Abstract
Background
In addition to their strong induction following stress, small heat shock proteins (Hsp) are also expressed during development in a wide variety of organisms. However, the precise identity of cell(s) expressing these proteins and the functional contribution of small heat shock proteins in such developmental context remain to be determined. The present study provides a detailed description of the Drosophila small heat shock protein Hsp23 expression pattern during embryogenesis and evaluates its functional contribution to central nervous system development.
Results
Throughout embryogenesis, Hsp23 is expressed in a stage-specific manner by a restricted number of neuronal and glial lineages of the central nervous system. Hsp23 is also detected in the amnioserosa and within a single lateral chordotonal organ. Its expression within the MP2 lineage does not require the presence of a functional midline nor the activity of the Notch signaling pathway. Transactivation assays demonstrate that transcription factors implicated in the differentiation of the midline also regulate hsp23 promoter activity. Phenotypic analysis of a transgenic line exhibiting loss of Hsp23 expression in the central nervous system suggests that Hsp23 is not required for development and function of this tissue. Likewise, its overexpression does not cause deleterious effects, as development remains unaffected.
Conclusions
Based on the presented data, we suggest that the tightly regulated developmental expression of Hsp23 is not actively involved in cell differentiation and central nervous system development per se but rather reflects a putative role in preventive "pre-stress" neuroprotection or in non-vital process(es) common to the identified cell lineages.
Background
The survival and perpetuation of a species depends on its capacity to cope with stress factors from its environment. One conserved manner by which all living organisms defend themselves at the cellular level when confronted with diverse types of stress is the induction of a defined class of polypeptides termed heat shock proteins (Hsp) [1]. The small heat shock proteins (sHsp) represent the least conserved subfamily of Hsp as their number and size (ranging from 12 to 40 kDa) vary from species to species. Studies in different experimental systems have revealed a variety of functions for the sHsp under stress conditions. These different roles, including basic chaperoning activity [2,3], cytoskeleton protection [4] and modulation of the apoptotic process [5] directly represent means of cellular defense against environmental aggression. Contrasting with the classical definition of heat shock proteins as polypeptides induced by stress, cell-specific expression of sHsp in the absence of stress has been reported during the development of a wide range of organisms such as Caenorhabditis elegans [6], Drosophila melanogaster [7-9], Xenopus laevis [10], Mus musculus [11-13] and man [14]. Even if functional roles have been demonstrated for certain high molecular weight Hsps in non-stress related processes such as RTK signaling [15] and spermatogenesis [16-18], only preliminary experimental evidence so far support such requirement for sHsp under non-stress conditions [19]. Their peculiar cell-specific pattern of expression has lead to the hypothesis that sHsp may be implicated in differentiation mechanisms. While recent studies in cultured cells have provided support to this possibility [20], no such evidence has yet been provided for a multicellular organism.
In Drosophila, sHsps are expressed throughout many stages of the life cycle (reviewed in [21,22]). During oogenesis, Hsp27 displays a stage-specific intracellular localization within nurse and follicle cells [23] while Hsp23, Hsp26 and Hsp27 are respectively expressed in distinct cell types during the spermatogenic process [9,24]. During embryogenesis, Hsp27 associates to cells of the brain and of the ventral nerve cord while Hsp26 is found exclusively in the gonads [25]. Hsp23 also displays a cell-specific pattern of expression during embryonic neurogenesis [26,27] and has recently been shown to be strongly downregulated following the targeted expression of the glial "master" gene gcm [28]. Despite this increasing knowledge on the developmental expression of sHsps, the precise identity of cells expressing these proteins along with the in vivo function(s) played by sHsp in these developmental instances remain to be unveiled. The expression of Hsp23 within a highly characterized morphogenetic system (the embryonic nervous system) combined to the isolation of a P-element insertion in the promoter region of its gene, provided the opportunity to precisely define its expression pattern and evaluate its functional implication in a specific developmental process.
This study reports the expression of Hsp23 in neuronal (MP2, VUMs) and glial (midline glia) lineage of the CNS, as well as in a single chordotonal organ per hemisegment and in cells of the amnioserosa. We demonstrate that Hsp23 expression in the neuroectoderm is closely and autonomously linked to the acquisition of MP2 fate as it does not requires the presence of a functional midline and is expanded in a neurogenic mutant where additional MP2s are specified. In vitro transactivation assays support that the Single-minded, Tango and Drifter transcription factors, which are all involved in midline determination and differentiation, may also regulate hsp23 promoter activity. Finally, we evaluate a putative functional contribution of Hsp23 to embryonic neurogenesis through phenotypic analysis of a P-element insertion line resulting in an inhibition of Hsp23 expression in the CNS. The absence of detectable phenotype in the ventral nerve cord of homozygous embryos suggests that the loss of Hsp23 is not detrimental to CNS formation. Furthermore, the failure to observe any differentiation or functional defects following targeted misexpression of Hsp23 indicates that its biological activity is related to non-vital features which are distinct from the normal developmental program.
Results
The MP2 neuronal lineage expresses Hsp23 beginning at stage 11
To clearly define the profile of hsp23 expression during embryogenesis, the distribution of its transcripts and protein species were both assessed. Data obtained by immunohistochemistry and in situ hybridization yielded an identical spatiotemporal pattern of expression associated to restricted cell populations of the embryo.
