antibodies to FLAG revealed that both GATA1 and GATA2 are co-immunoprecipitated by FOG1.
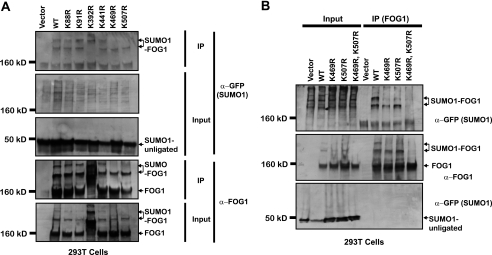
Using the online SUMO-site identification program SUMOsp (43), we found that FOG1 has six lysine residues that lie within a canonical SUMO-conjugating enzyme recognition sequence, ψKXE, where ψ represents a large hydrophobic amino acid. These sites are located at residues 88, 91, 392, 441, 469, and 507. The sites 392, 469, and 507 are conserved in the human homolog of FOG1 (data not shown). Interestingly, one of these sites is conserved in the murine FOG2 protein as well (lysine 471, with homology to the site at FOG1 lysine 507), implying that this mode of regulation might also function in FOG2-dependent contexts. To determine which of these sites is SUMOylated in vivo, we generated the following mutant versions of FOG1: K88R, K91R, K392R, K441R, K469R, and K507R. We co-expressed wild-type or mutant FOG1 in 293T cells in combination with the plasmid expressing GFP-tagged mouse SUMO1. Immunoprecipitation with anti-FOG1 antibody revealed that only two mutants, K469R and K507R, lost the SUMOylated bands (Fig. 4A). All other mutants behaved similarly to wild-type FOG1. Interestingly, the other conserved lysine, Lys-392, when mutated to arginine, demonstrated a major shift migration rate through the SDS-PAGE gel. As the mutation to arginine results in no change in charge, this result implies that protein folding may have been impacted, perhaps indicating a structural role for this residue. We generated a mutant of FOG1 containing both the K469R and K507R lysine to arginine mutations and found that, after co-expression of this doubly mutant version of FOG1 with GFP-SUMO1, FOG1 is no longer SUMOylated (Fig. 4B).
FIGURE 4.
FOG1 is SUMOylated on lysines K469R and K507R. Constructs in which all six putative SUMOylated lysines were mutated to arginine in FOG1 cDNA were co-expressed along with a GFP-SUMO1 fusion protein. Immunoprecipitation was performed on whole cell lysates with a FOG1 antibody, and input and immunoprecipitates (IP) were run for Western blot with anti-FOG1 or anti-GFP (A). After identification of lysines 469 and 507 as the SUMOylated lysines, a construct containing mutations in which both lysines were mutated to arginine was made and used in immunoprecipitation experiments along with constructs for wild-type FOG1 and both single lysine 469 and 507 single mutants. Western blot analysis using antibodies against FOG1 or GFP was performed for these FOG1 versions (B).
FOG1 SUMOylation Is Regulated by Phosphorylation
Many proteins are regulated dynamically through multisite post-translational modification, allowing for concerted control by diverse biological inputs (44). In particular, coordinate control by phosphorylation and SUMOylation of neighboring sites has been characterized for many proteins (45–47). In fact, the Lys-507 SUMOylation site in FOG1 fits the pattern of an extended SUMOylation motif that is regulated by phosphorylation. To test whether FOG1 is phosphorylated, we treated nuclear extracts from MEL cells with calf intestinal phosphatase or λ-phosphatase followed by Western blotting with anti-FOG1 antibody. Phosphatase treatment of MEL nuclear extracts by either λ-phosphatase or calf intestinal phosphatase results in a shift of both bands to a lower molecular weight providing evidence that FOG1 is in fact phosphorylated (Fig. 5A).
FIGURE 5.
FOG1 SUMOylation is regulated by Phosphorylation. Phosphatase treatment of MEL nuclear extracts with calf intestinal phosphatase or λ-phosphatase results in faster migration of FOG1 in SDS-PAGE as detected by Western blot using anti-FOG1 antibody (A). Nuclear extracts from G1E and G1ER cells differentiated with β-estradiol for 24 h were treated with λ-phosphatase and blotted with an antibody directed against FOG1 (B). Cellular extracts from 293T cells transfected with a construct containing HA-tagged FOG1 alone or with FLAG-tagged GATA1 or FLAG-tagged GATA2 were treated with λ-phosphatase or mock-treated before being run for Western blot with anti-HA (C). We generated constructs with either a FLAG-tagged wild-type FOG1 (WT) or a FLAG-tagged mutant version of FOG (2D) in which three residues (two serine and one threonine) C-terminal to the second SUMOylated lysines were mutated to aspartic acid in a FOG1 cDNA and performed immunoprecipitation experiments as above. Western blot analysis using antibodies against FOG1 or GFP for these single mutants was performed (D). A schematic shows the location of SUMOylated lysines and phosphorylation sites relative to other structural features of FOG1 (E).
