Abstract
StAR (steroidogenic acute regulatory protein) mediates the transport of cholesterol from the outer to the inner mitochondrial membrane, the process of which is the rate-limiting step for steroidogenesis. Transcriptional regulation of the proximal promoter of the human StAR gene has been well characterized, whereas analysis of its distal control region has not. Recently, we found that SF-1 (steroidogenic factor 1) induced the differentiation of mesenchymal stem cells (MSCs) into steroidogenic cells with the concomitant strong induction of StAR expression. Here, we show, using differentiated MSCs, that StAR expression is regulated by a novel distal control region. Using electrophoretic mobility shift (EMSA) and chromatin immunoprecipitation (ChIP) assays, we identified novel SF-1 binding sites between 3,000 and 3,400 bp upstream of StAR. A luciferase reporter assay revealed that the region worked as a strong regulator to exert maximal transcription of StAR. ChIP analysis of histone H3 revealed that upon SF-1 expression, nucleosome eviction took place at the SF-1 binding sites, not only in the promoter but also in the distal SF-1 binding sites. Chromosome conformation capture analysis revealed that the region upstream of StAR formed a chromatin loop both in the differentiated MSCs and in KGN cells, a human granulosa cell tumor cell line, where SF-1 is endogenously expressed. Finally, SF-1 knockdown resulted in disrupted formation of this chromatin loop in KGN cells. These results indicate that the novel distal control region participate in StAR activation through SF-1 dependent alterations of chromatin structure, including histone eviction and chromatin loop formation.
Keywords: Chromatin Immunoprecipitation (ChiP), Chromatin Structure, Ovary, Transcription Enhancers, Transcription Regulation, SF-1, StAR, Granulosa Cells, Mesenchymal Stem Cells
Introduction
StAR (steroidogenic acute regulatory protein) mediates the transport of cholesterol from the outer to the inner mitochondrial membrane, the process of which is the rate-limiting step for steroidogenesis. In humans, mutations in StAR are associated with lipoid congenital adrenal hyperplasia, which is characterized by impaired adrenal and gonadal steroid synthesis (1). Targeted disruption of Star in the mouse results in a phenocopy of human lipoid congenital adrenal hyperplasia (2, 3).
Transcriptional regulation of StAR expression is controlled by many transcription factors, including steroidogenic factor 1 (SF-1, NR5A1 also known as Ad4BP), C/EBPβ, GATA-4, and cAMP response element modulator (4). SF-1 plays a pivotal role in the regulation of reproductive endocrine functions at multiple levels during adrenal and gonadal development and differentiation, including regulating the expression of steroidogenic genes (5–7). SF-1-null mice show a complete loss of adrenal glands and gonads, emphasizing the critical role of SF-1 as a key regulator for adrenal and gonadal development (8). SF-1 plays a crucial role in regulating StAR expression (9–14). Several SF-1 binding motifs have been shown to be present in the StAR promoters of various species, and SF-1 can efficiently activate StAR transcription, as demonstrated in transfection assays (10–18). The −1.3 kb human StAR promoter has at least three SF-1 binding sites that are functionally important for basal as well as for cAMP-stimulated transcription (13). However, it has been unclear whether regions further upstream and downstream of the gene participate in its transcriptional regulation.
Chromatin structure plays a critical role in eukaryotic gene transcriptional regulation. Transcription factors bind to their target DNA elements and recruit chromatin remodeling or modifying proteins to alter chromatin structure. The dynamics of chromatin structure are tightly regulated through multiple mechanisms, including histone modification, chromatin remodeling, histone variant incorporation, and histone eviction (19). Several studies have shown a relationship between chromatin structure and StAR expression. In the Leydig tumor cell line, MA-10, histone modifications (i.e. H3 acetylation and H3K9 methylation) occurred at the proximal StAR promoter and in the coding region, but not distal to the StAR promoter (20). StAR mRNA accumulation induced by cyclic AMP (or tropic hormone) stimulation was associated with increased acetylation of histone H3 within the proximal StAR promoter in human, monkey, cow, and pig (21–23) However, there is no report regarding histone eviction or changes of chromatin architecture upon induction of StAR expression.
Adult stem cells from bone marrow, referred to as mesenchymal stem cells or marrow stromal cells (MSCs),2 are defined as multipotent cells that have the ability to differentiate into multiple mesodermal cell types (24). Recently, we found that introduction of SF-1 into MSCs resulted in the differentiation of the cells into a steroidogenic cell lineage in which StAR was markedly induced.
In this study, in the differentiated MSCs, as well as in KGN cells, a human granulosa cell tumor cell line, we used EMSA and ChIP assays to identify novel SF-1 binding sites between 3,000 and 3,400 bp upstream from the transcription start site (TSS) of human StAR. We characterized the novel distal SF-1 binding sites and analyzed changes of chromatin structure, including histone eviction and chromatin loop formation within the region upstream of StAR that is associated with the activation of transcription.
EXPERIMENTAL PROCEDURES
Reagents
8-Br-cAMP was purchased from Sigma. TnT-coupled reticulocyte lysate systems, Dual-Luciferase reporter assay system, the pGL4.11-basic, pGL4.24 containing minimal promoter, pRL-TK and pGL4.74 (hRluc/TK) vectors were purchased from Promega. [γ-32P]ATP (111 TBq/mmol) was purchased from PerkinElmer Life Science. TRIzol reagent, SuperScript III reverse transcriptase, Lipofectamine RNAiMAX, Lipofectamine and Lipofectamine Plus regents were purchased from Invitrogen. The Adeno-X expression system 1, Adeno-X rapid titer kits, Ex Taq Hot start version, and Advantage2 polymerase mix were purchased from Takara Bio (Otsu, Japan). Power SYBR Green PCR master mix was purchased from Applied Biosystems (Foster City, CA). Control siRNA-A (catalog no. sc-37007) and SF-1 siRNA (catalog no. sc-37901) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). pCMV-Tag-2 and -3 vectors and the QuikChange site-directed mutagenesis kit were purchased from Agilent Technologies (Santa Clara, CA). KOD-Plus, KOD-Fx, and pTA2 vectors were purchased from Toyobo (Osaka, Japan). A bacterial artificial chromosome clone containing the human StAR gene in vector RP11-90P-5 was purchased from Advanced GenoTechs (Tsukuba, Japan). All other reagents were obtained from commonly used suppliers.
Antibodies
Dynabeads protein G and M-280 sheep anti-mouse IgG were purchased from Invitrogen. Anti-FLAG M2 (F1804) antibody was purchased from Sigma. Anti-Myc tag (antibody 9132), anti-RNA polymerase II carboxy-terminal domain repeat YSPTSPS (8WG16; antibody 817), and anti-histone H3 (antibody 1791) antibodies were purchased from Abcam (Cambridge, MA).
Cell Culture, Transient Transfection, and Luciferase Assay
human MSC (hMSC) lines, hMSC-hTERT-E6/E7 cells (kindly provided by Dr. Junya Toguchida, Kyoto University, Kyoto, Japan (25)), UE7T-13 cells (26), HEK293, and Phoenix cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and gentamycin. KGN cells (kindly provided by Dr. Toshihiko Yanase, Fukuoka University, Fukuoka, Japan (27)) were maintained in DMEM/F-12 supplemented with 10% FBS and gentamycin. HEK293 and KGN cells were transfected using Lipofectamine and Lipofectamine Plus reagents according to the manufacturer's instructions (Invitrogen). Luciferase assays were performed as described previously (28). Each data point represents the mean of at least four independent experiments.
For siRNA experiments, an SF-1-targeting siRNA or a control scrambled siRNA was transfected into KGN cells that had been preseeded in a 60-mm dish with Lipofectamine RNAiMAX according to the manufacturer's instructions. The final siRNA concentration in the medium was 20 nm. Four days after transfection, RT-PCR or chromosome conformation capture (3C) assays were performed. For the combined luciferase assay, the reporter vectors and the siRNA were co-transfected using Lipofectamine Plus reagent, and luciferase assays were performed 48 h after transfection.