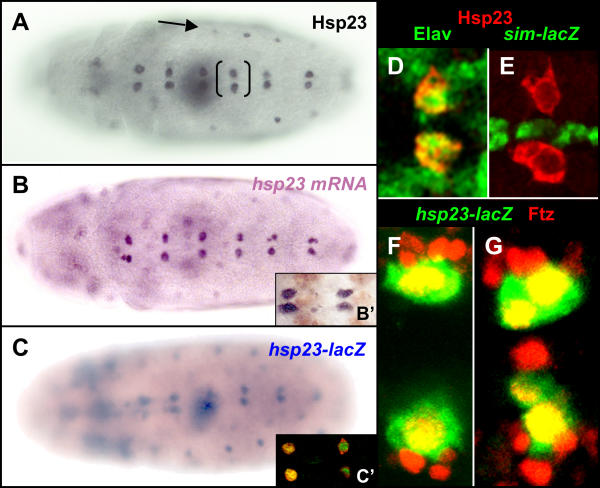
During early embryogenesis, maternally-contributed hsp23 transcripts and protein display a ubiquitous distribution (data not shown) which fades away to become undetectable at gastrulation. Onset of zygotic Hsp23 expression is first observed at early stage 11 through the accumulation of Hsp23 protein (Fig. 1A) and transcripts (Fig. 1B) in specific cells of the embryo. Prominent expression is detected in a segmentally-repeated population of cells located on each side of the midline (brackets, Fig. 1A). Co-expression of the neural marker Elav (Fig. 1D) indicates that these cells are neurons of the central nervous system. The origin (mesectoderm versus neuroectoderm) of these Hsp23-positive neurons was tested in a single-minded-lacZ fly line, in which all cells of mesectodermal origin are marked by β-Galactosidase (β-Gal) expression [29]. Exclusion of β-Gal from these Hsp23-expressing neurons (Fig. 1E) indicates that they arose from the neuroectoderm. Interestingly, these neurons displayed constant peculiar morphological features: they were large and rounded, and rapidly underwent cell division after onset of Hsp23 expression. Their intra-segmental position with regards to the midline (Fig. 1E) and to the domain of Engrailed expression (Fig. 1B') further suggested that these might represent neuroblasts of the MP2 lineage, as these undergo their sole mitotic division during stage 11 [30]. Their MP2 identity was confirmed by co-localization of the Ftz molecular marker [31,32] with the reporter gene from a hsp23(1.8)-lacZ transgenic fly line, which reflects endogenous Hsp23 expression within the MP2 lineage (Fig. 1C,1C'). The co-localization between Ftz and the reporter for hsp23 promoter activity is observed both before (Fig. 1F) and after (Fig. 1G) the MP2 mitotic division.
Figure 1.
Hsp23 expression in stage 11 embryos. (A) Detection of Hsp23protein in a lateral [arrow] and midline associated population [brackets]. (B) In situ hybridization for hsp23 mRNA showing distribution identical to that of panel A. (B') High magnification view of the midline region of two abdominal segments showing the relative position of hsp23-positive cells (purple) with regards to the expression of Engrailed (brown). (C) The expression of β-Galactosidase under the control of a 1.8 kb proximal fragment of the hsp23 promoter displays a similar distribution to that shown in panels A and B. (C') Confocal view similar to panel B' showing the co-localization of β-Galactosidase and endogenous Hsp23 in the midline associated cells. (D-G) Confocal high magnification views of a similar region indicated by brackets in A. (D) Co-expression of the neuronal marker Elav (green) and Hsp23 (red). (E) Absence of the midline marker sim-lacZ (green) in Hsp23-positive cells (red). (F and G) Co-localization of hsp23 promoter activity (green) and Ftz (red) in the MP2 neuroblasts both before (F) and after (G) their mitotic division. Ventral views of stage 11 embryos are shown in all panels with the anterior oriented toward the left.
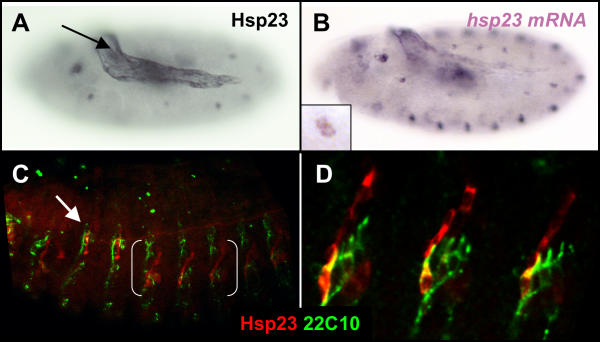
Lower levels of Hsp23 protein and transcripts are also detected in a single cell located on the lateral region of each segment (arrow, Fig. 1A). This cell rapidly divides (inset, Fig. 2B) and ultimately yields an elongated lateral structure (Fig. 2C). The dorso-ventral position and striking morphology of this structure strongly suggested that Hsp23 could be expressed by all cells of a single lateral chordotonal lineage (lch). Co-localization with the 22C10 antibody [33] (Fig. 2D), which recognized a highly organized pattern of motorneurons including all lch neurons, confirmed this hypothesis and thereby indicated that the single lateral cell seen at stage 11 (Fig. 1A,1B,1C; Fig. 2A,2B) is a single chordotonal organ precursor. It is noteworthy to point out that the single Hsp23-positive chordotonal organ in abdominal segment is lateral whereas in the thoracic segments T2 and T3, where lateral chordotonal organs are absent, Hsp23 associates with one of the dorsal chordotonal organs (white arrow, Fig. 2C).
Figure 2.
The amnioserosa and cells of a single chordotonal organ express Hsp23. (A) Hsp23 expression in cells of the amnioserosa (arrow) and in lateral cells (out of focus – see Fig. 1A) of a stage 11 embryo. (B) Similar distribution shown by hsp23 mRNA transcripts at stage 11. Inset shows hsp23 mRNA detected in two adjacent cells of the lateral region, reminiscent of a mitotic division. (C) Confocal view of a stage 13 embryo displaying Hsp23 expression (red) in cells of a single chordotonal organ per abdominal segments (3 segments indicated by brackets) and in a single dorsal chordotonal organ in segments T2 (white arrow) and T3. Neurons of the peripheral nervous system are identified using the 22C10 molecular marker (green) (D) Confocal high magnification of bracket region in C displaying co-expression of Hsp23 (red) in neuronal cells of lateral chordotonal organs (green). Lateral view is shown in all panels.
In addition to the MP2 and lch lineages, Hsp23 expression at stage 11 is also detected in the amnioserosa (arrow, Fig. 2A) and in uncharacterized cells of the cephalic region (out of focus in Fig. 1A,1B,1C; 2A,2B). Based on the amount of knowledge acquired on the morphogenetic development of the ventral nerve cord, we focused our analysis on the cells located therein and excluded from the present study the Hsp23-positive cephalic populations.
