Abstract
MazG nucleoside triphosphate pyrophosphohydrolase (NTP-PPase, EC 3.6.1.8) from the avirulent Mycobacterium tuberculosis H37Ra contains a spontaneous mutation on a highly conserved residue, resulting in an A219E substitution (MtMazG[A219E]). In this work, we show that mycobacterial MazG from either the virulent M. tuberculosis H37Rv (MtMazG) or the fast-growing Mycobacterium smegmatis (MsMazG) is a potent NTP-PPase capable of hydrolyzing all canonical (d)NTPs, as well as the mutagenic dUTP and 8-oxo-7,8-dihydro-2′-dGTP. However, this hydrolysis activity is diminished by the MtMazG[A219E] mutation. Moreover, deletion of mazG in M. smegmatis rendered the mycobacteria defective in response to oxidative stress. Importantly, expression of the wild-type MtMazG, but not the A219E mutant, restored cell viability under oxidative stress. Intriguingly, under oxidative stress, both the mazG-null and MtMazG[A219E]-expressing M. smegmatis strains failed to elevate relA, while retaining their ability to up-regulate sigE, suggesting a specific role for the MazG NTP-PPase activity in oxidative stress-triggered, transcriptional activation of relA. The MtMazG is a homotetramer with each subunit containing a single MazG core domain flanked by two regions, both of which are essential for NTP-PPase activity. Taken together, these results demonstrate that the mycobacterial MazG is a potent NTP-PPase and that this activity is required to maintain the full capacity of the mycobacteria to respond to oxidative stress. Our work implicates a role for the MazG activity in the virulence of M. tuberculosis.
Keywords: Bacteria, Enzyme Catalysis, Enzyme Mechanisms, Enzyme Mutation, Oxidative Stress, MazG, Mycobacterium, NTP Pyrophosphohydrolyase
Introduction
Mycobacterium tuberculosis H37Ra is an avirulent relative of the typical laboratory strain H37Rv, both of which are derived from the parent strain H37 that was originally isolated from a 19-year-old patient with chronic pulmonary tuberculosis by Edward R. Baldwin in 1905 (1). Our systematic comparison of the complete genomic sequences of H37Rv and H37Ra identified multiple H37Ra-specific single nucleotide polymorphisms (SNPs) affecting about 30 genes including the virulence determinants phoP, nrp, and pks12 (2), consistent with the widely accepted notion (3–7) that the virulence attenuation of H37Ra is due to multiple mutations. One of these mutations, the A219E substitution located in a highly conserved sequence of H37Ra MazG, prompted the current study aiming to reveal the function of the corresponding prototype enzyme in M. tuberculosis (MtMazG) and compare its role with that of Escherichia coli (EcMazG) in adaptation to stress response (8).
The MazG protein belongs to the all-α nucleoside triphosphate pyrophosphohydrolase (NTP-PPase,3 EC 3.6.1.8) superfamily that hydrolyzes in vitro all canonical nucleoside triphosphates into monophosphate derivatives and pyrophosphate (PPi). The physiological substrates of MazG, however, remain elusive (9, 10). Based on the resolved structure of MazG derived from Sulfolobus solfataricus and substrate modeling, it is proposed that the MazG family enzymes might perform house-cleaning function by degrading abnormal (d)NTPs, such as dUTP, 2-hydroxy-dATP, and 8-oxo-dGTP (9, 10). These modified nucleotides are usually produced under oxidative damage and are mutagenic, i.e. their incorporation into genomic DNA causes mispairing and subsequently leads to increased mutation rates. However, to date, this house-cleaning function of MazG remains to be validated experimentally because of the lack of physiological substrates (9, 10).
Accumulating evidence suggests a role for EcMazG in protecting cells from certain toxins. EcMazG regulated programmed cell death by interfering with the function of the MazEF complex, a well characterized toxin-antitoxin (TA) module (8, 11). MazEF was the first identified chromosomal TA module, characterized by the labile antitoxin, MazE, and its cognate stable toxin, MazF (12). MazF is an ACA sequence-specific mRNA endoribonuclease that can selectively degrades mRNA via cleavage between the nucleotide residues A and C, thereby inducing a programmed cell death process under stress conditions (13, 14). In E. coli, mazG is cotranscribed with mazEF, while the MazEF proteins inhibit the MazG enzymatic activity both in vivo and in vitro (8). It was proposed that the EcMazG regulates the MazEF TA module by lowering the cellular (p)ppGpp level, thus protecting the cell from the toxicity of MazF (8). However, it remains to be answered how MazG regulates the cellular (p)ppGpp level and whether such regulation concerns relA, which encodes the (p)ppGpp synthetase.
By employing the transposon insertion approaches, previous mouse model studies observed attenuated infection from an H37Rv mazG mutant, thereby implicating a role for MtMazG in virulence (15). Despite such advance, however, little is known concerning the enzymatic and physiological functions of MtMazG. In this work, we show that mycobacterial MazG is a potent NTP-PPase capable of hydrolyzing the mutagenic dUTP and 8-oxo-dGTP and is required to maintain Mycobacterium smegmatis full capacity to respond to oxidative stress, supporting the previously proposed “house cleaning” function of this family protein. However, these functions are diminished in the spontaneous A219E mutant found in H37Ra. Furthermore, mycobacterial mazG is an oxidative stress-inducible gene and its NTP-PPase activity is involved in transcriptional activation of relA in response to oxidative stress. In contrast to EcMazG, mycobacterial MazG is not involved in starvation response. Unlike mycobacterial MazG, the EcMazG cannot complement for M. smegmatis mazG under oxidative stress condition. Taken together, our findings demonstrate that MtMazG is distinct from EcMazG with respect to the properties of enzymes and the possible function in stress response. MtMazG is the first MazG of its kind to be thoroughly characterized, and might be relevant to the virulence of M. tuberculosis.
EXPERIMENTAL PROCEDURES
Reagents and Enzyme
Restriction enzymes and Pfu DNA polymerase were purchased from Fermentas. T4 DNA ligase, calf-intestinal alkaline phosphatase, and nucleoside triphosphates were obtained from New England Biolabs. The 8-oxo-dGTP was purchased from Trilink BioTechnologies, Inc. PiPer pyrophosphate assay kit from Molecular Probes, Inc. was chosen for pyrophosphate detection.
Cloning and Expression of mazG
Using the genomic DNA as template, the prototype mazG gene from M. tuberculosis H37Rv and its mutant type from H37Ra were PCR-amplified with primer TBF22 and TBR22. (All primers used in this study are listed in supplemental Table S1.) The PCR products were digested with NdeI and XhoI and cloned into the corresponding site of pET22b(+) with a fused hexahistidine tag at the C-terminal end of MazG. The resulting vector pET22b-mazG was used to transform competent E. coli BL21(DE3) cells, which were grown at 37 °C with vigorous shaking to A600 of 0.7 in Luria-Bertani (LB) medium containing 100 μg/ml ampicillin. Expression of MazG was induced with isopropyl-1-thio-β-d-galactopyranoside (0.5 mm) for 12 h at 22 °C. The mazG of E. coli, M. smegmatis, and the truncated variants, including its site mutants introduced by overlapping PCR, were cloned and expressed in the same way.