G1ER cells exposed to β-estradiol for 24 h possessed a shift in the migration pattern of unSUMOylated FOG1 compared with G1E cells (Fig. 2B), exactly opposite that observed after phosphatase treatment of MEL cell extracts above. G1ER cells differentiated in β-estradiol for 6 h resembled G1E cells treated for a similar time, demonstrating that the changes were not directly due to the presence of GATA1. Treatment of nuclear extracts from G1E and G1ER cells differentiated with β-estradiol for 24 h with λ-phosphatase as above revealed that the differentiation-dependent mobility shift in G1ER cells was due to FOG1 hyperphosphorylation (Fig. 5B).
To determine whether GATA1 and GATA2 could directly affect FOG1 phosphorylation, we co-expressed HA-tagged FOG1 alone or with FLAG-GATA1 or FLAG-GATA2, made cellular extracts, and incubated them alone or with λ-phosphatase. Co-expression of either GATA1 or GATA2 resulted in a phosphatase-sensitive shift in FOG1 gel migration (Fig. 5C) similar to that seen in G1ER cells after differentiation, indicating that in this context GATA factors can lead to increased FOG1 phosphorylation.
To determine which sites are phosphorylated, we used MEL cells co-expressing the bacterial biotin ligase, BirA, and FL-Bio-FOG1 to tandem affinity purify full-length FOG1 from MEL cells either undifferentiated or differentiated for 3 days in the presence of DMSO. Mass spectrometry was used to assign phosphorylated residues. Using trypsin digestion, the peptide coverage of FOG1 from undifferentiated MEL cells was 73%, whereas for FOG1 from differentiated MEL cells it reached 85.4%. Using another enzyme, AspN, we only achieved 7.3% peptide coverage but did obtain a novel peptide not recovered by trypsin digestion. We found that indeed FOG1 is heavily phosphorylated (supplemental Table S1), whereas no evidence of acetylation, methylation, or ubiquitination was found. In total, 20 different phosphorylated peptide species spanning the entire protein were identified. Many sites were conserved in all mammalian versions of FOG1 (supplemental Table S1), and the majority of sites were found in both undifferentiated and differentiated MEL cells.
We noted that the peptide C-terminal to the second SUMOylated lysine, Lys-507, was phosphorylated as expected. In addition, a number of the conserved potential phosphorylation sites on FOG1 were located between two SUMOylated lysines, Lys-469 and Lys-507, perhaps implying a novel mechanism for cross-talk between these two post-translational modifications (Fig. 5E). Strikingly, a number of other phosphorylation sites were also found in close proximity to other domains known to be important in FOG1 function, including three near the canonical CTBP binding site (residues 811–815) located at Ser-803, -805, and -822. Submission of FOG1 sequence to motif analysis (48) revealed a number of predicted kinases, which might be responsible for phosphorylation of the serine or threonine residues near these important sites (supplemental Table S1).
We were interested in testing whether the phosphorylation sites in the region just C-terminal to the Lys-507 SUMOylation site in FOG1 might regulate addition of SUMO moieties to this site, thus linking chromatin occupancy regulation to other post-translational modification-mediated cues. We generated a version of FOG1 in which two serines (residues 511 and 512) were mutated to aspartic acid to mimic phosphorylation (known as FOG1 2D). We then tested the effect of these mutations on FOG1 SUMOylation with a GFP-SUMO1 fusion protein as described above (Fig. 5D). We found that mutation of these residues to aspartic acid increased the amount of FOG1 that was SUMOylated, supporting a role for phosphorylation of these sites in the regulation of FOG1 SUMOylation. In three independent experiments, we found a 30% (p ≤ 0.05) increase in the relative amount of FOG1 that was SUMOylated in the presence of the 2D mutations.