Plasmids
The pCMV/FLAG- or pCMV/Myc-tagged SF-1 expression vectors (pCMV/FLAGSF-1 or pCMV/MycSF-1) and the retroviral plasmid for expressing FLAG-tagged SF-1 (pQCXIP/FLAGSF-1) vector have been described previously (29).
Human StAR upstream regions consisting of various 5′ ends cloned into the pGL4.11 luciferase basic vector were generated as follows. The 5′-flanking regions (−5,979/−95 and −2,459/+45) were amplified by PCR using KOD-Fx and genomic DNA from KGN cells as a template, and PCR products were ligated into pTA2 and pBlueScript. KpnI/HindIII (−2,292/−1,278) and HindIII (−1283/+33) fragments from pBS/hStAR(−2,459/+45) were ligated into pGL4.11, which was designated as pGL4/hStAR(−2292/+33). Subsequently a KpnI fragment (−5,979/−2287) from pTA2/hStAR(−5,979/−90) was ligated into KpnI-digested pGL4/hStAR(−2,292/+33), which was designated as pGL4/hStAR(−5,979/+33). HindIII (−1,283/+33) and BamH I (−4,925/+33) fragments from pGL4/hStAR(−5979/+33) were ligated into pGL4.11, which were designated as pGL4/hStAR(−1,283/+33) and pGL4/hStAR(−4,925/+33), respectively. Other deletion constructs were generated by PCR with KOD-Plus using pGL4/hStAR(−5,979/+33) as a template. Transcriptional activities were measured using constructs in which each SF-1 binding region, i.e. −15 kb, −5.5 ∼ −5.9 kb, and −3.0 ∼ −3.5 kb fragments upstream of StAR, was inserted into pGL4.24, a reporter vector with minimal promoter activity.
The −15 kb region was amplified by PCR with KOD-Fx using genomic DNA from KGN cells as a template. The other regions (−5,966/−5,521 and −3,494/−3,009) were amplified by PCR with KOD-Plus using pGL4/hStAR(−5,979/+33) as a template. Nucleotide numbering is relative to the TSS of human StAR. Each PCR product was ligated into pTA2 and then digested with SpeI and HindIII. Each SpeI/HindIII fragment was inserted into NheI/HindIII digested pGL4.24, Mutations of SF-1 binding sites in the StAR promoter (−3,402/+33) were created by PCR using either primers with nucleotide substitutions or by using the QuikChange site-directed mutagenesis kit. Primers used for PCR are shown in supplemental Table 1. The nucleotide sequences of all PCR products were confirmed by DNA sequencing.
Preparation and Infection of Adenovirus and Retrovirus
For adenovirus preparation, the pCMV/MycSF-1 vector and pCMV/MycLRH-1 vector (29) were subcloned into the pShuttle vector to generate pShuttle/MycSF-1 and pShuttle/MycLRH-1, respectively. These shuttle plasmids were used to generate a recombinant adenoviral vector.
The production of adenovirus expressing MycSF-1 (Adx-MycSF-1) and MycLRH-1 (Adx-MycLRH-1) were performed according to the manufacturer's recommended protocol (Takara Bio). The titration of the adenovirus was carried out using Adeno-X rapid titer kits.
One day after plating, cells were infected with adenovirus expressing LacZ (Adx-LacZ), MycSF-1 (Adx-MycSF-1) or MycLRH-1 (Adx-MycLRH-1) at a multiplicity of infection of 40 and incubated for 1 to 4 days. Retrovirus preparation and infection was performed as described previously (29).
In Vitro Translation and EMSAs
EMSAs using in vitro-synthesized proteins were performed as described previously (30). Briefly, 1 μg of the pCMV-FLAGSF-1 plasmid was incubated at 30 °C for 60 min with the TnT-coupled reticulocyte lysate system using T3 RNA polymerase. Oligonucleotides used for EMSA studies are listed in supplemental Table 2. One microliter of in vitro-translated product was added to the binding mixture with a γ-32P-labeled probe (10 fmol). For competition analysis, a 200-fold molar excess of competitor DNA was added to the binding mixture. After completion of binding, the mixture was subjected to 6% PAGE, and the gel was then dried and autoradiographed.
RT-PCR and Real-time RT-PCR
Total RNA from cultured cells was extracted using TRIzol reagent. RT-PCR (31) and real-time RT-PCR (24) was performed as described previously. The primers used for PCR have been described previously (24). Primers used for real-time PCR are shown in supplemental Table 3.
ChIP Assay
For ChIP assays, cells (1–2 × 106) were cross-linked with 1% formaldehyde in PBS for 10 min at room temperature, and then glycine was added to a final concentration of 0.125 m to quench reactive aldehydes. After washing cells with cold PBS, cells were harvested in nuclear extract buffer (50 mm Tris-HCl (pH 8.0), 2 mm EDTA, 0.1% Nonidet P-40, 10 mm NaCl, 1.5 mm MgCl2, 10% glycerol, and 1 mm PMSF). Nuclear pellets were collected by centrifugation (1,500 × g for 5 min) and resuspended in SDS lysis buffer (50 mm Tris-HCl (pH 8.0), 10 mm EDTA, and 1% SDS). Samples were sonicated using a Biorupter (Cosmo Bio Co., Tokyo, Japan). After centrifugation (20,000 × g for 5 min) to remove debris, the supernatants were diluted 10-fold in dilution buffer (16.7 mm Tris (pH 8.0), 1.2 mm EDTA, 167 mm NaCl, 1.1% Triton X-100, 0.01% SDS, and protease inhibitor mixture). An aliquot of the diluted supernatants was used as input, and the remaining supernatants were subjected to ChIP.
Protein G or M-280 sheep anti-mouse IgG-coated paramagnetic beads were incubated with antibodies of interest in 0.1% BSA-PBS for 6 h at 4 °C with rotation and washed with low salt washing buffer (20 mm Tris (pH 8.0), 2 mm EDTA, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, and 1 mm PMSF). The ChIP samples (1–2 mg protein) were incubated with the antibody-bound Dynabeads overnight at 4 °C with rotation. Beads were washed sequentially with low salt washing buffer, high salt washing buffer (20 mm Tris (pH 8.0), 2 mm EDTA, 500 mm NaCl, 1% Triton X-100, 0.1% SDS, and 1 mm PMSF), three times with LiCl wash buffer (10 mm Tris (pH 8.0), 1 mm EDTA, 250 mm LiCl, 1% Nonidet P-40, 1% deoxycholate, and 1 mm PMSF) and with TE buffer (10 mm Tris (pH 8.0) and 1 mm EDTA). After removing TE, beads were mixed with elution buffer (1% SDS, 100 mm NaHCO3, 200 mm NaCl, and RNase A) and incubated overnight at 60 °C to reverse cross-linking. Samples were then treated with proteinase K. DNA was recovered after phenol/chloroform treatment and precipitated with ethanol, using glycogen as a carrier.
Each sample was analyzed by real-time PCR (Power SYBR Green PCR master mix) using an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Results are presented as percentages of input DNA. Each data point represents the mean of at least three independent experiments. Primer sequences are shown in supplemental Table 3.