Purification of MazG
All operations were carried out at 4 °C. Cells from 200 ml of culture were harvested by centrifugation (7000 × g, 20 min) and resuspended in 10 ml of binding buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, 10% glycerol, 10 mm imidazole, 5 mm dithiothreitol). The cells were disrupted by ultrasonication followed by centrifugation at 20,000 × g for 20 min to remove cell debris and insoluble fraction. The resulting supernatant was applied to a 10-ml column with 1 ml of Ni-Sepharose Fast Flow (GE Healthcare Life Sciences) equilibrated with binding buffer. MazG protein eluted with 100 and 200 mm imidazole was pooled and concentrated by a 30-kDa cutoff ultrafilter (Millipore). The concentrated product was loaded onto Hiload 16/60 Superdex 200 perp grade column (GE Healthcare Life Sciences) and eluted with buffer containing 20 mm Tris-HCl, pH 7.9, 100 mm NaCl, 1 mm dithiothreitol. The final product was concentrated to 5 mg/ml maintained in 20 mm Tris-HCl, pH 7.9, 100 mm NaCl, with 20% glycerol, divided into small aliquots, and stored at −30 °C. The protein concentration was determined by bicinchoninic acid (BCA) method (16).
Enzyme Assay
The NTP-PPase activity of MazG was assayed by measuring the hydrolyzed product, PPi, by an enzyme-coupled colorimetric assay (Molecular Probes) with detection limit of 0.2 μm (17). The standard NTP-PPase assay was carried out in 20 μl reaction buffer (50 mm glycine-NaOH, pH 9.6, 100 mm NaCl, 5 mm Mg2+) containing 1 μg of MtMazG and appropriate amount of nucleoside triphosphates at 37 °C for 10 min. The reaction was stopped by heating at 70 °C for 10 min, and 5 μl of reaction product was applied for PiPer pyrophosphate assay (Molecular Probes) according to the manufacturer's instructions in the “Assaying for Pyrophosphate” setting. The reaction without MtMazG or substrates was carried out as a background control. Reaction conditions including temperature, pH, Mg2+ concentration, amount of MtMazG and reaction time, were optimized (supplemental Fig. S1). To optimize the pH, both the Tris-HCl buffer for pH 6.8–8.8, and the glycine-NaOH buffer for pH 9.2–11.0 were used.
CD Spectrometry
The CD (circular dichroism) spectra were measured on a Jasco J-715 spectropolarimeter in the far-ultraviolet region (260–190 nm) in steps of 1 nm. Records on protein solutions (20 μg/ml in H2O) employing a cell with path length of 1 cm at 18 °C were obtained. The mean residue ellipticity, represented in millidegrees, was calculated using a mean residue molecular mass of 110 Da. To see the CD spectra changes upon addition of Mg2+, MgCl2 solution was added to the protein samples immediately before measurement. Each spectrum reported is an average of five scans. Blank spectra of aqueous buffer were used to correct the observed spectra.
Construction of mazG-null Mutant and Complementation in M. smegmatis mc2 155
The following procedures were carried out to construct the ΔmazG::hyg null mutation in M. smegmatis mc2 155 (18). Two homology arms (0.83 kb and 0.99 kb for left and right arm, respectively) overlapping the mazG gene at its 75 and 99 extremities were PCR-amplified from genomic DNA by using primers MKO1/MKO2 and MKO3/MKO4, respectively. The PCR products were digested and cloned into the cosmid vector pYUB854 in their original genome orientation separated by the res-hyg-res cassette. The resulting cosmid was digested by PacI and inserted into the genome of mycobacteriophage phAE87 and was subsequently packaged by the Packagene® Lambda DNA Packaging System (Promega). The recombinant transducing phage was used to construct the mazG-null mutant. The dUTP-biotin labeled probe (Fermentas) was used for Southern blot analysis of the PstI digested chromosomal DNA on the Hybond-N+ nylon membrane (GE Amersham Biosciences), according to the standard method (19).
For complementation of the mazG mutant in M. smegmatis, mazG gene from H37Rv, H37Ra, M. smegmatis and E. coli were PCR-amplified using the primers CTBF/CTBR (for both H37Rv and H37Ra), CMSF/CMSR, and CECF/CECR, respectively. The native promoter region (425 bp region from 5509332 to 5509756 of M. smegmatis genome, see supplemental Fig. S2) of the mazG operon was amplified with the primers ProF/ProR. The PCR products were enzyme digested and subsequently cloned into the integrative plasmid pMV306. The resulting plasmids containing the mazG gene under control of its native promoter were used to complement the M. smegmatis mazG-null mutant.
Survival Assay under Oxidative Stress
M. smegmatis strains were grown in 7H9 broth supplemented with 0.2% glycerol and 0.05% Tween 80 at 37 °C. For determination of survival following hydrogen peroxide (H2O2) exposure, log phase cultures (A600 at 0.4–0.5) were diluted in fresh medium to an A600 of 0.2, and exposed to 5 mm hydrogen peroxide (20). Cultures were incubated by rolling at 37 °C, and aliquots were removed from each culture for plating at the indicated time points. Colonies were counted after 3 days by plating in triplicate at three dilutions onto 7H11 plates containing appropriate antibiotics.
Quantitative Real-time PCR
RNA was extracted from H2O2-treated and untreated log phase cultures (A600∼0.5) using the RiboPure-Bacteria kit (Ambion). cDNA was synthesized using the SuperScript III First Strand kit (Invitrogen) with random hexamers primer. Target gene transcript levels in H2O2-treated log phase cultures were measured by quantitative real-time PCR with SYBR Green as label, normalized to sigA transcript levels (21), and expressed as a fold change from untreated cultures.
Analytical Gel Filtration
Hiload 16/60 Superdex 200 perp grade column (GE Healthcare Life Sciences) was employed to determine the molecular mass of MazG. The sample was eluted by 50 mm phosphate buffer, pH 8.0, 150 mm NaCl, flow rate was set to 1 ml/min and elution of the protein was monitored at 280 nm. Aldolase (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and chymotrypsinogen A (25 kDa) were taken for calibration.
RESULTS
Cloning, Expression, and Purification of MazG from M. tuberculosis, M. smegmatis, and E. coli
The mazG genes containing the sequence of either the prototype H37Rv (Rv1021) or the mutated H37Ra (MRA_1029) were cloned into plasmid pET22b(+) and expressed in E. coli BL21(DE3) cells. The His-tagged MazG proteins (36.3 kDa) eluted from the nickel-affinity column were subsequently purified by Superdex 200 chromatography to nearly SDS-PAGE homogeneity. The EcMazG and the MsMazG (MSMEG_5422), including the truncated variants of MtMazG (see following), were all similarly expressed and purified (Fig. 1A).