Neither the Stability nor Cellular Localization of KR Mutant FOG1 Is Altered in Erythroid Cells
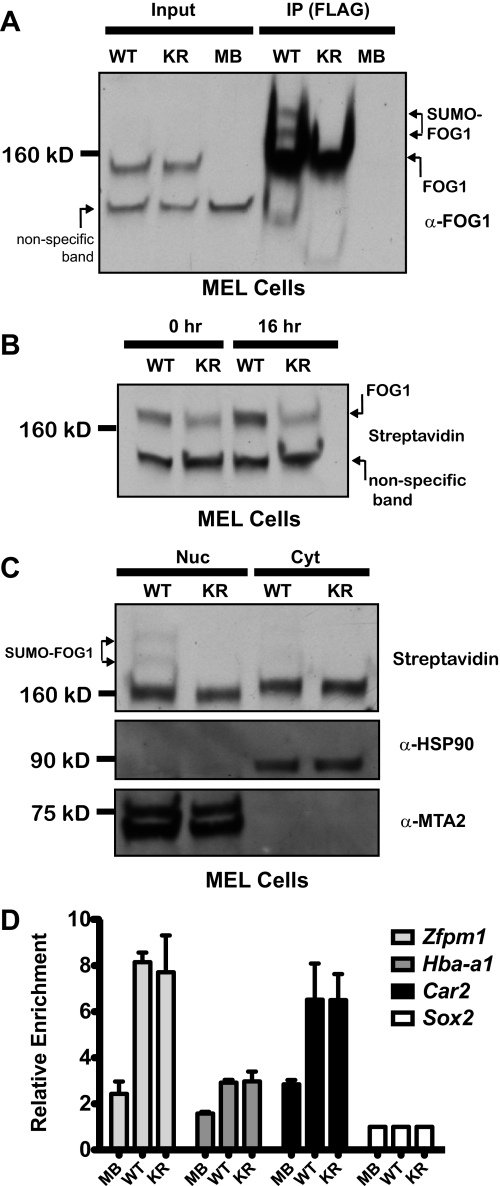
To determine whether mutagenesis of the lysine residues shown to be SUMOylated in the heterologous cell system would abrogate SUMOylation of FOG1 in a more relevant cellular context, we generated clones of MEL cells stably expressing a FL-Bio-tagged version of FOG1 containing both the K469R and K507R mutations (hereafter referred to as KR) and used them with the cells expressing (WT) or no tagged FOG1 (MB) described above. After immunoprecipitation, the slower migrating bands corresponding to SUMOylated FOG1 were observed in cells expressing wild type but not mutant FOG1 (Fig. 6A).
FIGURE 6.
KR mutant FOG1 is not altered in cellular localization, stability, or chromatin occupancy. Nuclear extracts from MEL cells expressing either a wild-type FL-Bio-tagged version of FOG1 (WT), a FL-Bio-tagged version of FOG1 containing both the K469R and K507R mutations (KR), or no tagged FOG1 (MB), were immunoprecipitated using anti-FLAG antibody (M2) conjugated to agarose. These extracts were blotted with streptavidin-HRP (A). Whole cell lysates were prepared from MEL cells expressing either a wild-type FOG1 (WT) or a mutant FOG1 (KR) after 0 or 16 h of cycloheximide treatment. Tagged FOG1 protein levels were assessed by blotting with Streptavidin-HRP (B). Nuclear (Nuc) and cytoplasmic (Cyt) extracts were prepared from MEL cells expressing either a wild-type FOG1 (WT) or a mutant FOG1 (KR), and tagged FOG1 protein levels were assessed by blotting with Streptavidin-HRP (C). Chromatin occupancy by FOG1 at select genes using biotin ChIP of wild-type (WT) and KR mutant FOG1 (KR) tagged FOG1 relative to cells expressing no tagged FOG1 (MB). Quantitative PCR of select genes (Zfpm1, Hba-a1, and Car2, with Sox2 as the negative control) in undifferentiated MEL cells (D).
We used cycloheximide to inhibit new FOG1 synthesis and assess turnover after 20 h in these cell lines. At 20 h post-cycloheximide treatment, we did not observe a difference in levels of the mutant FOG1 versus wild type, implying that there was no change in the stability of FOG1 in the absence of SUMOylation (Fig. 6B). Cellular fractionation studies in Fig. 2A implied that SUMOylation does not affect the nuclear localization of FOG1. To assess this more directly, we generated nuclear and cytoplasmic extracts from WT and KR cells and visualized FOG1 localization by Western blot using streptavidin. We did not detect a difference in the ratio of nuclear to cytoplasmic FOG1 in wild-type or KR mutant FOG1, demonstrating that SUMOylation is not required for nuclear localization (Fig. 6C). Additionally, we examined the subnuclear localization of WT and KR mutant FOG1 (KR) transfected into 293T using immunofluorescence and confocal microscopy. We found that both versions of FOG1 possess similar localization within the nucleus, exhibiting a diffuse pattern that is more intense at the nuclear periphery and excluded from nucleoli (supplemental Fig. S2).
GATA1 is SUMOylated (15), and recently this post-translational modification was shown to be important for GATA1 chromatin occupancy at select sites (49). To determine whether SUMOylation of FOG1 affects chromatin occupancy, we used clones containing either WT or KR mutant FL-Bio-tagged (KR) FOG1 or parental cells containing no FL-Bio-tagged protein (MB) to perform BioChIP followed by quantitative PCR (36, 50). We examined FOG1 binding at a previously defined GATA1-binding site found in the first intron of the Zfpm1 gene (51) and two novel sites in the Hba-a1 promoter and a regulatory region in the first intron of the Car2 gene. FOG1 bound these sites, and we observed no difference in occupancy by WT or KR mutant FOG1 (Fig. 6D). GATA1 co-occupancy at FOG1 binding sites was verified using antibody based ChIP of GATA1 in wild-type MEL cells for Zfpm1 and Hba-a1 (data not shown). These data show that FOG1 chromatin occupancy is not affected in the absence of SUMOylation sites.