An additional neuronal population expresses Hsp23 at stage 13
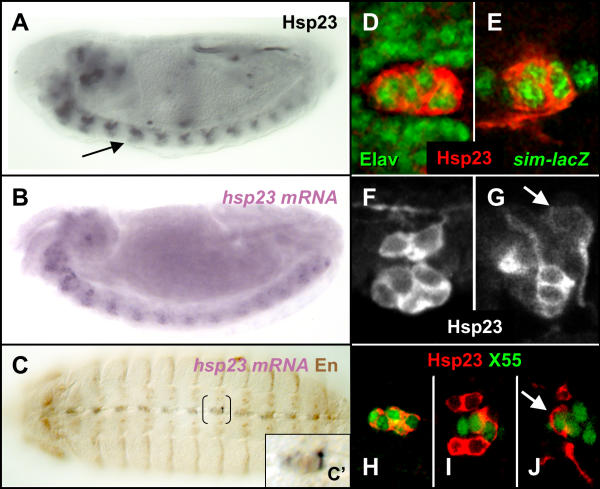
Modulation of hsp23 expression is detected at stage 13 when a novel group of ventral cells expressing Hsp23 protein (arrow, Fig. 3A) and mRNA (Fig. 3B) are observed within the ventral nerve cord (arrow, Fig. 3A). These represent neurons of the midline as characterized by the expression of the Elav (Fig. 3D) and sim-lacZ (Fig. 3E) molecular markers within these cells. Furthermore, their ventral position, peculiar morphology (grape-like cluster – Fig. 3F) and co-expression of Engrailed (Fig. 3C,3C') strongly suggest that they represent the Ventral Unpaired Median (VUMs) neurons. To verify this hypothesis, co-localization between Hsp23 and the X55 enhancer-trap was tested. The X55 line marks the VUMs, the midline neuroblasts (along with their respective support cells) and the posterior midline glia (MGP) [34]. Figure 3 (H to I) displays ventral confocal sections through a stage 13 nerve cord. The ventral cell population located at the midline exhibits co-localization of Hsp23 and β-Gal (Fig. 3H,3I) while the more dorsally located MP2 daughter cells (vMP2 and dMP2) located on either side of the midline do not express the X55 reporter gene (Fig. 3I). Intriguingly, the MGP located in the dorsalmost region of the nerve cord also expressed both Hsp23 and the X55 enhancer-trap (arrow, Fig. 3G,3J), suggesting that Hsp23 expression can also be found in midline glial cells of the CNS. The cell identity of the VUMs and MGP is further confirmed by the visualization of their respective dorso-anterior axonal and ventral cytoplasmic projections (Fig. 3G), which are both significantly labeled by the Hsp23 antibody.
Figure 3.
Onset of Hsp23 expression in the midline at stage 13. (A) Localization of Hsp23 in a distinct group of cells located ventrally in the nerve cord (arrow). (B) Similar distribution observed by in situ hybridization with hsp23 mRNA. (C) Co-expression of hsp23 mRNA (purple) and Engrailed protein in the novel ventral population of cells. (C') High magnification of region shown by brackets in C. (D-J) Confocal views. (D-E) Co-localization of Hsp23 (red) with Elav (D, green) and sim-lacZ (E, green), suggesting that these cells are neurons of the midline. (F, G) Sequential confocal sections. (F) Hsp23 expression in the two MP2 daughter cells and the ventral group of midline neurons. Note the detection of Hsp23 in the dorsal axonal projection of the vMP2 and dMP2. (G) Presence of Hsp23 in the ventral VUMs neurons and their dorso-anterior projection as well as in the posterior midline glia (MGP) that exhibits a distinctive ventral projection. (H-I) Sequential confocal sections showing the distribution of the X55 enhancer-trap (green) with regards to Hsp23 expression (red). (H) Co-localization in the midline VUMs. (I) Co-localization of X55 and Hsp23 in the VUMs but exclusion of X55 from the MP2 daughter cells located on each side of the midline. (J) Co-localization of Hsp23 and X55 in the MGP. Note that multiple posterior cells expressing X55 (presumptively identified as the MNB and its support cells) do not express Hsp23. Stage 13 embryos are shown in all panels. Ventral views are shown in panels C-E, H-J and lateral views in A, B, F, G. Anterior is towards the left in all panels.
Hsp23 expression becomes restricted to midline glial cells at the end of neurogenesis
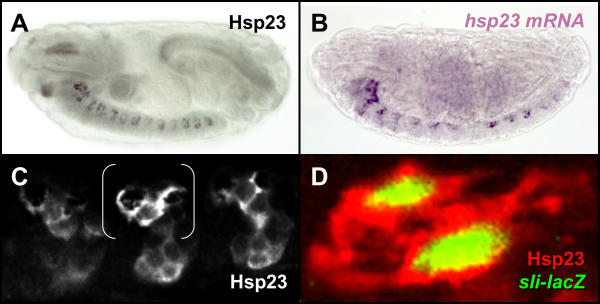
The observation that the MGP express Hsp23 at stage 13 combined to the detection at late stage 14 of Hsp23 protein (Fig. 4A) and mRNA (Fig. 4B) within three dorsal non-neuronal cells (Fig. 4A and data not shown) exhibiting the characteristic morphology of midline glia (Fig. 4C) suggested that this small heat shock protein was expressed by all surviving midline glial cells during late embryogenesis. The glial identity of these dorsal cells of the midline was confirmed using the slit-lacZ enhancer-trap line, a specific marker for midline glial cells (Fig. 4D).
Figure 4.
Hsp23 becomes restricted to midline glia by the end of embryogenesis. (A) Localization of Hsp23 in dorsal cells of the ventral nerve cord during late embryogenesis. (B) Detection of hsp23 transcripts in the same subset of cells. (C) Confocal high magnification view of Hsp23 expression in three abdominal segments. Declining Hsp23 expression can still be detected at lower levels in the ventral VUMs. Note the characteristic morphology of the dorsal midline glia (brackets). (D) Confocal high magnification of a similar region shown by brackets in C displaying co-localization of the midline glia marker slit-lacZ (green) and Hsp23 (red). Lateral views of stage 16 embryos are shown in all panels, anterior is towards the left.
Regulation of Hsp23 expression
The tight regulation of Hsp23 expression suggested that its induction might be regulated by specific signals confined to restricted cells and time-windows of embryogenesis. In an attempt to identify transcription factor(s) or signaling pathway(s) involved in hsp23 regulation, knowledge on developmental requirement for cells expressing Hsp23 (MP2, midline glia) was used to generate and test different hypothesis.