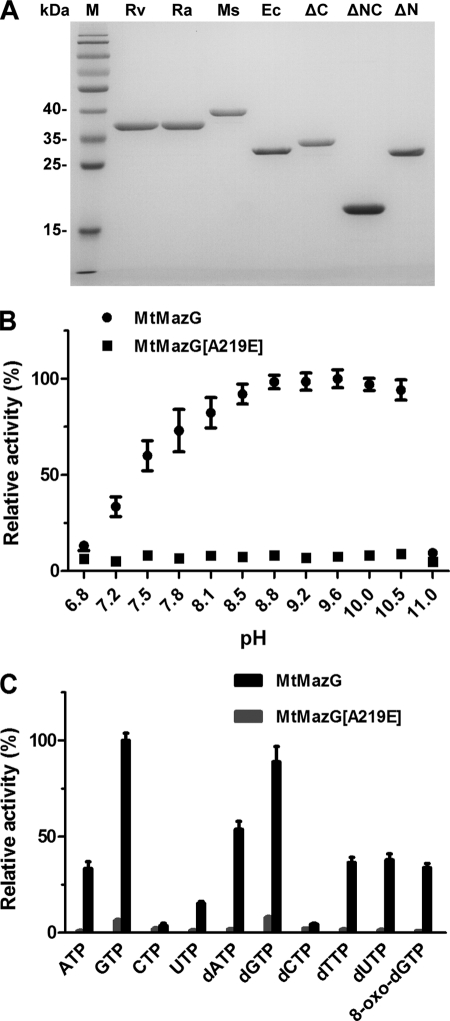
FIGURE 1.
Enzymatic properties of MtMazG. A, purified recombinant MazG proteins expressed in E. coli. Proteins were expressed and purified as described in “Experimental Procedures.” The protein purity was analyzed by 12% SDS-PAGE, about 1 μg of purified proteins were loaded. Rv, Ra, Ms, and Ec stand for H37Rv, H37Ra, M. smegmatis and E. coli MazG, respectively; ΔC, ΔN, and ΔNC stand for the C-terminal region, NE region, and dual-region truncated variants of MtMazG. B, optimal pH curve for MtMazG reactions. Reactions were conducted as described in “Experimental Procedures” with 1 mm GTP as substrate. The hydrolysis activity under each pH was normalized to the activity of that under pH 9.6 (100%, correspond to PPi formation ability of 600 ± 40 μm/10 min). C, substrate specificity of MtMazG. Relative hydrolysis activities toward various substrates (1 mm) at standard condition were normalized to that of GTP (100%, the same as panel B). All data were the mean ± S.E. from at least four independent experiments.
The NTP-PPase Activity of MtMazG and MsMazG
The NTP-PPase activity was assayed by measuring the PPi product, which was first converted to Pi by inorganic pyrophosphatase, and the Pi level was subsequently quantified by an enzyme coupled colorimetric method (see “Experimental Procedures”). Like other characterized MazG proteins (10, 22, 23), MtMazG was found to maintain more than 70% of its NTP-PPase activities between pH 7.8 and 10.5, with the optimum pH at 9.6 (Fig. 1B). Mg2+ acted as an optimal cofactor among the 7 divalent cations tested in our assay (supplemental Fig. S1A). The NTP-PPase activity of MtMazG was the highest at 37 °C and nearly abolished at 70 °C. The effects of pH and temperature on protein stability were also examined. Incubation of the enzyme in pH 9.6 at 37 °C for 10 min did not affect its NTP-PPase activity (supplemental Fig. S1B).
The general kinetic parameters of MtMazG were determined using 8 canonical (d)NTPs as the substrates (Table 1). It was found that MtMazG converted all of the tested substrates into their nucleoside monophosphate derivatives and PPi. No detectable Pi formed because the reaction strictly depended on inorganic pyrophosphatase. Of (d)NTPs examined, GTP and dGTP were the most preferred substrates for MtMazG (Fig. 1C), with Km values of 0.4 ± 0.06 mm for either of them, which is 1.5–4 times lower than that of other substrates. On the other hand, the CTP and dCTP hydrolysis activities of MtMazG were too low to allow for determination of their kinetic parameters (Fig. 1C). As shown, MsMazG exhibited the same optimal reaction conditions and substrate spectrum relative to MtMazG (Tables 1 and supplemental Table S2), indicating that MsMazG is functionally similar to MtMazG.
TABLE 1.
Kinetic parameters of MazG proteins analyzed in this study
Standard reaction assay was performed as described in “Experimental Procedures.” Kinetic parameters were determined with eight different concentrations of each substrate ranged from 0.1 to 2 mm (0.03∼1.5 mm for 8-oxo-dGTP). Values represent the mean ± S.E. of four independent experiments. Kinetic parameters for all tested substrates of MsMazG are listed in supplemental Table S2.
| Species | Enzyme | Mg2+ | Substrate | Vmax | kcat | Km | kcat/km |
|---|---|---|---|---|---|---|---|
| mm | nm/min/μg | min−1 | mm | min−1mm−1 | |||
| Mt | WT | 5 | GTP | 1.5 ± 0.1 | 55.2 ± 3.1 | 0.4 ± 0.1 | 139 |
| WT | 5 | dGTP | 1.4 ± 0.1 | 51.8 ± 2.1 | 0.4 ± 0.1 | 130 | |
| WT | 5 | dATP | 1.7 ± 0.2 | 60.4 ± 7.2 | 1.2 ± 0.3 | 52 | |
| WT | 5 | dTTP | 1.6 ± 0.1 | 58.4 ± 3.9 | 1.3 ± 0.2 | 44 | |
| WT | 5 | dUTP | 1.2 ± 0.2 | 45.3 ± 6.0 | 1.1 ± 0.3 | 43 | |
| WT | 5 | ATP | 1.3 ± 0.1 | 48.4 ± 5.1 | 1.6 ± 0.3 | 31 | |
| WT | 5 | UTP | 0.5 ± 0.04 | 16.8 ± 1.5 | 0.6 ± 0.2 | 26 | |
| E219 | 5 | GTP | 0.08 ± 0.01 | 2.8 ± 0.2 | 0.3 ± 0.1 | 10 | |
| WT | 20 | GTP | 2.3 ± 0.2 | 85.5 ± 6.4 | 0.6 ± 0.1 | 134 | |
| E219 | 20 | GTP | 0.8 ± 0.1 | 30.7 ± 3.7 | 1.2 ± 0.3 | 29 | |
| WT | 5 | 8-oxo-dGTP | 0.14 ± 0.01 | 5.2 ± 0.2 | 0.11 ± 0.02 | 47 | |
| E219 | 5 | 8-oxo-dGTP | NDa | ND | ND | ND | |
| Ms | WT | 5 | GTP | 3.1 ± 0.2 | 115 ± 6.2 | 0.9 ± 0.1 | 127 |
| E222 | 5 | GTP | 0.11 ± 0.02 | 4.1 ± 0.6 | 0.9 ± 0.3 | 4 | |
| WT | 5 | 8-oxo-dGTP | 0.16 ± 0.01 | 5.9 ± 0.2 | 0.16 ± 0.02 | 36 | |
| E222 | 5 | 8-oxo-dGTP | ND | ND | ND | ND | |
| Ec | WT | 5 | dATP | 1.9 ± 0.1 | 59.8 ± 2.3 | 0.34 ± 0.05 | 174 |
| E141 | 5 | dATP | 1.8 ± 0.1 | 57.8 ± 2.4 | 0.37 ± 0.05 | 156 | |
| WT | 5 | dUTP | 1.9 ± 0.1 | 60.3 ± 3.1 | 0.71 ± 0.1 | 85 | |
| WT | 5 | 8-oxo-dGTP | 0.23 ± 0.01 | 7.2 ± 0.4 | 0.36 ± 0.05 | 20 | |
| E141 | 5 | 8-oxo-dGTP | 0.22 ± 0.01 | 6.9 ± 0.3 | 0.42 ± 0.06 | 17 |
a ND, nondetectable.