3C Assays
Cells were treated as described for ChIP assays, up to the precipitation of nuclei. The nuclei were harvested and suspended in the restriction enzyme buffer (33 mm Tris acetate (pH 7.9), 10 mm magnesium acetate, 0.5 mm DTT, and 66 mm potassium acetate) containing 0.1% SDS and incubated at 37 °C for 1 h with shaking. Triton X-100 was added to 2% to sequester SDS, and samples were incubated at 37 °C for another 1 h with shaking. Samples were then digested with MspI overnight at 37 °C with shaking. SDS was added to 1.6%, and the samples were heated at 65 °C for 20 min to inactivate the restriction enzyme. Samples were then diluted with T4 DNA ligase buffer to ∼2.5 μg of DNA/ml. Triton X-100 was added to 1%, and samples were incubated at 37 °C for 1 h with shaking. T4 DNA ligase was added, and samples were incubated at 16 °C for 2 h. Thereafter, reverse cross-linking, protein digestion, and DNA purification were performed as described for ChIP assays. A portion (one-thirtieth) of each DNA sample was used in the PCR reaction. Primer sequences are shown in supplemental Table 4. For amplification from the ligation product, the reaction conditions were 38 cycles (94 °C for 20 s and 68 °C for 1 min) with Advantage 2. For amplification from genomic DNA, the reaction conditions were 27 cycles (94 °C for 20 s, 58 °C for 30 s, and 72 °C for 30 s) with Advantage 2. The products were subjected to electrophoresis in a 1.5% agarose gel, and the resulting bands were visualized by staining with ethidium bromide. At least three independent experiments were performed. As a template for the positive control, a bacterial artificial chromosome (BAC) clone (RP11-90P-5) was used. The MspI-digested DNA fragments of the bacterial artificial chromosome clone were ligated with T4 DNA ligase at a DNA concentration of 300 ng/μl. After purification of DNA, the samples were subjected to the PCR reaction.
Statistical Analysis
Values are given as means ± S.E. Data were analyzed by Student's t test. Statistical significance was accepted as p < 0.05.
RESULTS
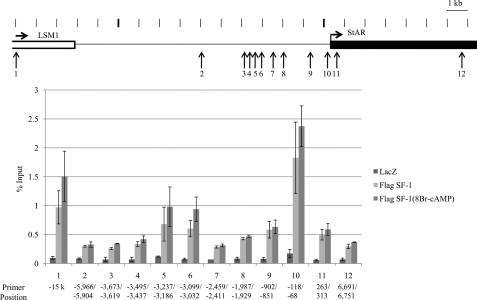
We reported previously that MSCs could be differentiated into steroidogenic cell-lineages, such as Leydig cells and adrenocortical cells, by the expression of SF-1 and treatment with 8-Br-cAMP (24). Such treatments induced expression of steroidogenesis-related genes, including StAR. In this study, we examined the recruitment of SF-1 over a wide region surrounding the human StAR gene. ChIP-on-Chip analysis revealed three SF-1 binding sites near the StAR locus (Chr 8p11) in human MSCs (hMSC-hTERT-E6/E7) stably transfected with FLAG-tagged SF-1 (SF-1-hMSC) (supplemental Fig. 1). The three SF-1 binding sites detected by ChIP-on-Chip along with an additional nine possible sites (Fig. 1) were further analyzed by the conventional ChIP assay using real-time PCR (ChIP-qPCR). Among the 12 sites examined, the sites, 1 (−15 kb), 5 (−3.5 kb), 6 (−3 kb), and 10 (TSS) were positive for SF-1 binding. Treatment of 8-Br-cAMP increased the binding of SF-1 to each of these binding sites. These results suggest that novel distal control regions may exist within the 15 kb region upstream of the TSS.
FIGURE 1.
ChIP analysis of the upstream region of the human StAR gene. The upper panel depicts the StAR gene locus. (Arrows indicate positions where quantitative PCR was performed.) The numbers correspond to the amplification products in the lower panel. DNA preparations from MSCs expressing LacZ (control), or FLAG-tagged SF-1 under unstimulated (FLAG SF-1) and stimulated (FLAG SF-1(8-Br-cAMP)) conditions were used as template. SF-1 strongly bound to the proximal promoter (bar 10) as well as to the −3.0 kb (bar 6), −3.5 kb (bar 5), and −15 kb (bar 1) regions under both stimulated and unstimulated conditions. Primer positions using real-time PCR were indicated at the bottom. The numbers refer to the transcription start site (+1).
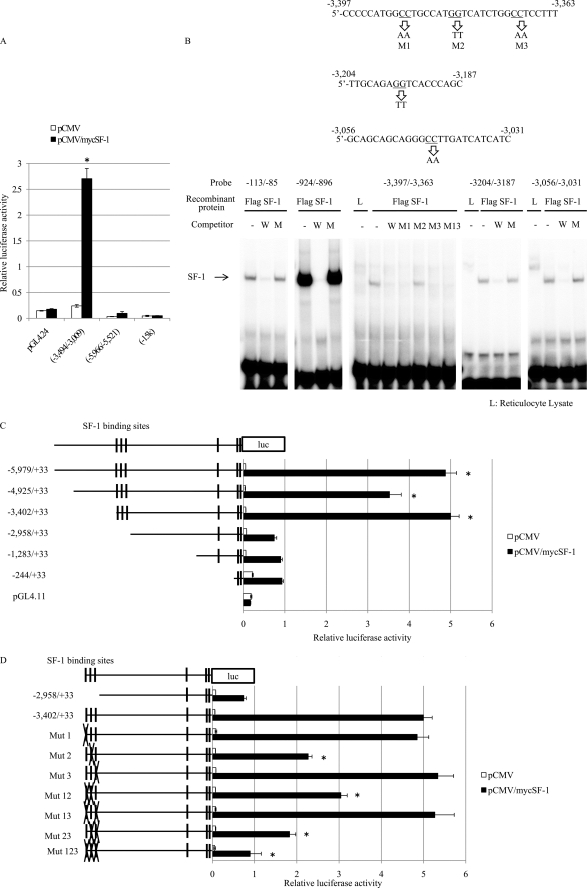
To determine whether these SF-1 binding sites function as transcriptional activator, we examined luciferase reporter activities using various constructs of the StAR upstream region. HEK293 cells were transiently co-transfected with luciferase reporter plasmids and an SF-1 expression vector, and the reporter activities were determined. As shown in Fig. 2A and supplemental Fig. 2, co-transfection of SF-1 strongly enhanced the luciferase activity of a reporter construct containing the fragment −3,494 to −3,009 bp, but not that of reporters containing the fragments −5,966 to −5,521 bp or −15 kb, either with or without 8-Br-cAMP treatment. Because the −3,494 to −3,009 bp region contains several consensus SF-1 binding sequences, we examined whether SF-1 binds to each putative binding site by EMSA using in vitro-translated SF-1 (Fig. 2B). The translated SF-1 bound to radiolabeled probes that contained sequences of the three novel SF-1 binding sites (−3,383/−3,375, −3,200/−3,193 and −3,046/−3,039). Probes containing −113/−86 (SF-1 site in the promoter) and the −924/−896 SF-1 sites were used as positive controls. SF-1 binding was prevented by the addition of an excess of the unlabeled probe but not by the addition of mutated probes. Consistent with previous reports, the band intensity of the −924/−896 site was much stronger than that of the −113/−86 site as well as those of other sites. The band intensity of each novel SF-1 site was similar to that of the −113/−86 site. These results suggest that each novel SF-1 site binds SF-1 with a similar affinity to the promoter SF-1 binding site (−113/−86). The complex was not detected when each probe was incubated with reticulocyte lysate without SF-1. There are seven more sites with possible SF-1 binding sequences (i.e. AGGCCA, AGGGCA, and AGGTCT) within the −3,494/−3,009 bp region. However, no SF-1 binding was detected when using radiolabeled probes for each of these sequences (data not shown). These results indicate that SF-1 binds to three sites within the −3,494/−3,009 bp region, at −3,383/−3,375, −3,200/−3,193 and −3,046/−3,039.
FIGURE 2.