SUMOylation of FOG1 Alters Its Interaction with CTBP Family Members
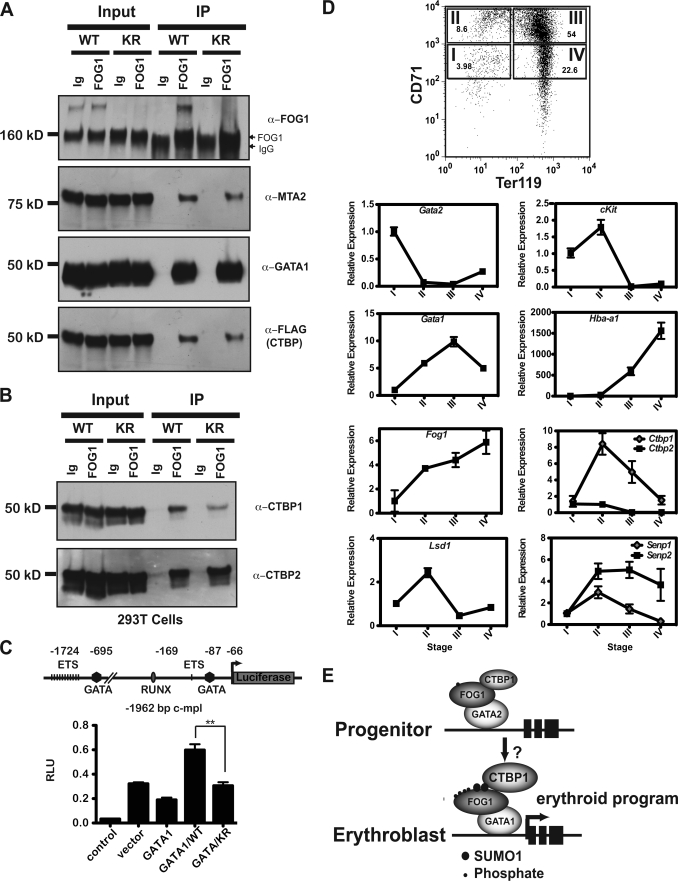
A common mechanism through which SUMO conjugation to a protein might affect its function is through alteration of physical interaction with its partners. To determine whether SUMOylation of FOG1 results in changes in its known interaction partners, GATA1, CTBP family members, or components of the NuRD complexes, we performed co-immunoprecipitation experiments. We co-expressed mutant FOG1, containing both K469R and K507R mutations (KR) or wild-type FOG1 (WT) with GATA1, CTBP1, and CTBP2. We found that similar amounts of GATA1 and GATA2 (data not shown) as well as the MTA2 and p66 (data not shown) components of the NuRD complex were recovered by immunoprecipitation of wild-type or KR mutant FOG1 with FOG1-specific antibody (Fig. 7A).
FIGURE 7.
SUMOylation of FOG1 alters its interaction with CTBP family members. A construct containing a GFP-SUMO1 fusion protein was co-expressed with constructs containing FOG1, either wild type (WT), or a version containing both the K469R and K507R mutations (known as KR), GATA1, CTBP1, and CTBP2. After immunoprecipitation was performed on whole cell lysates with a FOG1 antibody, input and immunoprecipitates (IP) were run for Western blot with antibodies to FOG1 and interaction partners MTA2 and GATA1 (A). Western blot analysis was performed for the interaction partners CTBP1 and CTBP2, first using an anti-FLAG antibody recognizing both members and then using antibodies specifically recognizing CTBP1 and CTBP2 (B). RNA was purified from cells sorted from different stages of differentiation and made into cDNA for quantitative PCR analysis of Gata2, c-kit, Gata1, Hba-a1, Fog1, Ctbp family members, Lsd1, and Senp1 and Senp2 (expression relative to β-actin levels) (C). A reporter construct consisting of 1992 bp of the murine c-mpl promoter linked to the firefly luciferase cDNA was cotransfected with an empty expression plasmid, a plasmid expressing FLAG-GATA-1, or plasmids expressing FLAG-GATA-1 with plasmids expressing wild-type HA-FOG-1 (WT) or KR mutant HA-FOG-1 (KR) as indicated. The positions of various binding sites relative to the natural transcriptional start site are indicated in the diagram of the reporter construct. Luciferase activity (RLU) was measured for triplicate samples after 24 h and is indicated on the y axis as the average ±S.E. D, shown is a model depicting increased post-translational modification of FOG1 during erythroid development and the effects on its CTBP binding (E).