dUTP and 8-oxo-dGTP Hydrolysis Activity of Mt-, Ms-, and EcMazG
It is noteworthy that MtMazG and MsMazG are able to hydrolyze dUTP (Table 1), the most common mutagenic NTP produced as a byproduct during thymine nucleotide biosynthesis (9, 24). Both enzymes exhibited a moderate affinity toward dUTP with a Km of ∼1 mm, similar to most of their tested canonical NTP substrates and of their homolog from Thermotoga maritima (23, 25). EcMazG is capable of hydrolyzing dUTP as well. Although it exhibits lower substrate affinity (with a Km of 0.7 mm) than that with canonical NTP substrates (Table 1), it is comparable to or better than that of MtMazG.
Given that MtMazG can hydrolyze dUTP, we subsequently tested the hydrolysis activity of MtMazG toward 8-oxo-dGTP, a major mutagenic substrate for DNA synthesis arising from oxidative damage (Table 1) (26–28). It was found that both MtMazG and MsMazG exhibited substantially high substrate affinity to 8-oxo-dGTP, with a Km of 0.11 and 0.16 mm, respectively. The Km value for 8-oxo-dGTP hydrolysis reaction of EcMazG was 0.36 mm, which is not significantly different from that with other substrates (Table 1). The order of kcat/Km for Mt-, Ms, and EcMazG is: 47, 36, and 20 min−1 mm−1, respectively.
The A219E Mutation in MtMazG Nearly Abolishes Its NTP-PPase Activity
Under the aforementioned optimal conditions for MtMazG, the hydrolytic activities of MtMazG[A219E] were examined using various substrates (Table 1 and Fig. 1C). MtMazG[A219E] hydrolyzed GTP with kcat that is 20-fold lower than that of the wild type. However, the mutant enzyme Km is reduced about 25% relative to that of MtMazG. Thus, the catalytic efficiency of MtMazG[A219E] is down to 7% of the wild-type level (Fig. 2B and Table 1). The same results were obtained from the experiments performed at a range of pH 6.8–11 (Fig. 1B). In addition, the corresponding mutation (A222E) in MsMazG resulted in a decrease of the hydrolysis activity to levels similar to those observed with the A219E mutation in MtMazG, suggesting that MsMazG and MtMazG share similar catalytic mechanism. No detectable 8-oxo-dGTP hydrolysis activities were observed with either MtMazG[A219E] or MsMazG[A222E], indicating that this mutation reduced 8-oxo-dGTP hydrolysis activity by a factor of greater than 35 times (Table 1).
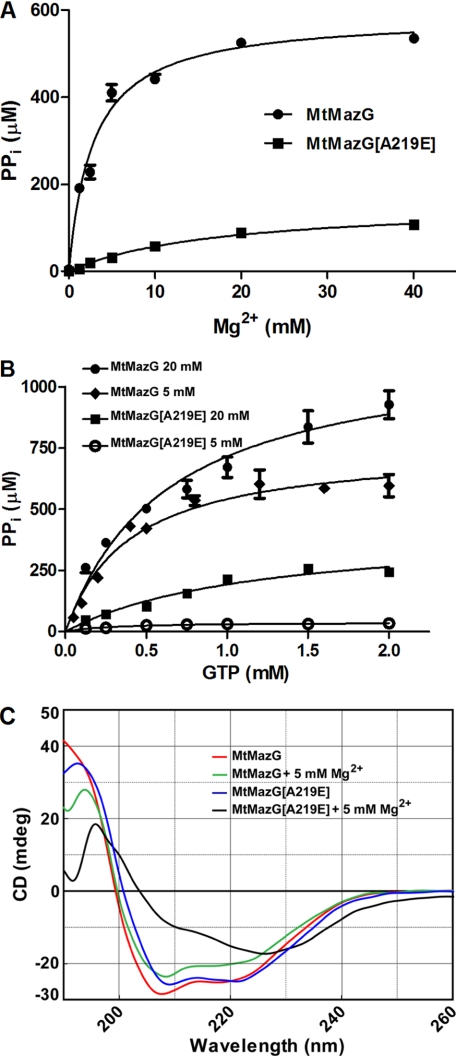
FIGURE 2.
Mg2+ requirement and CD spectrum study of MtMazG and MtMazG[A219E]. A, Mg2+ concentration-dependent activation of MtMazG and MtMazG[A219E]. With 0.5 mm GTP as substrate, the PPi formation ability (μm/10 min) of the MtMazG[A219E] variant and its prototype enzyme was monitored under increased Mg2+ concentration. The reaction mixture without substrates was used as a background control. B, Michaelis-Menten curves for MtMazG and MtMazG[A219E] under 5 mm and 20 mm Mg2+. GTP was chosen as substrate and its concentration were depicted. Each value (μm/10 min) is the mean ± S.E. from three independent experiments. C, CD spectra of MtMazG and MtMazG[A219E] measured at 18 °C in the presence or absence of 5 mm Mg2+. CD spectra with 10 or 15 mm Mg2+ exhibited a similar CD spectrum to that observed with 5 mm Mg2+. Each spectrum reported is an average of five scans.
A219E Mutation in MtMazG Affects Mg2+ Binding and Protein Structure
Residue Ala-141 of the EcMazG (corresponding to Ala-219 of MtMazG) is located near the catalytic center motif for Mg2+ and substrate binding (11), we reasoned that the A219 → E substitution of MtMazG might result in an increase of negative charge that alters Mg2+ binding. As revealed by the Mg2+ titration experiments, Mg2+ activated both MtMazG and MtMazG[A219E] (supplemental Fig. S3), while the MtMazG[A219E] exhibited a Kact for Mg2+ of 17.4 ± 4 mm, which is six times higher than that for MtMazG (2.8 ± 0.3 mm) (Fig. 2A). Increasing Mg2+ concentration from 5 to 20 mm enhanced the MtMazG[A219E] Vmax for the formation of PPi by nearly 10-fold (Fig. 2B and Table 1). However, even at 20 mm Mg2+, A219E exhibited the catalytic efficiency at levels no more than 25% of the wild type (Fig. 2B).