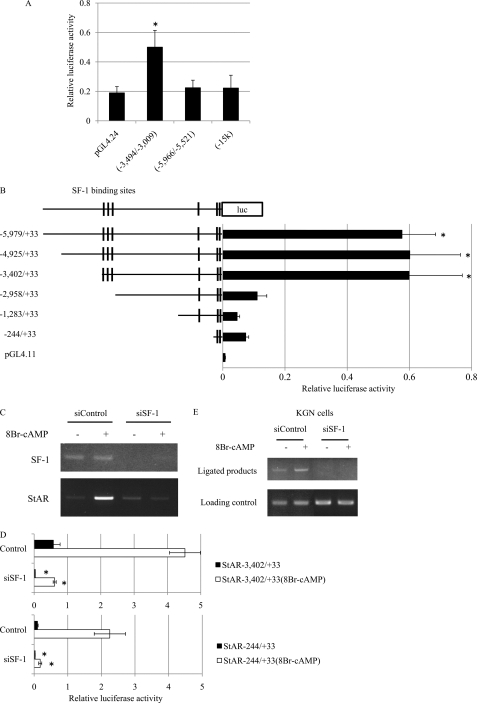
Transcriptional activity of the novel SF-1 binding sites. A, transcriptional activities of DNA fragments from the region upstream of the StAR gene. Each DNA fragment was cloned into a luciferase vector, and the reporter constructs, along with an empty vector pGL4.24, were co-transfected with control vector (pCMV) or an SF-1 expression vector (pCMV/MycSF-1) into 293 cells. Luciferase activities were measured 48 h after transfection. Only the −3,494/−3,009 DNA fragment showed strong transcriptional activity after expression of SF-1. Note that the −15 kb fragment showed no transcriptional activity, despite its ability to bind SF-1. *, significantly different from pGL4.24 at p < 0.05. B, EMSA analysis of SF-1 binding to the StAR upstream region. The upper panel shows probe sequences and the sites where mutations were introduced. The lower panel shows the results of EMSA. Probes for the proximal promoter (−113/−85), distal SF-1 binding site (−924/−896) and SF-1 sites (−3,397/3,363, −3,204/−3,187, and −3,056/−3,031) in the novel distal control region were used. FLAG-tagged SF-1 was prepared by in vitro translation. Unlabeled wild-type probes (W) and mutated probes (M1, M2, M3, M13, and M; see upper panel) were used as competitors. SF-1 bound strongly to the distal SF-1 binding site (−924/−896), whereas it bound weakly to the proximal promoter (−113/−85) and weakly to the three SF-1 sites in the distal control region. L, reticulocyte lysate. C, luciferase (luc) activities of StAR upstream region deletion constructs. Reporter constructs containing DNA fragments of different length from the region upstream of StAR were prepared and then co-transfected with control vector (pCMV) or a Myc-tagged SF-1 expression vector (pCMV/MycSF-1) into HEK293 cells. Luciferase activities were measured 48 h after transfection. A marked decrease of reporter activity was observed when the three SF-1 sites within the distal control region were deleted. *, significantly different from −2,958/+33 at p < 0.05. D, effects of mutations in the SF-1 sites within the distal control region on luciferase activity. Single, double, or triple mutations were introduced in the SF-1 sites present in the distal control region, and reporter activities were measured by a luciferase assay. Among the three SF-1 sites, mutation in the second SF-1 site (Mut2) was the most effective in reducing luciferase activity. Introduction of triple mutations caused a complete loss of the transcriptional activity. *, significantly different from −3,402/+33 at p < 0.05.
To examine transcriptional activity of the distal SF-1 binding sites, sequential 5′-deletion constructs (−5,979 to −235 bp) from the StAR upstream region were prepared. As shown in Fig. 2C, co-transfection of SF-1 increased the luciferase activities of all reporters. The −3,402/+33 construct showed the highest luciferase activity in HEK293 cells co-transfected with SF-1. Fig. 2C also shows that luciferase activity was dramatically reduced by deletion of the −3,402/−2,958 bp region. Treatment with 8-Br-cAMP augmented luciferase activities of all reporters (supplemental Fig. 3). The 8-Br-cAMP-induced activities also were dramatically reduced by deletion to −2,958 bp. These results strongly suggest that the region from −3,402 to −2,958 bp, which includes the three novel SF-1 binding sites, is essential for maximal transcriptional activity in both basal and cAMP-stimulated cells.
To assess which SF-1 binding sites are important for the reporter activity, constructs harboring mutations in each or in combinations of SF-1 binding sites were prepared. As shown in Fig. 2D and supplemental Fig. 4, a mutant construct designated Mut2, which has mutations in the −3,200/−3,193 site, showed marked reduction of luciferase activity in both basal and 8-Br-cAMP-stimulated cells. Mutations in other sites and in combinations of sites also showed reductions of reporter activities. These results suggest that the SF-1 binding site at −3,200/−3,193 bp may be the most important site for the transcriptional activation of StAR and that the other two sites also may involved in the activation of the gene.
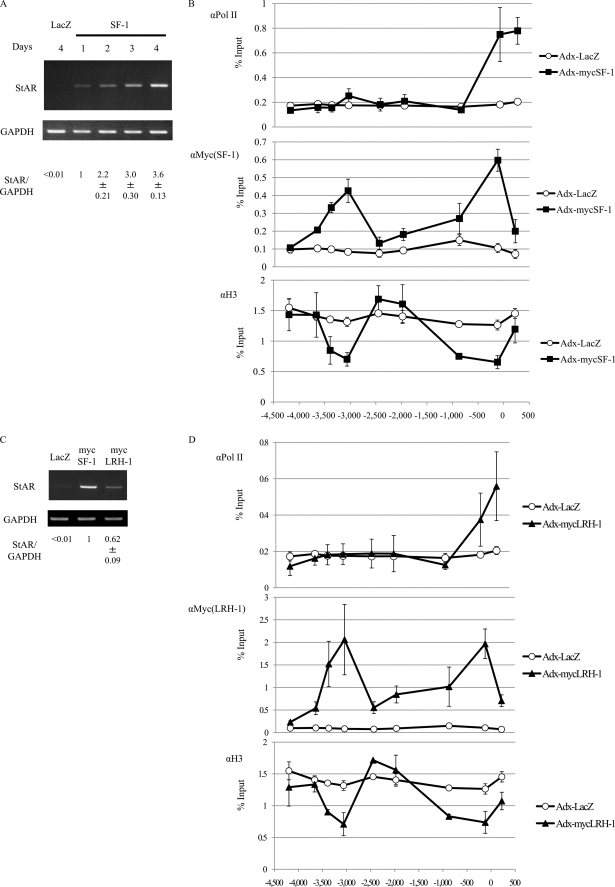
As shown in Fig. 3A, infection of UE7T-13 cells (a human MSC cell line) with adenovirus containing Myc-tagged SF-1 (Adx-MycSF-1) caused induction of StAR expression. The induction of StAR expression was apparent from 1 day after infection and continued to increase for up to 4 days. At the same time, other genes related to steroidogenesis also were induced in the MSCs by the infection of Adx-MycSF-1 (supplemental Fig. 5).
FIGURE 3.