In contrast to interactions with GATA1 and the proteins from the NuRD complex, FOG1 interaction with CTBP family members was modulated by mutation of SUMOylated lysines 469 and 507. Although wild-type FOG co-immunoprecipitated equivalent amounts of CTBP family members CTBP1 and CTBP2, as assessed by antibodies to the FLAG tag, mutant FOG1 pulled down less CTBP. When we probed for each of the two family members, we found that the diminished binding was due to a selective decrease in binding of CTBP1 by FOG1, as the levels of CTBP2 pulled down by KR mutant FOG1 remained unchanged (Fig. 7B). When the experiment was repeated in triplicate we found a 30% (p ≤ 0.05) relative decrease in CTBP1 binding by KR mutant FOG1. SUMOylation has been linked to the CTBP family through multiple lines of evidence; as CTBP1 is SUMOylated (52), both family members interact with components of the SUMOylation machinery (53) and have been shown to interact biochemically (54) or genetically (24) with SUMOylated hematopoietic transcription factors.
Transcriptional reporter assays performed using the c-mpl promoter as a model (55) supported a role for SUMOylation of FOG1 in its regulation of gene expression. GATA1 alone leads to a reduction in luciferase expression. Compared with vector alone, co-expression of GATA1 and wild-type FOG1 increased the expression of luciferase under the control of a 1992-bp DNA sequence located 5′ to the murine c-mpl transcriptional start site (Fig. 7C). In contrast, KR mutant FOG1 demonstrated a reduced ability to transactivate luciferase expression from the c-mpl promoter (p ≤ 0.01). Similar expression levels of the transfected proteins between different samples were verified by Western blot (data not shown).
As SUMOylation of FOG1 appears to selectively modulate binding to CTBP1, it was important to establish the pattern of co-expression of each CTBP family member, FOG1 and GATA1 and GATA2, during erythroid development. We used cDNA generated from primary fetal liver cells that had been sorted into Stages I-IV using antibodies directed to CD71 and Ter119 expression as described previously (38). Stage I is most analogous to BFU-E progenitors with high proliferative capacity and high Gata2 and c-Kit mRNA levels. These cells closely resemble G1E cells (31, 51). Stage II contains proerythroblast cells and most closely resembles G1ER cells differentiated for ∼24 h and undifferentiated MEL cells, whereas Stage III resembles fully differentiated G1ER and MEL cells. Erythroid-specific genes, such as the globin genes, become active in Stage II and continue to increase in expression through the remainder of the differentiation process. Levels of Fog1 and Gata1 increase steadily from Stage I to Stage III and then decline modestly in Stage IV. In contrast, Gata2 is only expressed appreciably in Stage I and then is rapidly repressed. During differentiation, Ctbp1 levels increase from Stage I to Stage II and then decrease slowly, remaining above basal levels. Ctbp2 transcript levels, in contrast to Ctbp1, remain low in Stages I and II and then decrease to almost undetectable levels Stages III and IV (Fig. 7D). These results indicate a differential pattern of mRNA expression for Ctbp1 and Ctbp2 during erythroid differentiation. In Stage II and III (when SUMOylation is increased at the analogous stage in the G1E system) the levels of both Fog1 and Ctbp1 increase, perhaps implying a necessity to regulate the interaction of these two molecules at the protein level through post-translational modification.
DISCUSSION
SUMOylation has emerged as an important post-translational mechanism for the regulation of transcription factor function (19). Several erythroid specific factors, including GATA1 (15, 49), are SUMOylated. Mechanisms through which SUMOylation regulates target molecules include alterations in nuclear and subnuclear localization, protein stability, or interactions with binding partners (21).
We have shown that the GATA1 coregulator, FOG1, is SUMOylated in erythroid and megakaryocyte cell lines as well as in primary erythroid cells. SUMOylation of FOG1 is low at the early erythroid progenitor stage and increases with erythroid differentiation. Co-expression of either HA-tagged hSUMO1 or GFP-mSUMO1 fusion with FOG1 in a heterologous cell line results in immunoprecipitation of SUMOylated FOG1. In addition, HA-tagged hSUMO3 can be immunoprecipitated by FOG1, demonstrating that both SUMO1 and SUMO2/3 can be efficiently attached to FOG1.