Given that MtMazG A219 is essential for catalysis but is unlikely located in the catalytic center, we suspect that it may play an important role in determining the overall conformation of the protein. To address this possibility, we performed CD spectra analysis with purified MtMazG and MtMazG[A219E] in the presence or absence of Mg2+. In the absence of Mg2+, both the wild type and the mutant enzyme exhibited a similar pattern of α-helices and β-strands (Fig. 2C). These results are consistent with the CD spectra pattern observed with the MazG family proteins in the previous studies (10, 25), suggesting that MtMazG and MtMazG[A219E] share nearly identical structures in the absence of Mg2+. However, whereas the addition of 5 mm Mg2+ resulted in minor changes in the CD spectra of the wild-type enzyme, it induced significant alterations to the A219E protein (Fig. 2C). Similar observations were made with the wild type and A219E in the presence of 10 or 15 mm Mg2+ (data not shown). These results suggest that upon Mg2+ binding, the negative charge of the E219 side chain induces rather drastic alterations in the secondary structures of MtMazG, and this altered conformation may explain the severe deficiency of MtMazG[A219E] in hydrolysis.
MazG Is Required for Full Oxidative Stress Response in M. smegmatis
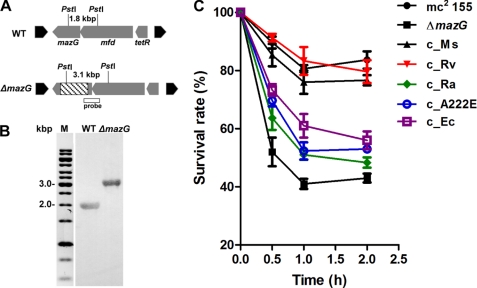
To explore the in vivo function of MtMazG and evaluate the physiological effect of A219E mutation, we constructed a ΔmazG::hyg null mutation in the fast-growing M. smegmatis mc2 155, which was confirmed by Southern blot (Fig. 3B). The wild-type and mazG-null strains were subject to oxidative stress, elicited by exposure to 5 mm H2O2 (20, 29). Nearly 80% of the wild-type bacteria were viable after the treatment for 1 h, whereas only 40% of the mutant bacteria survived under the same condition (Fig. 3C), revealing a defect for the mazG-null mutant in response to oxidative stress. This 2-fold decrease in viability by the mazG mutant under oxidative stress is significant, as comparable effect was observed on mycobacterial sigJ, sigE and ppk mutants in previous publications (29–31). Importantly, the attenuated phenotype in the mazG-null mutant can be fully complemented by the expression of the wild-type mazG gene of M. smegmatis under its native promoter (the characterization of the mazG operon and its promoter region is shown in supplemental Fig. S2).
FIGURE 3.
Southern blot analysis and phenotypic characterization of mazG mutant in M. smegmatis. A, schematic diagrams of WT and the mazG-deleted (ΔmazG) loci. The putative mazG operon (gray arrows) and its adjacent genes (black arrows) are depicted. The expected sizes for the restriction enzyme, PstI and the probe fragment used for Southern blot are as shown. B, Southern blot analysis of the WT and ΔmazG strains. A dUTP-biotin labeled fragment was used to probe PstI-digested chromosomal DNA separated by 0.8% agarose gel. Lane M contains a 1-kb DNA ladder (Fermentas). C, survival abilities under oxidative stress exerted by 5 mm H2O2. The strains used in this assay are wild type (mc2 155), mazG mutant (ΔmazG), and strains complemented by MsMazG (c_Ms), MtMazG (c_Rv), MtMazG[A219E] (c_Ra), MsMazG[A222E] (c_A222E), and EcMazG (c_Ec). The starting CFU of this survival assay was around 1∼2 × 107 (100%). All data were the mean ± S.E. from four independent experiments.
Of note, the mazG-null mutant showed no growth defects in vitro (supplemental Fig. S4). Moreover, no difference was observed between the wild-type and mazG-null M. smegmatis under starvation conditions (log-phase bacteria were placed in phosphate-buffered saline for 30 days (32)) (data not shown). This finding suggests that mycobacterial mazG, unlike its counterpart in E. coli (8), is not required for starvation response. Taken together, these results suggest that mazG is specifically required for M. smegmatis full capacity to respond to oxidative stress.
The Oxidative Stress-Response Property of MtMazG Is Dependent on Its NTP-PPase Activity
The mazG fusion genes from H37Rv and H37Ra under control of the native mazG promoter from M. smegmatis were separately integrated into the attB sites of the mazG-null mutant strain of M. smegmatis. As with MsMazG complementation, H37Rv mazG fully restored the ability of the mutant strain to survive under oxidative stress. However, expression of MtMazG[A219E] under the same condition improved cell viability only slightly (Fig. 3C). Similarly, the introduction of MsMazG[A222E], equivalent to the MtMazG[A219E], failed to complement (Fig. 3C). Thus, the NTP-PPase activity is required for Mt- or MsMazG to mediate full cellular response to oxidative stress.
Interestingly, in contrast to Mt- or MsMazG, EcMazG only marginally complement the mazG-null mutant (Fig. 3C). These findings suggest that MazG-mediated cellular response to oxidative stress is specific to mycobacteria.
Transcription of mazG Is Up-regulated under Oxidative Stress, and Induction of relA Is Repressed in mazG-null or MtMazG[A219E] Mutant
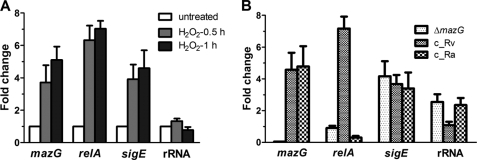
Quantitative real time PCR was used to investigate transcriptional changes of mazG under oxidative stress. It was found that expression of sigE and relA, two regulatory genes involved in oxidative stress response (30, 33), were increased about 3.9 and 6.3 times, respectively, after exposure to 5 mm H2O2 for 0.5 h (Fig. 4A). Transcription of mazG was up-regulated 3.7 times and 5.1 times after the treatment for 0.5 and 1 h, respectively (Fig. 4A), indicating that mazG is an oxidative stress response gene.
FIGURE 4.
Mycobacterial mazG is an oxidative stress-inducible gene and its NTP-PPase activity is required for induction of relA upon oxidative insult. qRT-PCR was used to measure target gene mRNA level in H2O2-treated log phase cultures. Transcript levels were normalized relative to sigA, and expressed as a fold change from untreated cultures. A, oxidative stress-induced transcript levels of mazG, relA, sigE, and 16 S rRNA in wild-type M. smegmatis. B, transcript levels of mazG, relA, sigE, and 16 S rRNA in mazG-null mutant (ΔmazG), and strains complemented with MtMazG[A219E] (c_Ra) or MtMazG (c_Rv), which were treated by 5 mm H2O2 for 30 min. All data were presented as mean ± S.E. from at least three independent experiments.
The sigE and relA mRNA levels in mazG-null mutant were also measured. Surprisingly, while the transcriptional activation of sigE under oxidative stress remained unchanged in the mazG-null strain, the induction of relA is severely repressed after exposure to H2O2 for 0.5 h (Fig. 4B). Consistent with this observation, the expression of 16 S RNA, a well known target for repression by RelA-mediated stringent response (34), was found up-regulated only in the mazG mutant strain under oxidative stress (Fig. 4B). Moreover, expression of MtMazG, but not the H37Ra-derived MtMazG[A219E], in mazG mutant strain restored relA transcription to the level of wild type (Fig. 4B). These results establish that induction of relA under oxidative stress is dependent on the NTP-PPase activity of mycobacterial MazG.