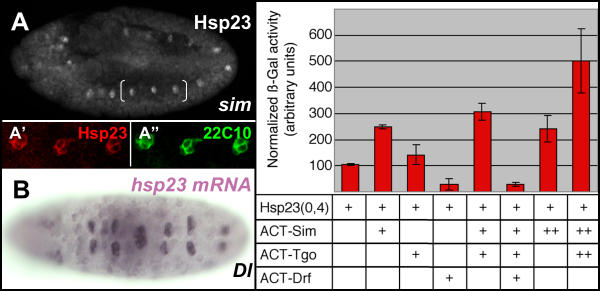
To verify whether Hsp23 expression in the MP2 lineage was dependent on an external signal emanating from midline, the distribution of Hsp23 was examined in stage 11 single-minded mutant embryos, where the development of the midline is impaired [29]. As shown in Fig. 5A, Hsp23 is still expressed in stage 11 mutant embryos in a group of 2 to 4 cells joined at the ventral midline (3 segments in brackets). These Hsp23-positive cells (Fig. 5A') represent cells of the MP2 lineage as judged by their co-expression of 22C10 (Fig. 5A"). The collapsed character of the MP2 cells reflects the absence of midline cells.
Figure 5.
Regulation of hsp23 developmental expression. (A) Neuroectodermal expression of Hsp23 is maintained in a stage 11 single-minded mutant. (A' and A") Confocal High magnification views of region in brackets in A displaying co-expression of Hsp23 (red, A') and 22C10 (green, A") in cells of the MP2 lineage. (B) Detection of hsp23 mRNA in additional MP2 neuroblasts generated in a delta mutant. Graph: Modulation of the proximal hsp23 promoter by the Single-minded (Sim), Tango (Tgo) and Drifter (Drf) transcription factors. Normalized β-Galactosidase activity obtained after transfection in S2 cells is shown. Note the capacity of Sim and Tgo to activate hsp23 promoter and the potent repressive effects of Drf.
The selection of neuroblasts from the neuroectoderm is mediated by lateral inhibition, a Notch signaling-mediated process [35]. To verify if activation of hsp23 transcription in the MP2 required Notch signaling, we probed Delta null embryos where no Notch signaling occurs during embryogenesis. The increase in the number of cells expressing hsp23 transcripts (Fig. 5B) is consistent with the specification of additional MP2 due to the absence of Notch signaling. This observation also supports the hypothesis that transcriptional activation of the hsp23 gene does not require Notch signaling. Coherent with these observations, additional chordotonal organ precursors expressing hsp23 transcripts are also observed in Delta mutant embryos (data not shown).
Molecular analysis of the hsp23 promoter for putative regulatory elements unveiled the presence of a CME box (CNS Midline Enhancer [36]: TACGTG, located at -329 of the transcription initiation site) which has readily been shown to be bound by the "master" determinant for midline identity, Single-minded (Sim), and its dimerization partner Tango (Tgo) in the direct regulation of at least another midline gene, Slit [37]. As Hsp23 is expressed by the VUMs and midline glia, the potential for these transcription factors to regulate the hsp23 promoter was tested in cell culture-based transactivation assays. Quantitative analysis of hsp23 transactivation shows that Sim and Tgo can synergize to activate the hsp23 promoter (Graph, Fig. 5). Interestingly, another transcription factor (Drifter) previously shown to be involved in the regulation of Slit [37] acted as potent repressor of hsp23 promoter in these assays.
Hsp23 expression is not required for embryonic neurogenesis
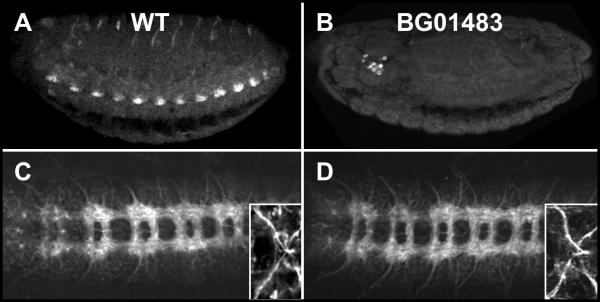
To assay the functional significance of Hsp23 expression during embryogenesis, we used a P-element line (BG01483) obtained from the Berkeley Drosophila Genome Project [38] carrying an insertion located at -149 of the transcriptional start site of hsp23. This insertion strongly abrogates the endogenous pattern of Hsp23 expression, restricting it to a distinct cluster of a few cells located in the posterior region of the brain, tentatively identified as the crystal cells (Fig. 6B). Such loss of Hsp23 provides an ideal context to assess whether this Hsp is necessary for global CNS organization and proper differentiation of cell types where it is usually expressed. Despite abrogating CNS-associated Hsp23 expression, the chromosome carrying this insertion is fully viable and maintained in a homozygous state, thereby suggesting that Hsp23 expression does not fulfill a vital role in the CNS. Furthermore, labeling BG01483 embryos with the commissural marker BP102 reveals that the absence of Hsp23 within the CNS does not disturb its ultra structure, as longitudinal commissures are still well fasciculated and transversal commissures are properly separated (Fig. 6D). This simple observation directly supports that functional VUMs and MG are present since both of these cell types interact to engineer proper transversal commissure separation [34]. 22C10 labeling further shows that motorneuron axons projections, including the VUMs projection through the anterior commissure, remain unaffected in BG01483 embryos (inset, Fig. 6D).
Figure 6.
Normal CNS development in the absence of Hsp23. (A, B) Ventro-lateral view of Hsp23 expression at stage 16. (A) Wild-type embryo. (B) Embryo homozygous for the P-element insertion BG01483. Note the absence of Hsp23 expression in the CNS. The Hsp23-positive cells located behind the embryonic brain are tentatively identified as the crystal cells of the immune system. (C, D) Ventral views of CNS ultra structure in stage 16 embryos using the BP102 antibody. Insets show the anterior VUMs axonal projections visualized with the 22C10 antibody. (C) Wild-type embryo. (D) Embryo homozygous for the P-element insertion BG01483. No differences are detected between C and D.