SUMOylation of FOG1 can be reduced by the SUMO-isopeptidase, SENP1, a molecule already implicated in fetal liver erythropoiesis, potentially broadening the biological role of this enzyme to include deSUMOylation of critical erythroid transcription factors. In addition, SUMOylation of FOG1 can be reduced by the SUMO-isopeptidase, SENP2. Co-expression of either GATA1 or GATA2 substantially increases the relative amount of SUMOylated FOG1. However, as G1ER cells expressing GATA1 display a substantial increase in FOG1 SUMOylation compared with G1E cells expressing GATA2, other factors must regulate this process in the erythroid context. As both SENP1 and SENP2 are increased at the mRNA level during erythroid differentiation (Fig. 7D), transcriptional down-regulation of these SUMO-isopeptidases does not appear to be responsible for increased FOG1 SUMOylation during differentiation. We have mapped the SUMOylation sites of FOG1 to lysines 469 and 507, located between zinc fingers 4 and 5 in a conserved region with no known function. Mutation of these lysines to arginines results in loss of SUMOylation of FOG1 in transient transfection assays as well as in MEL cell lines stably expressing a mutated version of FOG1. Although it is possible that these lysine residues could be modified by covalent attachment of additional moieties (e.g. acetylation (56)), no evidence of FOG1 acetylation at these residues was uncovered using peptide analysis.
We also found that FOG1 is extensively phosphorylated at a number of sites proximal to various known structural features, including the SUMOylated lysine residues. Similar to SUMOylation, FOG1 phosphorylation occurs in a differentiation-dependent manner and can be positively regulated by GATA1 or GATA2. Coordinate control by phosphorylation and SUMOylation of neighboring sites has been characterized for many proteins (45–47). One of the phosphorylation sites near the second SUMOylated lysine, which fits the pattern for the extended SUMOylation sequence defined before (45–47), can influence SUMOylation of FOG1. These data strongly imply a role for cross-talk by phosphorylation and SUMOylation in the regulation of FOG1. Although pathways involved in FOG1 phosphorylation are yet to be elucidated, it is attractive to speculate that they may be important in carefully regulating the stepwise transcription of erythroid genes in response to cellular and extracellular signals. Especially interesting is the existence of this same module in GATA1 (45–47) and GATA2,4 hinting at a large degree of synchronized regulation of the two GATA family members and FOG1 by similar or distinct cellular cues. A recent report demonstrated that SUMOylation of GATA1 is FOG1-independent (49). Conversely, FOG1 can be SUMOylated in the absence of GATA1 in GIE cells. GATA1 and GATA2 appear to be able to increase FOG1 SUMOylation directly in some contexts.
We found no evidence that SUMOylation alters the stability of FOG1 in half-life experiments using cycloheximide in MEL cells expressing tagged versions of wild-type or mutant FOG1. SUMOylation of FOG1 does not appear to have any effect on nuclear localization of FOG1, as shown through the use of KR mutant FOG1. Although SUMOylation of certain proteins has been shown to be involved in their localization to specific subnuclear domains (57), no evidence of this effect was observed with FOG1. Finally, mutation of SUMOylated lysines does not impact chromatin occupancy by FOG1 in contrast to results found for GATA1 (49).
FOG1 interacts with GATA1 along with at least two repression complexes. We found that the absence of SUMOylation does not appear to substantially alter the interaction of FOG1 with the NuRD complex (as shown by interaction with MTA2 and p66) or GATA1 itself but does change its binding with CTBP proteins. We show that the absence of the SUMOylated lysines leads to the reduction in CTBP binding, specifically affecting the binding of CTBP1. Previous data (53, 58–61) have indicated that the two CTBP family members may recruit different effector molecules and have different functions in the regulation of gene expression. We have shown the interaction of FOG1 and LSD1 as part of the CTBP complex (40). Examination of LSD1 chromatin occupancy data previously generated in our laboratory (50) revealed that the genes we have shown here to be occupied by FOG1 are co-occupied by LSD1. These data are the first to show that FOG1 and components of the CTBP-complex, LSD1, co-occupy chromatin sites in genes known to be regulated by GATA1.
FOG1 interacts with two putative repression complexes; that is, the CTBP-containing complex (11, 12) and the NuRD repression complex (13, 14). The interaction of FOG1 and the NuRD complex was recently shown to mediate both activating and repressing functions at select genes (62, 63), implying that further layers of regulation are required to specify different transcriptional modes at specific loci. It is tempting to speculate that increased post-translational modification of FOG1 occurring during the transition from highly proliferative progenitor cells to erythroblasts (Fig. 7E) serves as one mechanism for modulating the co-occupancy of CTBP family members and their associated effector molecules with GATA1 and the NuRD complex at differentially regulated genes.