MtMazG Is Structurally Different from EcMazG
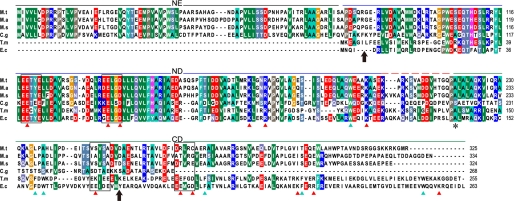
We investigated structural differences that may underlie the seemingly differential physiological functions exhibited by MtMazG and EcMazG. It was reported that EcMazG is composed of two core MazG domains connected in tandem, termed the N- and the C-terminal domains (11). Sequence comparison indicated that MtMazG had an extra N-terminal 80 residues (NE region) relative to that of EcMazG (Fig. 5). Furthermore, it seems that MtMazG only has one region highly homologous to the N-terminal domain of EcMazG, which contains key residues required for Mg2+ coordination and substrate binding. However, the MtMazG C terminus is unlike composed of a MazG-like core domain because secondary structure predictions reveal that this region contains only two α-helices, whereas a typical MazG-like core domain consists of five α-helices (10, 22). Taken together, MtMazG appears to organize by a single MazG-like core domain that is flanked with an N-terminal and a C-terminal region with unknown functions.
FIGURE 5.
Sequence alignment of MazG proteins. The sequences are from M. tuberculosis H37Rv (M.t), M. avium 104 (M.a), M. smegmatis mc2155 (M.s), Corynebacterium glucuronolyticum ATCC 51867 (C.g), T. maritima MSB8 (T.m), and E. coli MG1655 (E.c). The N-terminal (ND) and C-terminal domains (CD) of EcMazG and the N-terminal extra region (NE) of MtMazG are indicated. Four EEXX (E/D) motifs from EcMazG are indicated by blank rectangles. Key residues of EcMazG involved in Mg2+ coordination and substrates binding are indicated by red and cyan triangles, respectively (11). The Ala-219 residue is marked with an asterisk. Black arrows indicate the truncation sites for NE region and C-terminal region.
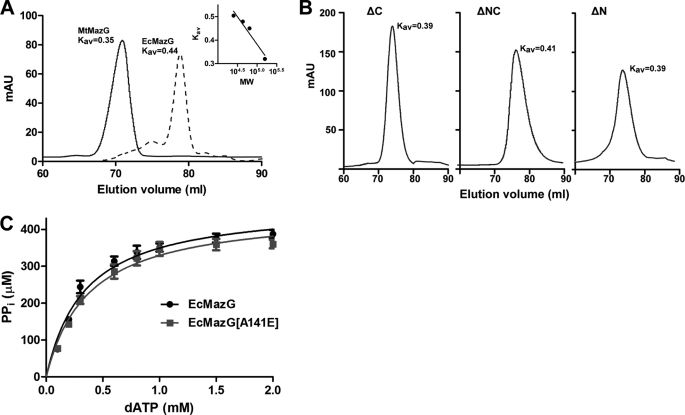
It has been shown that MazG proteins with a single core domain form tetrameric structures, whereas an analog with two core domains is dimeric (10, 11, 22). To determine the stoichiometric composition of MazG oligomer, MazG from different species were subject to size exclusion chromatographic analysis. MtMazG was eluted as a single species with a partition coefficient value (Kav) of 0.35, corresponding to the molecular mass of ∼140 kDa, while EcMazG was eluted as a dimer with a molecular mass of ∼64 kDa (Kav = 0.44) (Fig. 6A). These results suggest that MtMazG is a tetrameric complex under native conditions, which further supports the hypothesis that MtMazG contains a single core MazG domain.
FIGURE 6.
MtMazG is structurally different from EcMazG. A, size exclusion chromatographic analysis of MtMazG (solid) and EcMazG (dashed). Two independent elution curves are overlaid with Kav value for each peak indicated. The calibrated curve is shown in the inset. B, size exclusion chromatographic analysis for the oligerization conformation of MtMazG mutants with deletions in the NE region (ΔN), C-terminal region (ΔC), or dual-regions (ΔNC). The ΔN and ΔC exhibited an apparent molecular mass of ∼106 kDa, whereas ΔNC is about 84 kDa. C, Michaelis-Menten curves for EcMazG and its A141E variant. The assay was conducted as described previously (23). dATP was used as substrate with eight different concentrations ranging from 0.1 to 2.0 mm. Each value (μm/10 min) is the mean ± S.E. of three independent experiments.
Although MtMazG is structurally different from EcMazG, the A219 residue of MtMazG is conserved in the EcMazG protein. The corresponding A141E substitution protein of EcMazG (EcMazG[A141E]) was expressed and purified to nearly homogeneity (Fig. 1A), and its enzymatic activity was assayed as described (23). With dATP as the substrate, EcMazG[A141E] exhibited the wild-type level of NTP-PPase activity (Fig. 6C and Table 1). Its Kmvalue was 9% higher than that of the wild-type enzyme, while the Vmax remained unchanged. Altogether, the above results suggest that the MtMazG and the EcMazG differ substantially in their overall structures and the catalytic mechanisms.
The NE and C-terminal Regions of MtMazG Are Essential for NTP-PPase Activity
To study the functions of the NE and C-terminal regions of MtMazG, we performed deletion analysis by constructing MtMazGΔN (corresponding to residues 84–325; 27.7 kDa), MtMazGΔC (corresponding to residues 1–253; 27.9 kDa), and MtMazGΔNC (corresponding to residues 84–253; 19.8 kDa) (Fig. 5). The deletion sites were selected according to the following criteria. 1) The deleted regions should not exceed the target domains. 2) The truncation sites should not interrupt secondary structure elements, especially helices. 3) If possible, the deleted regions should not be conserved among MazG family proteins.
These truncated MtMazG proteins were expressed and purified (Fig. 1A). These truncations did not affect protein yields or solubility, and the tetrameric structures were well retained as shown by the size exclusion chromatographic analysis (Fig. 6B). However, no NTP-PPase activity was observed with any of three truncated proteins in the presence of GTP or 8-oxo-dGTP as substrate. These results suggest that both the MtMazG NE region and C-terminal region are essential for the NTP-PPase activity.
DISCUSSION
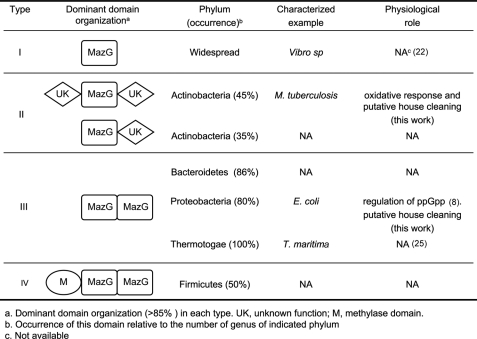
In this study, we characterized a novel MazG protein from Mycobacterium, a genus of the Actinobacteria phylum. The mycobacterial MazG appears to differ structurally and functionally from its homologs characterized to date, including MazG from E. coli (8, 23), T. maritima (25), Vibrio sp (22), and Mus musculus (designated RS21-C6) (35). Size exclusion chromatographic analysis demonstrated that the MtMazG forms a tetrameric structure with each subunit containing a single core MazG domain flanked by two additional regions (or domains). Both of these flanking regions are essential for the hydrolysis activity, as deletion of either leads to complete loss of NTP-PPase activity. This structural feature distinguishes MtMazG from all other currently characterized MazG family proteins (9, 10, 22). We found that nearly 80% of MazG homologs from Actinobacteria exhibited the MtMazG-like domain organization (Fig. 7), whereas in other phyla, such configuration is rarely observed.