Overexpression of Hsp23 during development is not detrimental
As Hsp23 expression is confined to given cell types during specific developmental stages, it was conceivable that this protein plays defined role(s) associated with the process of differentiation for each of these cells. To test for the putative implication of Hsp23 in the differentiation program and identify any detrimental effect of its overexpression on CNS formation and function, we overexpressed Hsp23 using the GAL4-UAS system. For this purpose, three different GAL4 drivers targeting distinct subpopulations of cells (actin-GAL4 / general, scabrous-GAL4 / neuroectoderm, elav-GAL4 / neuronal) were used. Increase in endogenous Hsp23 levels resulting from crossing out a UAS-hsp23 line (UAS-hsp23/4) to an actin-GAL4 driver is shown in Fig. 7A while spatial modulation of Hsp23 expression using an elav-GAL4 driver crossed to the same UAS-hsp23 line is shown in Fig. 7B. Although a marked increase in Hsp23 levels was observed in flies which carry any of these GAL4 driver and the UAS-hsp23 construct, no disruption of the CNS or PNS was observed based on the two independent criteria of fly viability and tissue ultra structure visualized with BP102 staining for the CNS (right inset, Fig. 7B) and Elav for the PNS (Fig. 7B). Hsp23 also retained its cytoplasmic localization when misexpressed and still showed high association with axonal / cellular projection. The capacity of Hsp23 to serve as a "cell morphology tracer" is best visualized in the lateral chordotonal organs clusters of the PNS (boxes, Fig. 7B). Thus, misexpression of Hsp23 in cells where it is normally absent is not detrimental to CNS and PNS formation and function.
Figure 7.
Overexpression of Hsp23 in the CNS and PNS. (A) Western blot showing the increase in Hsp23 protein level in flies carrying the actin-GAL4 chromosome in conjunction with the UAS-hsp23/4 insertion. For control purposes, levels of Hsp23 in heat-shocked S2 cells and in flies carrying the same UAS chromosome with the CyO balancer are also shown. (B) Stage 16 embryos of the elav-GAL4;UAS-hsp23/4 genotype. Cellular co-localization of Hsp23 (red) and Elav (green) in embryonic neurons. Note that perfect merging of the two signals is not obtained as the proteins differ in their intracellular localization (Elav: nuclear, Hsp23: cytoplasmic). Left inset shows ventro-lateral view with strong Hsp23 expression in the nerve cord. Right inset is a high magnification ventro-lateral view of the nerve cord showing unaffected separation of the transversal commissures (green, visualized with BP102) and general Hsp23 expression (red).
Discussion
The developmental expression of sHsp, which has now been observed in many species, differs from their stress-induced expression in two major ways. Under stress stimuli, most cells of the organism activate the transcription of all sHsp genes, leading to a massive and ubiquitous expression of all sHsp. In contrast, sHsp developmental expression displays cell type and stage specificity. Such a spatio-temporal regulation suggests that sHsp fulfill distinct function(s) in a cell-specific fashion within normal developmental processes. This study characterized the expression pattern of a Drosophila cytoplasmic sHsp (Hsp23) during embryonic neurogenesis and assessed its functional implication during development of the CNS. The combinatorial use of different detection methods (transcript / protein and promoter activity) allowed us to precisely define the pattern of Hsp23 expression while ruling out the possibility of cross-detecting other members of this conserved family. This is particularly relevant as scanning of the Drosophila genome [39] reveals at least twelve ORF containing the alpha-crystallin domain, hallmark of the sHsp (CG4167, CG4183, CG4190, CG4460, CG4461, CG4463, CG4466, CG4533, CG7409, CG13133, CG14207, CG32041). The CNS cell lineages and their respective time window for Hsp23 expression were first identified. Analysis of loss of function and overexpression for Hsp23 suggests that this sHsp does not fulfill a vital function during embryonic CNS development.
Hsp23: mediator of cell contact or morphology?
In the CNS, Hsp23 is first detected at stage 11 in the MP2 neuroblast, which is subsequent to its specification and delamination from the neuroectoderm (stage 8; [31]). This delay suggests that hsp23 activation is an event occurring downstream of MP2 fate acquisition. The presence of Hsp23 in the extra MP2 neuroblasts specified in a Delta mutant (Fig. 5B) is consistent with this idea. Intriguingly, onset of hsp23 transcription in this particular lineage correlates in time with the occurrence of its sole division (stage 11) raising the possibility that similar signals may induce both events. After its division, the vMP2 and dMP2 daughter cells will establish the medial lateral commissural tract by sending out growth cones.
At stage 13, the ventrally-located VUM neurons and the posterior midline glia (MGP) also begin to express Hsp23. As observed in the MP2 lineage, Hsp23 expression follows cell determination in both lineages. The VUMs are first specified in the dorsal region of the nerve cord and must undergo a ventral migration to occupy their final position [40]. During late stage 12 and stage 13 the trailing axons of the VUMs (now located in a dorso-anterior position with regards to their soma) serve as guidance cue for the migrating middle midline glial cells. In the MGP, induction of Hsp23 correlates with its contact to the commissure, which marks the end of its anterior migration [34]. In the last stages of embryogenesis, Hsp23 expression becomes restricted to the three surviving midline glia of the CNS (Fig. 1D to 1I and 3E) at a time when they ensheath the transversal commissures of the nerve cord.
Therefore, the timing of Hsp23 expression in both of these lineages favors the hypothesis of Hsp23 implication in cell anchoring or in mediation of cell contact (VUMs / MGM or MGP / commissure) rather than a role in the migratory process itself. Such role(s) would be reminiscent of its mammalian counterpart (Hsp25/27) that is involved in cytoskeleton modulation [4].
Hsp23 is regulated in a cell-autonomous fashion
The association of Hsp23 with different cell types at different time points during CNS development prompted us to assess whether its expression was induced through a common inductive cue or by different cell-specific mechanisms. As two of three Hsp23-expressing lineages derive from the mesectoderm, such origin could favor the expression of Hsp23 by potentiating its promoter to further be modulated by cell-specific factors. While in vivo analysis clearly demonstrated that the acquisition of midline identity was a requirement for Hsp23 expression in the presumptive VUMs and midline glia, transactivation assays using the midline "master gene" Single-minded and its dimerization partner Tango revealed that both of these proteins can act as activators of hsp23 promoter activity in cultured cells (Fig. 5). The absence of Hsp23 expression in different midline lineages however suggests that additional factors must be implicated in the regulation of the hsp23 gene during CNS development. Interestingly, the Sim-Tgo heterodimer has previously been shown to act as a "priming" agent allowing promoters to be modulated by additional cell-specific factors such as Drifter and Dichaete, both of which are present in cells of the midline [37]. Furthermore, dichaete mutants exhibit midline glia defects [41] while its gene product has been shown to regulate cell fate in the intermediate neuroblast column of the early neuroectoderm by preventing the expression of medial column identity genes [42]. This suggests that Dichaete could positively regulate hsp23 expression in the context of the midline (in the midline glia) while acting as a negative regulator within neuroectodermal lineages of the intermediate column.