Supplementary Material
Acknowledgments
We thank A. Cantor for biotin-tagging constructs and luc2-c-mpl and MEL cells expressing BirA and for advice on affinity purification and chromatography, J. Palvimo for GFP-SUMO1 and GFP-SUMO1GA vectors, E. Yeh for FLAG-SENP1, and L. Zon for the pEBB-HA-SUMO1 and pEBB-HA vector. We also thank Grigoriy Losyev and the Brigham and Women's Hospital Flow Cytometry Core for Flow Activated Cell sorting and R. Tomaino and S. Gygi for performing LC-MS/MS and for providing advice and assistance in data collection and analysis. We thank Jianlong Wang, Jianlin Chu, Tyler Moran, Andrew Woo, Thomas Akie, and Stephan Chan for technical advice and helpful discussions. In addition we thank Emery Bresnick and Stephan Chan for critical comments in preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HL32259 (to S. H. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
J. W. Snow, J. Kim, C. R. Currie, J. Xu, and S. H. Orkin, unpublished observations.
- FOG1
- Friend of GATA 1
- GATA1
- GATA-binding protein 1
- GATA2
- GATA-binding factor 2
- SUMO
- small ubiquitin-like modifier
- LSD1
- lysine-specific demethylases 1
- MEL
- murine erythroleukemia
- CTBP
- C-terminal-binding protein
- NuRD
- nucleosome remodeling domain
- EGFP
- enhanced GFP
- NEM
- N-ethylmaleimide.
REFERENCES
- 1.Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. (1989) Nature 339, 446–451 [DOI] [PubMed] [Google Scholar]
- 2.Evans T., Felsenfeld G. (1989) Cell 58, 877–885 [DOI] [PubMed] [Google Scholar]
- 3.Weiss M. J., Orkin S. H. (1995) Exp. Hematol. 23, 99–107 [PubMed] [Google Scholar]
- 4.Orkin S. H. (1992) Blood 80, 575–581 [PubMed] [Google Scholar]
- 5.Pevny L., Simon M. C., Robertson E., Klein W. H., Tsai S. F., D'Agati V., Orkin S. H., Costantini F. (1991) Nature 349, 257–260 [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara Y., Browne C. P., Cunniff K., Goff S. C., Orkin S. H. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12355–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shivdasani R. A., Fujiwara Y., McDevitt M. A., Orkin S. H. (1997) EMBO J. 16, 3965–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vyas P., Ault K., Jackson C. W., Orkin S. H., Shivdasani R. A. (1999) Blood 93, 2867–2875 [PubMed] [Google Scholar]
- 9.Vyas P., Crispino J. D. (2007) Curr. Opin. Pediatr. 19, 9–14 [DOI] [PubMed] [Google Scholar]
- 10.Tsang A. P., Visvader J. E., Turner C. A., Fujiwara Y., Yu C., Weiss M. J., Crossley M., Orkin S. H. (1997) Cell 90, 109–119 [DOI] [PubMed] [Google Scholar]
- 11.Turner J., Crossley M. (1998) EMBO J. 17, 5129–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox A. H., Liew C., Holmes M., Kowalski K., Mackay J., Crossley M. (1999) EMBO J. 18, 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K. E., Guyot B., Heck A. J., Vyas P., de Boer E., Grosveld F., Strouboulis J. (2005) EMBO J. 24, 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong W., Nakazawa M., Chen Y. Y., Kori R., Vakoc C. R., Rakowski C., Blobel G. A. (2005) EMBO J. 24, 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collavin L., Gostissa M., Avolio F., Secco P., Ronchi A., Santoro C., Del Sal G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8870–8875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W., Kitidis C., Fleming M. D., Lodish H. F., Ghaffari S. (2006) Blood 107, 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamonica J. M., Vakoc C. R., Blobel G. A. (2006) Blood 108, 3736–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez M. S., Dargemont C., Hay R. T. (2001) J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 19.Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 20.Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 21.Dohmen R. J. (2004) Biochim. Biophys. Acta 1695, 113–131 [DOI] [PubMed] [Google Scholar]
- 22.Seeler J. S., Bischof O., Nacerddine K., Dejean A. (2007) Curr. Top Microbiol. Immunol. 313, 49–71 [DOI] [PubMed] [Google Scholar]
- 23.Siatecka M., Xue L., Bieker J. J. (2007) Mol. Cell. Biol. 27, 8547–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perdomo J., Verger A., Turner J., Crossley M. (2005) Mol. Cell. Biol. 25, 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shyu Y. C., Lee T. L., Ting C. Y., Wen S. C., Hsieh L. J., Li Y. C., Hwang J. L., Lin C. C., Shen C. K. (2005) Mol. Cell. Biol. 25, 10365–10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Lloris R., Osses N., Jaffray E., Shen L. N., Vaughan O. A., Girwood D., Bartrons R., Rosa J. L., Hay R. T., Ventura F. (2006) FEBS Lett. 580, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 27.Motohashi H., Katsuoka F., Miyoshi C., Uchimura Y., Saitoh H., Francastel C., Engel J. D., Yamamoto M. (2006) Mol. Cell. Biol. 26, 4652–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chun T. H., Itoh H., Subramanian L., Iñiguez-Lluhí J. A., Nakao K. (2003) Circ. Res. 92, 1201–1208 [DOI] [PubMed] [Google Scholar]