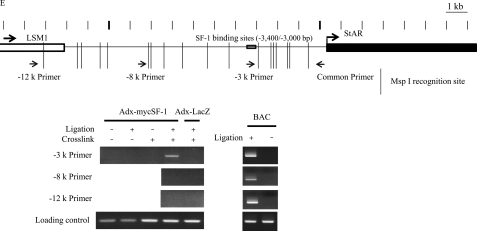
SF-1-dependent histone eviction and chromatin loop formation at the StAR locus in differentiated MSCs expressing SF-1. A, induction of StAR mRNA in MSCs by adenovirus-mediated expression of SF-1. Human MSCs were infected with adenoviruses expressing LacZ or SF-1. StAR expression was examined by RT-PCR and real-time RT-PCR at the indicated time points after infection. Results of real-time RT-PCR (mean ± S.E.) were shown as a StAR/GAPDH ratio. StAR mRNA levels were normalized against GAPDH. A ratio at day 1 after SF-1 transfection was arbitrarily defined as 1. Pol, polymerase. B, ChIP analysis of the region upstream of StAR. Human MSCs were infected with adenoviruses expressing LacZ (Adx-LacZ) or Myc-tagged SF-1 (Adx-MycSF-1). DNA templates for ChIP were extracted from cells 48 h after infection. Antibodies for RNA polymerase II (αRNAPII), Myc (αMyc(SF-1)) and histone H3 (αH3) were used for the ChIP assay. Histone eviction was observed both at the promoter and at the distal control regions. C, induction of StAR mRNA in MSCs by adenovirus-mediated expression of SF-1 or LRH-1. Human MSCs were infected with adenoviruses expressing LacZ, SF-1, or LRH-1. StAR expression was examined by RT-PCR and real-time RT-PCR at 2 days after infection. The numbers at the bottom show the results of real-time RT-PCR (mean ± S.E.). StAR mRNA levels were normalized against GAPDH. A ratio at day 1 after SF-1 transfection was arbitrarily defined as 1. D, ChIP analysis of the region upstream of StAR. Human MSCs (UE7T-13 cells) were infected with adenoviruses expressing LacZ (Adx-LacZ) or Myc-tagged LRH-1 (Adx-MycLRH-1). DNA templates for ChIP were extracted from cells 48 h after infection. Antibodies for RNA polymerase II, Myc (αMyc(SF-1/LRH-1)) and histone H3 (αH3) were used for the ChIP assay. Histone eviction was observed both at the promoter and distal control regions by the expression of LRH-1, as in the case for SF-1 expression. E, 3C analysis of the StAR promoter and distal control region. The upper panel shows a schematic of StAR with the position and orientation of the 3C primer pairs indicated. The common primer at the TSS and the −3 k primer were designed to amplify a novel ligation product formed between the restriction fragments from the promoter and the distal control (−3,000/−3,400) regions. The −8 k primer and the −12 k primer were designed to detect loop formation between the promoter and the regions 8 kb and 12 kb upstream of StAR, respectively. The lower panel shows the results of 3C analysis and the presence of a specific (common primer/-3 k primer) 3C product, only under conditions in which formaldehyde cross-linking and ligation of the samples was performed. No product was observed with the primer pairs: common primer/−8 k primer, common primer/−12 k primer. No 3C product was observed from SF-1-expressing cells without ligation or cross-linking of DNA samples. No 3C product was observed in DNA preparations from LacZ-expressing control cells. The loading control showed that all of the samples contained similar amounts of DNA. Ligation products from a StAR bacterial artificial chromosome were used as positive controls for each amplification.
Next, various ChIP-qPCRs were performed using UE7T-13 cells, 2 days after Adx-SF-1 infection, to examine changes in chromatin structure during the initial induction of StAR expression by SF-1. One of the important mechanisms for altering chromatin structure involves covalent modifications of the histone tails. The “histone code” hypothesis (32) predicts that different modifications at specific amino acid residues in histones or combinations of these modifications are translated into functionally distinct nuclear processes. For example, histone acetylation, which is a positive mark of transcription, neutralizes the charge on the basic histone proteins leading to relaxation of the protein/DNA interactions. H3K9 and H3K27 methylations have been linked to transcriptional repression because of their recognition by heterochromatin protein 1 and polycomb proteins, respectively. In contrast, H3K4 methylation recruits chromatin-remodeling enzymes that lead to a relaxed chromatin structure permissive for transcription (33). Therefore, analysis of these modifications is quite important for the understanding of mechanisms of StAR gene transcription. Recruitment of RNA polymerase II (RNAPII), SF-1, and unmodified histone H3 (Fig. 3B), along with acetylated histone H3 (H3Ac) and methylated H3K4, H3K27, and H3K9 (supplemental Fig. 6), were surveyed within the region from −5 kb to the TSS of StAR by ChIP-qPCR. As shown in Fig. 3B, RNAPII and SF-1 were recruited to the proximal StAR promoter site near the TSS, as expected. It is interesting that marked reduction of histone H3 binding to the promoter region was observed after the introduction of SF-1. Similar results were obtained when using anti-histone H2B antibody (data not shown). In agreement with this, reduction of binding was observed for most of the modified histones in SF-1 expressing cells (supplemental Fig. 6). These observations suggest that nucleosome eviction might occur at the StAR proximal promoter site in association with SF-1 binding to the site.
With respect to the distal control region between −3,500 and 3,000 bp from the TSS, nucleosome eviction (reduction of H3 binding) also was observed in association with SF-1 binding to this region (Fig. 3B). Similar results were obtained using anti-histone H2B antibody (data not shown). These results suggest that histone eviction occurred at both the −3 kb and the proximal promoter regions in the presence of SF-1.
In the case of LRH-1 (liver receptor homologue-1), which belong to same NR5A family as SF-1, infection of UE7T-13 cells with adenovirus containing Myc-tagged LRH-1 (Adx-MycLRH- 1) also caused induction of StAR expression (Fig. 3C). At the same time, LRH-1 bound to the same distal control region and promoter of StAR and induced recruitment of RNAP II to the promoter site and histone eviction at these sites (Fig. 3D). These results suggest that LRH-1 and SF-1 have similar functions in the induction of StAR transcription. Therefore, we hypothesize that both regions might be occupied by the same SF-1 molecules to form a DNA loop with a protein-DNA complex, which may promote histone eviction at both sites.
To demonstrate close proximity of the two sites on the chromosomal architecture, 3C assays were performed. As shown in Fig. 3E, a ligation product was detected in the SF-1 expressing cells (Adx-MycSF-1) only when the cells were cross-linked and ligated, whereas no ligation product was detected in control cells (Adx-LacZ). No PCR product was observed between the proximal promoter site and the −10 to −8 kb site or the −16 to −12 kb site. Taken together, we propose that SF-1 simultaneously binds to the proximal promoter site and the −3 kb distal control region to form a DNA loop and a protein-DNA complex, which then promotes histone eviction from these sites as well as transcription of StAR.
To confirm the importance of the −3 kb distal control region in cells expressing endogenous StAR and SF-1, we examined KGN cells, a human cell line derived from a granulosa cell tumor. Fig. 4A and supplemental Fig. 7 show that the −3,494/−3,009 bp region, but not the −5,966/−5,521 bp and −15 kb regions, showed a strong transcriptional activity in KGN cells with or without 8-Br-cAMP treatment. Consistent with these results, reporter activities of various deletion constructs showed the importance of the distal control region (−3,402/−2,958 bp) for StAR expression in KGN cells (Fig. 4B). The 8-Br-cAMP treatment also augmented the reporter activities of all constructs from 10- to 20-fold compared with basal activities (supplemental Fig. 8). The −3,402/−2,958 bp region also was required for maximal activity in cAMP-stimulated KGN cells. Luciferase activity of the Mut2 construct was reduced markedly in KGN cells under both stimulated and unstimulated conditions, suggesting the importance of the SF-1 site at −3,200/−3,193 bp (supplemental Fig. 9).
FIGURE 4.