To test whether hsp23 expression in the MP2 neuroblast resulted from a combination of intracellular transcriptional programs integrated with external activating signals, we examined its distribution in absence of Notch (in a delta mutant) or Egfr (in a single-minded mutant) signaling. In both instances, Hsp23 remained expressed in the MP2, suggesting that its regulation in this lineage is cell autonomous. Independence to Egfr signaling was also observed in the neuronal lineage of the midline (VUMs), as both gain and loss of function of the two main effectors of this cascade (PointedP2 [43] and Yan [44]) did not prevent nor ubiquitously activate Hsp23 expression (data not shown). Analysis of Notch signaling in the midline was impaired by the early requirement of Notch for the establishment of the mesectoderm [45].
No vital function of Hsp23 during neurogenesis
The expression of Hsp23 within restricted cell types raised the possibility that it may be required within these lineages for appropriate cell differentiation. To test this hypothesis, we examined a P-element line where Hsp23 expression was drastically abrogated. Examination of CNS ultra structure and individual cell differentiation failed to provide any detectable phenotype, thereby suggesting that Hsp23 function is dispensable for embryonic CNS establishment and function. This simple conclusion is directly correlated by the fact that the chromosome bearing the P-element insertion is homozygous viable. In addition, specific lineages which usually express Hsp23 remain apparently unaffected in the absence of this protein as they retain the expression of respective identity molecular markers such as 22C10 (MP2 and the VUMs – data not shown) and completion of specific in vivo function such as separation of transversal commissures (VUMs and midline glial cells).
In experiments designed to evaluate if Hsp23 overexpression could impair neurogenesis, flies overexpressing Hsp23 displayed normal CNS structure and developed to adulthood without any obvious detrimental signs. Both loss and gain of function data therefore support that Hsp23 function is not required for embryonic CNS establishment and function.
Conclusion
The data gathered so far on Hsp23 expression within the CNS converge to a common theme identified for sHsp developmental expression: cell-specificity. As expression often relates to function, a requirement for Hsp23 during CNS establishment could be expected. However, none of the observations made during the course of this study supports that Hsp23 fulfills a vital function within the identified lineages. Not only have overall CNS establishment and function (through fly survival) remained unaffected, but we have also examined the development and function of specific lineages using a battery of molecular markers. Although the possibility that a subtle phenotype has eluded the current analysis cannot be formally discarded, the data support that Hsp23 is not required for CNS development. The additional possibility that Hsp23 requirement is masked by other member(s) of the sHsp family displaying functional redundancy appears unlikely as it would require that the complementing protein be expressed in a similar spatiotemporal pattern and possesses an activity affecting identical intracellular process(es).
Another appealing hypothesis is that Hsp23 expression within the CNS could serve as a protective mechanism seeded by evolution in cells carrying out vital functions for CNS development. The MP2 daughter cells are pioneer neurons for the MP1/MP2 longitudinal commissural tract while the VUMs, through their axonal projections, serve to guide the midline glial cells for the proper separation of transversal commissural tracts. Therefore, the constitutive expression of a chaperone protein such as Hsp23 making these cells more resistant to environmental insults would undoubtedly be beneficial to the organism. In vitro, Hsp23 has been shown to be a powerful chaperone which, in addition to prevent protein aggregation, can also help in protein refolding both within the reticulocyte lysate system and in microinjected Xenopus oocytes (Morrow et al., in preparation). While sHsp levels are upregulated during the normal ageing process [46], an in vivo increase of sHsp expression has been reported in genetically selected lines for increased longevity [47]. The beneficial effects of sHsp expression have also been observed at neuronal synapses, where targeted expression of distinct Hsps has been shown to confer neuroprotection [48,49]. Furthermore, ongoing studies in our and other laboratories demonstrate that overexpression of different sHsp confers beneficial effects in vivo [50] and G. Morrow, et al, submitted).
The identification of intracellular role(s) of Hsps in normal development, whether it be to act as simple chaperones with or without substrate specificity, or to fulfill unsuspected functions, remains an important step in order to fully grasp the implication of these highly conserved polypeptides on non-stress-related processes.
Methods
Drosophila strains
Flies were raised on standard Drosophila medium at 25°C. The Oregon-R was used as wild type strain and the w1118 strain for transgenic generations. The following mutants and enhancer-trap lines were used: BG01483 (insertion of P(GT1) in hsp23 promoter – BDGP), X55 enhancer-trap [34], slit1.0-lacZ [29], sim-lacZ [36], DeltaX. Ectopic protein expression was achieved by the transactivation GAL4-UAS system [51] using the following lines: scabrous-GAL4, actin-GAL4, elav-GAL4. The UAS-hsp23 and hsp23(1.8)-lacZ transgenic lines were developed during this study and are described below.
Immunohistochemistry and in situ hybridization
Standard procedures for whole mount immunohistochemistry [52] and in situ [53] were used for all reactions, with the exception of antibody staining using the anti-Hsp23, where the methanol treatment for embryo devitellination was kept to a maximum of 30 seconds and directly followed by washes in PBS 0.2% Tween-20. After the primary antibody, embryos were either incubated with biotinylated (Vector) or fluorochrome-associated secondary antibodies (Alexa 488 or Cy3 – Molecular Probes). For biotinylated secondary antibodies, signal was revealed using the Vectastain ABC kit (Vector) according to the manufacturer protocol. Following the staining reaction, embryos were dehydrated and mounted in DPX (Fluca). Fluorescently-labeled embryos were directly mounted in Vectashield (Vector) and visualized on a LSM 310 laser scanning confocal microscope (Zeiss). The following primary antibodies were used at the indicated dilutions: rabbit anti-Hsp23 1362 (1/1000) produced against a recombinant Hsp23 (Tanguay, unpublished), mouse anti-Eng 4D9 (1/50), mouse BP102 (1/50), mouse 22C10 (1/50), mouse anti-Elav (1/50), mouse anti-βGal J1E7 (1/50), rabbit anti-βGal (Promega – 1/500), rabbit anti-Ftz (1/200). Mabs BP102, anti-Elav, J1E7 and 22C10 were obtained from the Developmental Study Hybridoma Bank (DSHB – University of Iowa) developed under the auspices of the NICHD. For in situ hybridization, Digoxigenin-labeled hsp23 RNA probe (Roche Molecular Biochemicals) was generated from a partial hsp23 cDNA clone containing the complete hsp23 open reading frame. The clone was PCR-amplified from genomic DNA using the following primers: hsp23_F 5'-CAGCTAAAGCGAAAGTAACC-3' and hsp23_R 5'-TCTCGGAACGAGTCCTCTAC-3'.