FIGURE 7.
Domain associations of MazG family proteins and their relationships to species evolution. MazG family proteins from 832 species were selected for analysis based on protein sequence homology. Dominant domain associations in each type are depicted, and their occurrence relative to total genus number is illustrated.
Genome-wide analysis of the MazG family proteins from 832 species has led us to classify them into I-IV major subgroups based on their determined/predicted structural organizations (Fig. 7 and supplemental Table S3). Type I, represented by iMazG from Vibrio sp (22), contains a single MazG core domain. Type II, found predominantly in Actinobacteria, is composed of a single core MazG domain and one or two flanking regions. It has two subclasses depending on whether it contains either both N- and C-terminal flanking regions or a single C-terminal segment. Type III has double core-domain in tandem and is found in Proteobacteria. Finally, in Firmicutes, type IV is organized by a double core-domain in tandem and a methylase domain at the N terminus.
MazG family proteins are long believed to function as house-cleaning enzymes involved in degrading non-canonical NTPs, such as dUTP, 8-oxo-dGTP and other mutagenic NTPs, and thus preventing their incorporation into DNA or RNA (9, 10, 22). However, direct evidence supporting this putative function is lacking. In this study, we showed, for the first time, that MtMazG and EcMazG can hydrolyze both dUTP and 8-oxo-dGTP (Table 1). MtMazG exhibited a Km of 0.11 mm toward 8-oxo-dGTP, suggesting a role for this enzyme to recognize the damaged nucleotide preferentially in comparison to canonical NTPs. However, this affinity is about 230-fold lower than that exhibited by E. coli MutT, which hydrolyzes 8-oxo-dGTP with a Km of 0.48 μm (36). It thus appears unlikely that 8-oxo-dGTP is a physiological substrate of MtMazG. Given that only a few out of more than 20 different types of damaged nucleotides have been discovered are available (37), it remains a daunting challenge to identify the MtMazG physiological substrate(s) by in vitro enzyme assay at the present time. Despite the lack of physiological substrates, our current study has advanced the understanding of the function of mycobacterial MazG by linking this enzyme to cellular regulatory pathway in response to oxidative stress. We have shown that in M. smegmatis, MazG is an oxidation-inducible gene product (Fig. 4) that is required for the full capacity of mycobacteria to respond to oxidative stress (Fig. 3C). These observations are in line with the house-cleaning hypothesis, suggesting that mycobacterial MazG, induced by oxidative stress, could conceivably act to degrade the oxidation-induced damaged nucleotides. Impairment of such hydrolysis activity would compromise cell viability, which was observed in M. smegmatis that was deleted of MazG or expressed a mutant form of MazG (MtMazG[A219E]) with poor catalytic function (Fig. 3C).
Another interesting finding of our study is that induction of relA under oxidative stress is dependent on the NTP-PPase activity of mycobacterial MazG (Fig. 4B). This observation is in agreement with a previous report that E. coli MazG-mediated, starvation-induced programmed cell death was associated with altered cellular ppGpp levels (8), suggesting an interplay between EcMazG and RelA. As a key player in stress-induced physiological adaptation (34), RelA is required for mycobacterial survival under in vitro starvation and hypoxia conditions, as well as long-term persistence of M. tuberculosis in mice (32, 38). Therefore, RelA dysfunction may affect bacterial survival under stress conditions as observed in the mazG-null mutant. However, although RelA is involved in oxidative response, the relA-deleted M. smegmatis did not show survival defect under oxidative stress (32, 39). This suggests that induction of relA may not be required for cellular oxidative stress response directly. Alternatively, redundant RelA-independent pathways might be sufficient for cellular oxidative stress response under the experimental conditions. It is also possible that RelA is involved in adaptation following oxidative stress. It is worth noting that mycobacterial and E. coli MazG appear to act differently in cellular stress response. We have shown that in contrast to mycobacterial MazG, EcMazG cannot complement M. smegmatis mazG-null mutant in response to oxidative stress (Fig. 3C). However, unlike EcMazG (8), MsMazG is not required for cell viability under the starvation conditions (data not shown).
We have validated our previous prediction (2) that the A219E substitution in H37Ra MazG is a loss-of-function mutation both in vitro and in vivo. Whereas it remains to be elucidated how the A219E substitution inactivates the MazG hydrolyase activity, it can be argued that a likely underlying cause is the mutation-imposed conformational changes given that A219E induced drastic alteration of secondary structures of MtMazG upon Mg2+ binding (Fig. 2C). Surprisingly, however, Mg2+ at high concentration, while inducing profound alteration in secondary structures, was able to increase the activity of MtMazG[A219E], albeit to a limited extent (Fig. 2B). The mechanistic basis for this partial activation of the MtMazG[A219E] hydrolytic activity by high levels of Mg2+ remains to be explored.
Based on the findings that MsMazG and MtMazG have similar enzymatic properties (Table 1), and on the conserved function between these two enzymes in maintaining cell viability under oxidative stress (Fig. 3C), it is likely that MtMazG and MsMazG share similar physiological role, at least to some extent. Apart from this study uncovering a role for MtMazG in oxidative stress response, little is known about the physiological functions of this enzyme. A previous genetic screening study found that the H37Rv mazG mutant was attenuated in the mouse infection model (15), implying that mazG is a potential virulence factor in M. tuberculosis. Rengarajan et al. (40) reported that the transcription of H37Rv mazG was up-regulated 2.7 times after infection in INF-γ activated macrophage cells. To survive inside the macrophages, M. tuberculosis has acquired abilities to withstand the bactericidal host defenses, such as oxidative and nitrosative stress, starvation, hypoxia and acidification (41, 42). It was suggested that to a certain extent, the loss of virulence of the avirulent strain H37Ra may be related to its survival ability inside the macrophages (43). Our work has shown that the H37Ra-derived A219E mutation diminishes the MazG hydrolase activity (Figs. 1 and 2) as well as its capacity to respond to oxidative stress (Fig. 3). Based on these findings, we speculate that H37Ra may lose its virulence partly due to the A219E mutation-caused reduction of MazG hydrolase activity and viability under oxidative stress within the macrophage environment. If validated, this line of research would elucidate MazG as a potentially critical factor that contributes to the virulence of M. tuberculosis.