SF-1 dependent transcriptional activation and chromatin loop formation through the novel distal control region in KGN cells. A, transcriptional activities of DNA fragments from the upstream region of StAR in KGN cells, which express endogenous SF-1. Each DNA fragment was cloned into a luciferase vector, and the reporter constructs, along with an empty vector pGL4.24, were transfected into KGN cells. Luciferase (luc) activities were measured 48 h after transfection. Only the DNA fragment from −3,494/−3,009 showed strong transcriptional activity in KGN cells. Note that DNA from −15 kb showed no transcriptional activity despite its ability to bind SF-1. *, significantly different from pGL4.24 at p < 0.05. B, luciferase activities of deletion constructs of the StAR upstream region in KGN cells. Reporter constructs containing DNA fragments of different length from the StAR upstream region were prepared and then transfected into KGN cells. Luciferase activities were measured 48 h after transfection. A marked decrease of reporter activity was observed when the three SF-1 sites within the distal control region were deleted. *, significantly different from −2,958/+33 at p < 0.05. C, effects of siRNA on the levels of endogenous SF-1 and StAR mRNA. Synthetic siRNA for SF-1 (siSF-1) or control (siControl) was introduced into 8-Br-cAMP-stimulated or un-stimulated KGN cells. Expression of SF-1 was almost totally suppressed by siSF-1. StAR expression also was suppressed, even in the presence of 8-Br-cAMP. D, effects of siRNA on the distal control region and promoter activities of StAR. The reporter constructs StAR(−3,402/+33) (involves both the distal control region and the promoter) and StAR(−244/+33) (involves only the promoter site) were used to examine effects of siRNA. Introduction of siSF-1 strongly suppressed the luciferase activities of both reporters. Strong suppression was also observed under 8-Br-cAMP-stimulated conditions. *, significantly different from the negative control at p < 0.05. E, effects of siRNA on chromatin loop formation. Chromatin loop formation between the distal control region and the promoter of the StAR gene was disrupted by the introduction of siSF-1 into KGN cells. No product was observed by 3C analysis when the cells were treated with siSF-1, whereas the amplified product was detected when cells were treated with control siRNA.
To determine whether the loop formation is dependent on SF-1, we used siRNA to knockdown endogenous SF-1 expression in KGN cells. This resulted in a significant decrease of StAR expression under both cAMP-stimulated and unstimulated conditions (Fig. 4C). This also was confirmed by the reporter assay where co-transfection of siRNA with luciferase reporter constructs (−244/+33 and −3,402/+33) resulted in a dramatic decrease of reporter activities compared with the control (Fig. 4D). As shown in Fig. 4E, depletion of SF-1 resulted in the abolishment of the 3C assay PCR product, indicating that SF-1 is essential for the loop formation between the proximal promoter and the −3,400/−3,000 bp distal control region. These results strongly suggest that endogenously expressed SF-1 plays a critical role in the transcriptional activation of StAR through the formation of a DNA loop between its distal control region (−3,494/−3,009 bp) and the proximal promoter by altering chromatin structure.
To examine whether the SF-1 binding sites in the region upstream of StAR are conserved evolutionarily, we examined and compared the sequences of the distal control region to different species whose genomes have been sequenced. As shown in Fig. 5A, there are many putative SF-1 binding sites in the region upstream of StAR in human, rat, and mouse, except cow. Surprisingly, the entire distal control regions among human, rat, and mouse were highly conserved (Fig. 5B), suggesting that the region also may be important for achieving maximum transcription of StAR in other mammals.
FIGURE 5.
The distal control region is evolutionarily conserved. A, schematic representation of the region upstream of StAR from human, rat, mouse, and cow. Putative SF-1 binding sites are shown. Asterisks indicate position of the functional SF-1 binding sites for transcriptional activation. Evolutionarily conserved regions are boxed. B, comparison of sequences of the distal control region from human, rat, and mouse. The functional SF-1 binding sites for transcriptional activation are boxed.
DISCUSSION
Accumulating evidence indicates that chromatin remodeling is linked tightly to differentiation, and it is difficult to understand such chromatin remodeling during differentiation by analyzing only the proximal promoter regions of genes. We performed genome-wide analysis of SF-1 binding sites in human mesenchymal stem cells during SF-1-induced differentiation by a ChIP-on-Chip analysis and then focused to the region around the StAR gene, which functions as a rate-limiting protein for steroidogenesis. We identified novel SF-1 binding sites in the region upstream of the human StAR gene. However, it was not evident whether these binding sites are simply platforms or docking stations for SF-1 binding or functional sites that have a role in SF-1 mediated transcription. We demonstrated that the −3 kb upstream SF-1 binding sites are bona fide transcriptional activators that are critical for achieving maximum transcriptional activation of StAR. On StAR activation, we found that nucleosomes were evicted from the SF-1 binding sites in both the proximal promoter and the −3 kb region during SF-1-induced differentiation of hMSCs. Previous reports describe that many promoters of actively transcribed genes are depleted of nucleosomes (34, 35) or are associated with nucleosome eviction (36). In vitro studies suggest that the binding of transcription factors to a nucleosome-DNA complex can induce nucleosome eviction (37–39). This model was supported by in vivo studies demonstrating that the binding of Pho4 to the PHO5 promoter in yeast (40, 41), the binding of progesterone receptor to the mouse mammary tumor virus promoter (42), as well as the recruitment of nuclear factor of activated T-cells and c-Rel to the proximal promoter of the interleukin-2 gene (43), resulted in nucleosome eviction. Reduced nucleosome occupancy also has been observed at distal enhancers controlled by specific transcription factors (44, 45). Recently, a genome-wide native ChIP study revealed that histone variants H2A.Z and H3.3 are preferentially incorporated into active promoter, enhancer, and insulator regions (46). H2A.Z and H3.3 double variants also have been shown to form less stable nucleosomes. These unstable nucleosomes may lead to histone eviction, thereby facilitating the access of transcription factors to promoters and other regulatory sites in vivo (46).
Nucleosome eviction also could be regulated by histone modification or modifier such as histone acetylation or acetyltransferase, respectively. Reinke and Hörz (47) showed that induction of PHO5 in the absence of remodeling resulted in localized increases in acetylation, demonstrating that histones at the PHO5 promoter are indeed modified by the HAT activity of SAGA in a targeted fashion. A lack of hyperacetylation in the absence of Gcn5 therefore might be the reason for the observed delay in remodeling (47). It was not clear whether the histone at the StAR promoter was modified or not by introduction of SF-1 into MSCs, because the amount of histone was reduced by nucleosome eviction (supplemental Fig. 6). However, histone modification or histone modifier itself might be participating in the regulation of nucleosome eviction at SF-1 binding site of the StAR upstream region.
Recruitment of SF-1 to StAR was observed within 24 h after adenovirus-mediated introduction of SF-1 into MSCs (data not shown). At the same time, levels of nucleosomes bound to the StAR promoter and distal control region began to decrease. Therefore, it is likely that recruitment of SF-1 to these sites may induce recruitment of cofactors as well as histone chaperones, which in turn may lead to histone replacement and/or histone eviction. However, it should be noted that histone eviction is not always observed at SF-1 binding sites; rather, it may occur at SF-1 binding sites of very actively transcribed genes. Indeed, in other SF-1 target genes, histone eviction was not observed when the introduction of SF-1 resulted in weak induction of gene transcription (data not shown).
SF-1 transcripts first appear on embryonic day 9 in the murine urogenital ridge, the probable source of steroidogenic cells of both adrenals and gonads (48). Consistent with its proposed role in regulating StAR, SF-1 is expressed before StAR, which its transcripts first appear at embryonic day 10.5 (49). SF-1 is expressed highly in the adrenal and gonads (50) where StAR gene is exclusively expressed. These observations also support the concept that SF-1 is a major transcriptional regulator for StAR expression.
Transcriptional regulation of StAR has been well characterized in various species. Mapping of the StAR promoter in various species demonstrated the importance of the first 150 nucleotides from the transcription start site in the regulation of StAR (12–15, 51–53). The proximal region of the human StAR promoter also is believed to be sufficient for activation of StAR (12, 13, 18, 54, 55). Herein, we provide the first evidence that the distal control region participates in the activation of the human StAR gene by chromatin loop formation. In other species, the putative SF-1 binding sites also exist in the region upstream of StAR, and the distal control region is evolutionarily conserved at least in human, rat, and mouse (Fig. 5). Chromatin loop formation at the StAR locus could, therefore, be necessary for achieving maximum transcription of StAR in different species.