- 29.Garriga-Canut M., Orkin S. H. (2004) J. Biol. Chem. 279, 23597–23605 [DOI] [PubMed] [Google Scholar]
- 30.Woo A. J., Moran T. B., Schindler Y. L., Choe S. K., Langer N. B., Sullivan M. R., Fujiwara Y., Paw B. H., Cantor A. B. (2008) Mol. Cell. Biol. 28, 2675–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss M. J., Yu C., Orkin S. H. (1997) Mol. Cell. Biol. 17, 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamitani T., Nguyen H. P., Kito K., Fukuda-Kamitani T., Yeh E. T. (1998) J. Biol. Chem. 273, 3117–3120 [DOI] [PubMed] [Google Scholar]
- 34.Hang J., Dasso M. (2002) J. Biol. Chem. 277, 19961–19966 [DOI] [PubMed] [Google Scholar]
- 35.Chrétien S., Varlet P., Verdier F., Gobert S., Cartron J. P., Gisselbrecht S., Mayeux P., Lacombe C. (1996) EMBO J. 15, 4174–4181 [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J., Iyer V. R. (2005) Curr. Protoc. Mol. Biol., Chapter 21, Unit 21.10 [DOI] [PubMed] [Google Scholar]
- 38.Zhang J., Socolovsky M., Gross A. W., Lodish H. F. (2003) Blood 102, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay D., Dasso M. (2007) Trends Biochem. Sci. 32, 286–295 [DOI] [PubMed] [Google Scholar]
- 40.Snow J. W., Orkin S. H. (2009) J. Biol. Chem. 284, 29310–29319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orkin S. H., Harosi F. I., Leder P. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng J., Kang X., Zhang S., Yeh E. T. (2007) Cell 131, 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Y., Zhou F., Fu C., Xu Y., Yao X. (2006) Nucleic Acids Res. 34, W254–W257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X. J., Grégoire S. (2006) Mol. Cell 23, 779–786 [DOI] [PubMed] [Google Scholar]
- 45.Yang S. H., Jaffray E., Senthinathan B., Hay R. T., Sharrocks A. D. (2003) Cell Cycle 2, 528–530 [DOI] [PubMed] [Google Scholar]
- 46.Anckar J., Sistonen L. (2007) Biochem. Soc. Trans. 35, 1409–1413 [DOI] [PubMed] [Google Scholar]
- 47.Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee H. Y., Johnson K. D., Fujiwara T., Boyer M. E., Kim S. I., Bresnick E. H. (2009) Mol. Cell 36, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saleque S., Kim J., Rooke H. M., Orkin S. H. (2007) Mol. Cell 27, 562–572 [DOI] [PubMed] [Google Scholar]
- 51.Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., Weiss M. J. (2004) Blood 104, 3136–3147 [DOI] [PubMed] [Google Scholar]
- 52.Lin X., Sun B., Liang M., Liang Y. Y., Gast A., Hildebrand J., Brunicardi F. C., Melchior F., Feng X. H. (2003) Mol. Cell 11, 1389–1396 [DOI] [PubMed] [Google Scholar]
- 53.Chinnadurai G. (2007) Int. J. Biochem. Cell Biol. 39, 1593–1607 [DOI] [PubMed] [Google Scholar]
- 54.Gómez-del Arco P., Koipally J., Georgopoulos K. (2005) Mol. Cell. Biol. 25, 2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H., Yu M., Akie T. E., Moran T. B., Woo A. J., Tu N., Waldon Z., Lin Y. Y., Steen H., Cantor A. B. (2009) Mol. Cell. Biol. 29, 4103–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X. J., Seto E. (2008) Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heun P. (2007) Curr. Opin. Cell Biol. 19, 350–355 [DOI] [PubMed] [Google Scholar]
- 58.Quinlan K. G., Nardini M., Verger A., Francescato P., Yaswen P., Corda D., Bolognesi M., Crossley M. (2006) Mol. Cell. Biol. 26, 8159–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan K. G., Verger A., Kwok A., Lee S. H., Perdomo J., Nardini M., Bolognesi M., Crossley M. (2006) Mol. Cell. Biol. 26, 8202–8213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuppuswamy M., Vijayalingam S., Zhao L. J., Zhou Y., Subramanian T., Ryerse J., Chinnadurai G. (2008) Mol. Cell. Biol. 28, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao L. J., Subramanian T., Chinnadurai G. (2008) Oncogene 27, 5214–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miccio A., Wang Y., Hong W., Gregory G. D., Wang H., Yu X., Choi J. K., Shelat S., Tong W., Poncz M., Blobel G. A. (2010) EMBO J. 29, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gregory G. D., Miccio A., Bersenev A., Wang Y., Hong W., Zhang Z., Poncz M., Tong W., Blobel G. A. (2010) Blood 115, 2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.