DNA constructions
hsp23 promoter – lacZ chimeric genes
The pBR322-Dm202.7 genomic clone of the 67B genomic region was used to excise a 3.3 kb XbaI-SalI fragment that contains 2.2 kb of hsp23 promoter along with its entire coding region. This fragment was subcloned in the pBluescript II SK(-) vector (Stratagene) at identical sites, yielding the pBS23-(3.3) vector. Removal of the hsp23 coding region was achieved through partial digestion of pBS23-(3.3) using the XbaI site and an EcoRI site located 32 nucleotides downstream of the TATAA box in the promoter followed by treatment with T4 DNA polymerase and recircularization of the vector. The resulting vector, pBS23-(2.2), was submitted to partial digestion with a combination of SalI and either EcoRI or PstI. The truncated versions of the promoter (0.4 to 1.8 kb) were blunted and recircularized. Two versions of the hsp23 promoter, of 0.4 and 1.8 kb in length, were subsequently excised from pBS23-(0.4) or pBS23-(1.8) using XhoI and XbaI and subcloned into the XhoI and NheI sites of a modified version of the original pCaSpeR-AUG-βGAL (pCAβ) vector, yielding the pCAβM23-0.4 and pCAβM23-1.8 vectors respectively carrying the hsp23(0.4)-LacZ and hsp23(1.8)-LacZ chimeric genes. The modified pCAβ, thereafter named pCaSpeR-AUG-βGAL-MCS (pCAβM), was generated by inserting the multiple cloning site from the pMEP vector (Invitrogen) using the flanking BamHI and KpnI sites into the similar sites of pCAβ, thereby enabling the use of the XbaI-compatible NheI site for directional subcloning.
UAS-hsp23 vector and generation of transgenic lines
The UAS-hsp23 construct was generated by inserting a EcoRI – XbaI fragment from pTZ1888.25 [54] containing the full length Hsp23 coding sequence into respective sites of the pUAST vector (Brand and Perrimon, 1993). Transgenic lines carrying either the pUAS-hsp23 or pCAβM23-1.8 constructs were generated by standard injection procedures [55] of Qiagen Endofree prepared DNA along with the pHSΠ [56] helper plasmid into a w1118 strain. For targeted expression of Hsp23, the UAS-hsp23/4 line that carries a homozygous insertion on the third chromosome was used in all experiments.
Protein extracts and western blot
GAL4-UAS induction of Hsp23 was tested on adult heads of both control and targeted genotypes dissected in PBS and mechanically homogenized in 100 μl SDS-PAGE buffer [23]. Proteins were separated by SDS-gel electrophoresis [57] and transferred to nitrocellulose membranes (Gelman). Hsp23 was detected using a specific monoclonal antibody for this sHsp (7B12- [8]) diluted 1/100. Chemiluminescent detection was achieved using the POD kit according to the manufacturer's protocol (Boehringer).
Cell culture transactivation assays
Transactivation of the hsp23 promoter was assessed by transient transfection of S2 Drosophila cells using different combinations of a reporter vector for hsp23 promoter activity (pCAβM23-0.4) and a normalization vector (pCMV-Su9Luciferase – a derivative of pGEM-Su9Luciferase which encodes for a mitochondrially-targeted luciferase) [58] along with expression vectors driving the expression of different proteins under the control of the constitutive actin promoter (actin-sim, actin-tgo, actin-drf; [37]). 500 ng of each vector (except for the normalization vector – 200 ng) were used for each transfection combination and the total of DNA per transfection reaction was adjusted to 2.2 μg using the empty pCaSpeR-ACT(R) plasmid. Transfection reactions were carried out in 30 mm petri dishes on 2 × 106 S2 cells using the FuGene 6 transfection reagent (Roche). After 36 to 48 hours of expression, cells were lysed in 250 μl Passive Lysis Buffer (PLB – Promega) and the activity of both the Luciferase and βGal reporter proteins were assayed. Luciferase activity, which served to normalize any variation in transfection efficiency, was quantified with the Dual Luciferase Assay kit (Promega) using 10 μl of cell extract. The enzymatic activity of β-galactosidase was measured by a colorimetric assay using the following protocol: 50 μl of cell extract was added to 100 μl of PLB. 150 μl of 2× βGal assay buffer (200 mM NaPO4 pH 7.3, 2 mM MgCl2, 100 mM β-mercaptoethanol, 1.33 mg/ml O-Nitrophenyl-β-D-galactopyranoside (Calbiochem)) was then added and the mixed samples were incubated at 37°C for at least 30 minutes. The reaction was stopped by addition 500 μl of 1 M sodium carbonate and optic density recorded at a wavelength of 420 nm on a spectrophotometer. The reported data constitute an average of triplicates ± s.d.
Authors' contributions
Both authors participated in the conception and design of the study, as well as in drafting the manuscript. SM performed all the experiments. RMT provided the original observation of the cell-specific expression of Hsp23 during embryogenesis.
Acknowledgments
Acknowledgments
We wish to thank G. Boulianne, S. Crews, A. Giangrande, R. Jacobs, H. Krause, J.T. Lis and C. Thummel for providing fly lines and reagents used in this study. We are most grateful to H. Lipshitz and A. Karaiskakis (Toronto) for their help with the generation of transgenic fly lines and to A. Giangrande and members of her laboratory (R. Bernardoni, M. Kammerer and V. Van de Bor) for their useful advice on immunohistochemistry technique. RMT is also particularly thankful to Prof H. Jäckle (Göttingen) in whose lab (Inst. Genetics, München) he originally noted the cell specific expression of Hsp23 while on a sabbatical leave. Finally, we would like to thank G. Morrow and S. Gisselbrecht for critical reading of this manuscript. This study was supported by a Canadian Institute of Health Research operating grant to RMT and a Medical Research Council of Canada doctoral research award to SM.
Contributor Information
Sébastien Michaud, Email: smichau1@genetics.med.harvard.edu.
Robert M Tanguay, Email: robert.tanguay@rsvs.ulaval.ca.