It is likely that various transcription-regulatory factors that interact with SF-1 also play important roles in the chromatin loop formation. However, results of knockdown experiments indicate that SF-1 plays a central role in the loop formation (Fig. 4, C and E). As in the case of StAR, SF-1 binding sites are present in the promoters of most, if not all, steroidogenesis-related genes. Therefore, it is possible that there are unidentified SF-1 binding sites in the upstream regions of other steroidogenesis related genes and that SF-1 interacts with these sites and with promoter SF-1 sites to form chromatin loops that exert maximal transcription of the genes.
EMSA experiments revealed that SF-1 binding to the sites in the enhancer region (−3,402 to −2,958 bp) was relatively weak. Consistent with previous reports, SF-1 binding to the site in the promoter region (−113/−85) also was weak, whereas SF-1 binding to −924/−896 site was very strong (Fig. 2B). Previous reports also indicated that the contribution of the −924/−896 site to SF-1-dependent transcription of StAR was small, whereas the SF-1 site in the promoter was essential for SF-1-dependent transcriptional activation. Therefore, despite low binding affinity of the SF-1 sites in the enhancer, as well as in the promoter region, all of these sites may play critical roles for StAR transcription.
It is well known that cAMP plays a critical role in steroidogenesis. This may be partly due to increased SF-1 protein levels (56, 57) and modification(s) of SF-1 itself (58) or modifications of co-activators interacting with SF-1 (59, 60). Actually, our data indicate the importance of cAMP stimulation for the induction of StAR transcription. On the other hand, knockdown of endogenous SF-1 resulted in the abolishment of StAR expression, even under cAMP stimulated conditions, suggesting that cAMP may play a role for steroidogenesis secondary to SF-1.
The nuclear receptor family in mammals, NR5A, consists of two major members, SF-1 and LRH-1. LRH-1 is known as a transcription factor that mainly participates in the homeostasis of bile acid production in the liver (61, 62). Recently, LRH-1 was found to be another regulator of steroidogenesis in the ovary (63). Conditional knock-out of LRH-1 in the ovary resulted in infertility due to ovulation failure (64). We also have demonstrated that introducing LRH-1 into MSCs caused differentiation into a steroidogenic cell lineage (29) (supplemental Fig. 5). At the same time, LRH-1 bound to the distal control region and promoter of StAR and induced recruitment of RNAPII to the promoter site and histone eviction at these sites (Fig. 3D). This indicates that both members of the NR5A family can similarly induce changes to chromatin structure at the StAR gene promoter and the distal control region and suggests that SF-1 and LRH-1 work together in steroidogenic tissues, especially in the ovary, to regulate steroid hormone production.
In this study, we found novel SF-1 binding sites upstream of the StAR gene (−3,402 to −2,958 bp) that function as a strong transcriptional activator. Upon binding of SF-1 to these sites and to the promoter site, we found, for the first time, that histone eviction and chromatin loop formation took place and that transcription of StAR was activated in an SF-1-dependent manner. Identification of the novel StAR gene control region might inform future studies to identify gene mutations in this enhancer region in lipoid congenital adrenal hyperplasia patients. Analysis of a mouse with mutations at the upstream sites of StAR gene would be the ultimate proof in the future.
Supplementary Material
Acknowledgments
We are grateful to Drs. T. Yanase for providing the KGN cells and to J. Toguchida for providing the hMSC-hTERT-E6/E7 cells. We also thank Y. Inoue, H. Fujii, Y. Yamazaki, and Y. Usami for technical assistance.
This work was supported by a grant from Health and Labor Sciences Research; a grant from the Ministry of Education, Culture, Sports, Science and Culture; the Yamaguchi Endocrine Research Foundation; and the Precursory Research for Embryonic Science and Technology program from the Japan Science and Technology Agency.

The on-line version of this article (available at http://www.jbc.org) contains ”Experimental Procedures,” Tables 1–4, and Figs. 1–9.
- MSC
- mesenchymal stem cell
- hMSC
- human MSC
- 3C
- chromosome conformation capture
- RNAPII
- RNA polymerase II
- TSS
- transcription start site
- BAC
- bacterial artificial chromosome.
REFERENCES
- 1.Bose H. S., Sugawara T., Strauss J. F., 3rd, Miller W. L. (1996) N. Engl. J. Med. 335, 1870–1878 [DOI] [PubMed] [Google Scholar]
- 2.Caron K. M., Soo S. C., Wetsel W. C., Stocco D. M., Clark B. J., Parker K. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11540–11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasegawa T., Zhao L., Caron K. M., Majdic G., Suzuki T., Shizawa S., Sasano H., Parker K. L. (2000) Mol. Endocrinol. 14, 1462–1471 [DOI] [PubMed] [Google Scholar]
- 4.Manna P. R., Wang X. J., Stocco D. M. (2003) Steroids 68, 1125–1134 [DOI] [PubMed] [Google Scholar]
- 5.Luo X., Ikeda Y., Parker K. L. (1994) Cell 77, 481–490 [DOI] [PubMed] [Google Scholar]
- 6.Luo X., Ikeda Y., Lala D. S., Baity L. A., Meade J. C., Parker K. L. (1995) Endocr. Res. 21, 517–524 [DOI] [PubMed] [Google Scholar]
- 7.Parker K. L., Schimmer B. P. (1997) Endocr. Rev. 18, 361–377 [DOI] [PubMed] [Google Scholar]
- 8.Sadovsky Y., Crawford P. A., Woodson K. G., Polish J. A., Clements M. A., Tourtellotte L. M., Simburger K., Milbrandt J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10939–10943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark B. J., Combs R. (1999) Endocrinology 140, 4390–4398 [DOI] [PubMed] [Google Scholar]
- 10.Manna P. R., Eubank D. W., Lalli E., Sassone-Corsi P., Stocco D. M. (2003) J. Mol. Endocrinol. 30, 381–397 [DOI] [PubMed] [Google Scholar]
- 11.Rust W., Stedronsky K., Tillmann G., Morley S., Walther N., Ivell R. (1998) J. Mol. Endocrinol. 21, 189–200 [DOI] [PubMed] [Google Scholar]
- 12.Sandhoff T. W., Hales D. B., Hales K. H., McLean M. P. (1998) Endocrinology 139, 4820–4831 [DOI] [PubMed] [Google Scholar]
- 13.Sugawara T., Kiriakidou M., McAllister J. M., Kallen C. B., Strauss J. F., 3rd (1997) Biochemistry 36, 7249–7255 [DOI] [PubMed] [Google Scholar]
- 14.Wooton-Kee C. R., Clark B. J. (2000) Endocrinology 141, 1345–1355 [DOI] [PubMed] [Google Scholar]
- 15.Caron K. M., Ikeda Y., Soo S. C., Stocco D. M., Parker K. L., Clark B. J. (1997) Mol. Endocrinol. 11, 138–147 [DOI] [PubMed] [Google Scholar]