Supplementary Material
Acknowledgments
We thank Prof. Zhen-Qiang Pan at Mount Sinai School of Medicine, Petros Karakousis at Johns Hopkins University School of Medicine, and Benfang Lei at Montana State University for discussions and critical comments on the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (No. 30830002 and No. 30970077), Shanghai Rising-Star Program, Science and Technology Commission of Shanghai Municipality (No. 09QA1403600), the State Key Development Programs for Basic Research of China (973 Program No. 2009CB522605), and the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S3.
- NTP-PPase
- nucleoside triphosphate pyrophosphohydrolase
- PPi
- pyrophosphate
- CD
- circular dichroism
- TA
- toxin-antitoxin
- 8-oxo-dGTP
- 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate
- Mt
- M. tuberculosis
- Ms
- M. smegmatis
- Ec
- E. coli
- WT
- wild type.
REFERENCES
- 1.Streeken W., JR, Gardner L. U. (1946) Am. Rev. Tuberc. 54, 62–66 [DOI] [PubMed] [Google Scholar]
- 2.Zheng H., Lu L., Wang B., Pu S., Zhang X., Zhu G., Shi W., Zhang L., Wang H., Wang S., Zhao G., Zhang Y. (2008) PLoS One 3, e2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosch R., Philipp W. J., Stavropoulos E., Colston M. J., Cole S. T., Gordon S. V. (1999) Infect Immun 67, 5768–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Q., Kripke K., Arinc Z., Voskuil M., Small P. (2004) Tuberculosis 84, 188–196 [DOI] [PubMed] [Google Scholar]
- 5.He X. Y., Zhuang Y. H., Zhang X. G., Li G. L. (2003) Microbes Infect 5, 851–856 [DOI] [PubMed] [Google Scholar]
- 6.Lari N., Rindi L., Lami C., Garzelli C. (1999) Microb Pathog 26, 281–286 [DOI] [PubMed] [Google Scholar]
- 7.Mostowy S., Cleto C., Sherman D. R., Behr M. A. (2004) Tuberculosis 84, 197–204 [DOI] [PubMed] [Google Scholar]
- 8.Gross M., Marianovsky I., Glaser G. (2006) Mol. Microbiol. 59, 590–601 [DOI] [PubMed] [Google Scholar]
- 9.Galperin M. Y., Moroz O. V., Wilson K. S., Murzin A. G. (2006) Mol. Microbiol. 59, 5–19 [DOI] [PubMed] [Google Scholar]
- 10.Moroz O. V., Murzin A. G., Makarova K. S., Koonin E. V., Wilson K. S., Galperin M. Y. (2005) J. Mol. Biol. 347, 243–255 [DOI] [PubMed] [Google Scholar]
- 11.Lee S., Kim M. H., Kang B. S., Kim J. S., Kim G. H., Kim Y. G., Kim K. J. (2008) J. Biol. Chem. 283, 15232–15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aizenman E., Engelberg-Kulka H., Glaser G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Zhang J., Hoeflich K. P., Ikura M., Qing G., Inouye M. (2003) Mol. Cell 12, 913–923 [DOI] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka H., Hazan R., Amitai S. (2005) J. Cell Sci. 118, 4327–4332 [DOI] [PubMed] [Google Scholar]
- 15.Sassetti C. M., Rubin E. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker J. M. (1994) Methods Mol. Biol. 32, 5–8 [DOI] [PubMed] [Google Scholar]
- 17.Payandeh J., Fujihashi M., Gillon W., Pai E. F. (2006) J. Biol. Chem. 281, 6070–6078 [DOI] [PubMed] [Google Scholar]
- 18.Bardarov S., Bardarov Jr. S., Jr., Pavelka Jr. M. S., Jr., Sambandamurthy V., Larsen M., Tufariello J., Chan J., Hatfull G., Jacobs Jr. W. R., Jr. (2002) Microbiology 148, 3007–3017 [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 20.Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manganelli R., Dubnau E., Tyagi S., Kramer F. R., Smith I. (1999) Mol. Microbiol. 31, 715–724 [DOI] [PubMed] [Google Scholar]
- 22.Robinson A., Guilfoyle A. P., Harrop S. J., Boucher Y., Stokes H. W., Curmi P. M., Mabbutt B. C. (2007) Mol. Microbiol. 66, 610–621 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Inouye M. (2002) J. Bacteriol. 184, 5323–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzminova E. A., Kuzminov A. (2004) Mol. Microbiol. 51, 1279–1295 [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Zhang Y., Inouye M. (2003) J. Biol. Chem. 278, 21408–21414 [DOI] [PubMed] [Google Scholar]
- 26.Pursell Z. F., McDonald J. T., Mathews C. K., Kunkel T. A. (2008) Nucleic Acids Res. 36, 2174–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tassotto M. L., Mathews C. K. (2002) J. Biol. Chem. 277, 15807–15812 [DOI] [PubMed] [Google Scholar]
- 28.Sekiguchi M., Tsuzuki T. (2002) Oncogene 21, 8895–8904 [DOI] [PubMed] [Google Scholar]
- 29.Wu Q. L., Kong D., Lam K., Husson R. N. (1997) J. Bacteriol. 179, 2922–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sureka K., Dey S., Datta P., Singh A. K., Dasgupta A., Rodrigue S., Basu J., Kundu M. (2007) Mol. Microbiol. 65, 261–276 [DOI] [PubMed] [Google Scholar]
- 31.Hu Y., Kendall S., Stoker N. G., Coates A. R. (2004) FEMS Microbiol. Lett. 237, 415–423 [DOI] [PubMed] [Google Scholar]
- 32.Primm T. P., Andersen S. J., Mizrahi V., Avarbock D., Rubin H., Barry C. E., 3rd. (2000) J. Bacteriol. 182, 4889–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes N. D., Wu Q. L., Kong D., Puyang X., Garg S., Husson R. N. (1999) J. Bacteriol. 181, 4266–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braeken K., Moris M., Daniels R., Vanderleyden J., Michiels J. (2006) Trends Microbiol. 14, 45–54 [DOI] [PubMed] [Google Scholar]
- 35.Wu B., Liu Y., Zhao Q., Liao S., Zhang J., Bartlam M., Chen W., Rao Z. (2007) J. Mol. Biol. 367, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 36.Maki H., Sekiguchi M. (1992) Nature 355, 273–275 [DOI] [PubMed] [Google Scholar]
- 37.Slupphaug G., Kavli B., Krokan H. E. (2003) Mutat Res. 531, 231–251 [DOI] [PubMed] [Google Scholar]
- 38.Dahl J. L., Kraus C. N., Boshoff H. I., Doan B., Foley K., Avarbock D., Kaplan G., Mizrahi V., Rubin H., Barry C. E., 3rd. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10026–10031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stallings C. L., Stephanou N. C., Chu L., Hochschild A., Nickels B. E., Glickman M. S. (2009) Cell 138, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rengarajan J., Bloom B. R., Rubin E. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehrt S., Schnappinger D. (2009) Cell Microbiol. 11, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan J., Xing Y., Magliozzo R. S., Bloom B. R. (1992) J. Exp. Med. 175, 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackaness G. B., Smith N., Wells A. Q. (1954) Am. Rev. Tuberc. 69, 479–494 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.