- 16.LaVoie H. A., Garmey J. C., Veldhuis J. D. (1999) Endocrinology 140, 146–153 [DOI] [PubMed] [Google Scholar]
- 17.Reinhart A. J., Williams S. C., Clark B. J., Stocco D. M. (1999) Mol. Endocrinol. 13, 729–741 [DOI] [PubMed] [Google Scholar]
- 18.Sugawara T., Holt J. A., Kiriakidou M., Strauss J. F., 3rd (1996) Biochemistry 35, 9052–9059 [DOI] [PubMed] [Google Scholar]
- 19.Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 20.Hiroi H., Christenson L. K., Chang L., Sammel M. D., Berger S. L., Strauss J. F., 3rd (2004) Mol. Endocrinol. 18, 791–806 [DOI] [PubMed] [Google Scholar]
- 21.Christenson L. K., Stouffer R. L., Strauss J. F., 3rd (2001) J. Biol. Chem. 276, 27392–27399 [DOI] [PubMed] [Google Scholar]
- 22.Rusovici R., Hui Y. Y., Lavoie H. A. (2005) Biol. Reprod. 72, 862–871 [DOI] [PubMed] [Google Scholar]
- 23.Shimizu T., Sudo N., Yamashita H., Murayama C., Miyazaki H., Miyamoto A. (2009) Mol. Cell Biochem. 328, 41–47 [DOI] [PubMed] [Google Scholar]
- 24.Yazawa T., Mizutani T., Yamada K., Kawata H., Sekiguchi T., Yoshino M., Kajitani T., Shou Z., Umezawa A., Miyamoto K. (2006) Endocrinology 147, 4104–4111 [DOI] [PubMed] [Google Scholar]
- 25.Okamoto T., Aoyama T., Nakayama T., Nakamata T., Hosaka T., Nishijo K., Nakamura T., Kiyono T., Toguchida J. (2002) Biochem. Biophys. Res. Commun. 295, 354–361 [DOI] [PubMed] [Google Scholar]
- 26.Mori T., Kiyono T., Imabayashi H., Takeda Y., Tsuchiya K., Miyoshi S., Makino H., Matsumoto K., Saito H., Ogawa S., Sakamoto M., Hata J., Umezawa A. (2005) Mol. Cell Biol. 25, 5183–5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishi Y., Yanase T., Mu Y., Oba K., Ichino I., Saito M., Nomura M., Mukasa C., Okabe T., Goto K., Takayanagi R., Kashimura Y., Haji M., Nawata H. (2001) Endocrinology 142, 437–445 [DOI] [PubMed] [Google Scholar]
- 28.Mizutani T., Yamada K., Minegishi T., Miyamoto K. (2000) J. Biol. Chem. 275, 22512–22519 [DOI] [PubMed] [Google Scholar]
- 29.Yazawa T., Inanoka Y., Mizutani T., Kuribayashi M., Umezawa A., Miyamoto K. (2009) Endocrinology 150, 3885–3893 [DOI] [PubMed] [Google Scholar]
- 30.Yoshino M., Mizutani T., Yamada K., Tsuchiya M., Minegishi T., Yazawa T., Kawata H., Sekiguchi T., Kajitani T., Miyamoto K. (2002) Biol. Reprod. 66, 1813–1819 [DOI] [PubMed] [Google Scholar]
- 31.Mizutani T., Yamada K., Yazawa T., Okada T., Minegishi T., Miyamoto K. (2001) Mol. Endocrinol. 15, 1693–1705 [DOI] [PubMed] [Google Scholar]
- 32.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 33.Fischle W., Wang Y., Allis C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 34.Lee W., Tillo D., Bray N., Morse R. H., Davis R. W., Hughes T. R., Nislow C. (2007) Nat. Genet. 39, 1235–1244 [DOI] [PubMed] [Google Scholar]
- 35.Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 36.Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D. (2004) Nat. Genet. 36, 900–905 [DOI] [PubMed] [Google Scholar]
- 37.Adams C. C., Workman J. L. (1995) Mol. Cell Biol. 15, 1405–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owen-Hughes T., Workman J. L. (1996) EMBO J. 15, 4702–4712 [PMC free article] [PubMed] [Google Scholar]
- 39.Workman J. L., Kingston R. E. (1992) Science 258, 1780–1784 [DOI] [PubMed] [Google Scholar]
- 40.Adkins M. W., Howar S. R., Tyler J. K. (2004) Mol. Cell 14, 657–666 [DOI] [PubMed] [Google Scholar]
- 41.Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D. (2004) Mol. Cell 14, 667–673 [DOI] [PubMed] [Google Scholar]
- 42.Vicent G. P., Nacht A. S., Smith C. L., Peterson C. L., Dimitrov S., Beato M. (2004) Mol. Cell 16, 439–452 [DOI] [PubMed] [Google Scholar]
- 43.Chen X., Wang J., Woltring D., Gerondakis S., Shannon M. F. (2005) Mol. Cell Biol. 25, 3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 45.Zhao H., Kim A., Song S. H., Dean A. (2006) J. Biol. Chem. 281, 30573–30580 [DOI] [PubMed] [Google Scholar]
- 46.Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. (2009) Nat. Genet. 41, 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinke H., Hörz W. (2003) Mol. Cell 11, 1599–1607 [DOI] [PubMed] [Google Scholar]
- 48.Ikeda Y., Shen W. H., Ingraham H. A., Parker K. L. (1994) Mol. Endocrinol. 8, 654–662 [DOI] [PubMed] [Google Scholar]
- 49.Clark B. J., Soo S. C., Caron K. M., Ikeda Y., Parker K. L., Stocco D. M. (1995) Mol. Endocrinol. 9, 1346–1355 [DOI] [PubMed] [Google Scholar]
- 50.Morohashi K., Honda S., Inomata Y., Handa H., Omura T. (1992) J. Biol. Chem. 267, 17913–17919 [PubMed] [Google Scholar]
- 51.Manna P. R., Dyson M. T., Eubank D. W., Clark B. J., Lalli E., Sassone-Corsi P., Zeleznik A. J., Stocco D. M. (2002) Mol. Endocrinol. 16, 184–199 [DOI] [PubMed] [Google Scholar]
- 52.Silverman E., Eimerl S., Orly J. (1999) J. Biol. Chem. 274, 17987–17996 [DOI] [PubMed] [Google Scholar]
- 53.Sugawara T., Lin D., Holt J. A., Martin K. O., Javitt N. B., Miller W. L., Strauss J. F., 3rd (1995) Biochemistry 34, 12506–12512 [DOI] [PubMed] [Google Scholar]
- 54.Christenson L. K., Johnson P. F., McAllister J. M., Strauss J. F., 3rd (1999) J. Biol. Chem. 274, 26591–26598 [DOI] [PubMed] [Google Scholar]
- 55.Shea-Eaton W. K., Trinidad M. J., Lopez D., Nackley A., McLean M. P. (2001) Endocrinology 142, 1525–1533 [DOI] [PubMed] [Google Scholar]
- 56.Aesøy R., Mellgren G., Morohashi K., Lund J. (2002) Endocrinology 143, 295–303 [DOI] [PubMed] [Google Scholar]
- 57.Yazawa T., Mizutani T., Yamada K., Kawata H., Sekiguchi T., Yoshino M., Kajitani T., Shou Z., Miyamoto K. (2003) Endocrinology 144, 1920–1930 [DOI] [PubMed] [Google Scholar]
- 58.Hammer G. D., Krylova I., Zhang Y., Darimont B. D., Simpson K., Weigel N. L., Ingraham H. A. (1999) Mol. Cell 3, 521–526 [DOI] [PubMed] [Google Scholar]
- 59.Chen W. Y., Juan L. J., Chung B. C. (2005) Mol. Cell Biol. 25, 10442–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yazawa T., Inaoka Y., Okada R., Mizutani T., Yamazaki Y., Usami Y., Kuribayashi M., Orisaka M., Umezawa A., Miyamoto K. (2010) Mol. Endocrinol. 24, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., Maloney P. R., Willson T. M., Kliewer S. A. (2000) Mol. Cell 6, 517–526 [DOI] [PubMed] [Google Scholar]
- 62.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. (2000) Mol. Cell 6, 507–515 [DOI] [PubMed] [Google Scholar]
- 63.Hinshelwood M. M., Repa J. J., Shelton J. M., Richardson J. A., Mangelsdorf D. J., Mendelson C. R. (2003) Mol. Cell Endocrinol. 207, 39–45 [DOI] [PubMed] [Google Scholar]
- 64.Duggavathi R., Volle D. H., Mataki C., Antal M. C., Messaddeq N., Auwerx J., Murphy B. D., Schoonjans K. (2008) Genes Dev. 22, 1871–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.