References
- Tissieres A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974;84:389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. Embo J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil HR, Buchner J. Hsp26: a temperature-regulated chaperone. Embo J. 1999;18:6744–6751. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie JN, Gingras-Breton G, Tanguay RM, Landry J. Induction of Chinese hamster HSP27 gene expression in mouse cells confers resistance to heat shock. HSP27 stabilization of the microfilament organization. J Biol Chem. 1993;268:3420–3429. [PubMed] [Google Scholar]
- Bruey JM, Ducasse C, Bonniaud P, Ravagnan L, Susin SA, Diaz-Latoud C, Gurbuxani S, Arrigo AP, Kroemer G, Solary E, Garrido C. Hsp27 negatively regulates cell death by interacting with cytochrome c. Nat Cell Biol. 2000;2:645–652. doi: 10.1038/35023595. [DOI] [PubMed] [Google Scholar]
- Ding L, Candido EP. HSP25, a small heat shock protein associated with dense bodies and M-lines of body wall muscle in Caenorhabditis elegans. J Biol Chem. 2000;275:9510–9517. doi: 10.1074/jbc.275.13.9510. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Wolfner MF, Lis JT. Spatial and temporal pattern of hsp26 expression during normal development. Embo J. 1986;5:747–754. doi: 10.1002/j.1460-2075.1986.tb04277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin R, Valet JP, Tanguay RM. hsp23 and hsp26 exhibit distinct spatial and temporal patterns of constitutive expression in Drosophila adults. Dev Genet. 1993;14:69–77. doi: 10.1002/dvg.1020140109. [DOI] [PubMed] [Google Scholar]
- Michaud S, Marin R, Westwood JT, Tanguay RM. Cell-specific expression and heat-shock induction of Hsps during spermatogenesis in Drosophila melanogaster. J Cell Sci. 1997;110:1989–1997. doi: 10.1242/jcs.110.17.1989. [DOI] [PubMed] [Google Scholar]
- Lang L, Miskovic D, Fernando P, Heikkila JJ. Spatial pattern of constitutive and heat shock-induced expression of the small heat shock protein gene family, Hsp30, in Xenopus laevis tailbud embryos. Dev Genet. 1999;25:365–374. doi: 10.1002/(SICI)1520-6408(1999)25:4<365::AID-DVG10>3.3.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Gernold M, Knauf U, Gaestel M, Stahl J, Kloetzel PM. Development and tissue-specific distribution of mouse small heat shock protein hsp25. Dev Genet. 1993;14:103–111. doi: 10.1002/dvg.1020140204. [DOI] [PubMed] [Google Scholar]
- Armstrong CL, Krueger-Naug AM, Currie RW, Hawkes R. Constitutive expression of heat shock protein HSP25 in the central nervous system of the developing and adult mouse. J Comp Neurol. 2001;434:262–274. doi: 10.1002/cne.1176. [DOI] [PubMed] [Google Scholar]
- Tanguay RM, Wu Y, Khandjian EW. Tissue-specific expression of heat shock proteins of the mouse in the absence of stress. Dev Genet. 1993;14:112–118. doi: 10.1002/dvg.1020140205. [DOI] [PubMed] [Google Scholar]
- Jantschitsch C, Kindas-Mugge I, Metze D, Amann G, Micksche M, Trautinger F. Expression of the small heat shock protein HSP 27 in developing human skin. Br J Dermatol. 1998;139:247–253. doi: 10.1046/j.1365-2133.1998.02361.x. [DOI] [PubMed] [Google Scholar]
- Cutforth T, Rubin GM. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- Eddy EM. Role of heat shock protein HSP70-2 in spermatogenesis. Rev Reprod. 1999;4:23–30. doi: 10.1530/revreprod/4.1.23. [DOI] [PubMed] [Google Scholar]
- Yue L, Karr TL, Nathan DF, Swift H, Srinivasan S, Lindquist S. Genetic analysis of viable Hsp90 alleles reveals a critical role in Drosophila spermatogenesis. Genetics. 1999;151:1065–1079. doi: 10.1093/genetics/151.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timakov B, Zhang P. The hsp60B gene of Drosophila melanogaster is essential for the spermatid individualization process. Cell Stress Chaperones. 2001;6:71–77. doi: 10.1379/1466-1268(2001)006<0071:THGODM>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzik-Dumke U, Lohmann E. Sequence of the new Drosophila melanogaster small heat-shock-related gene, lethal(2) essential for life [l(2)efl], at locus 59F4,5. Gene. 1995;154:171–175. doi: 10.1016/0378-1119(94)00827-F. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Morange M. Hsp25 and the p38 MAPK pathway are involved in differentiation of cardiomyocytes. Dev Biol. 2000;218:146–160. doi: 10.1006/dbio.1999.9596. [DOI] [PubMed] [Google Scholar]
- Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones? Prog Mol Subcell Biol. 2002;28:79–101. doi: 10.1007/978-3-642-56348-5_5. [DOI] [PubMed] [Google Scholar]
- Tanguay R. M., Joanisse, D. R., Inaguma, Y., and Michaud, S. Small heat shock proteins: in search of functions in vivo. In: K B Storey, editor. Environmental Stress and Gene Regulation. Oxford, BIOS Scientific Publishers Ltd.; 1999. pp. 125–138. [Google Scholar]
- Marin R, Tanguay RM. Stage-specific localization of the small heat shock protein Hsp27 during oogenesis in Drosophila melanogaster. Chromosoma. 1996;105:142–149. doi: 10.1007/s004120050169. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Lis JT. Multiple, compensatory regulatory elements specify spermatocyte-specific expression of the Drosophila melanogaster hsp26 gene. Mol Cell Biol. 1990;10:131–137. doi: 10.1128/mcb.10.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterns of gene expression in Drosophila embryogenesis http://www.fruitfly.org/cgi-bin/ex/insitu.pl
- Arrigo AP, Tanguay RM. Expression of heat shock proteins during development in Drosophila. Results Probl Cell Differ. 1991;17:106–119. doi: 10.1007/978-3-540-46712-0_8. [DOI] [PubMed] [Google Scholar]
- Haass C, Klein U, Kloetzel PM. Developmental expression of Drosophila melanogaster small heat-shock proteins. J Cell Sci. 1990;96:413–418. doi: 10.1242/jcs.96.3.413. [DOI] [PubMed] [Google Scholar]
- Egger B, Leemans R, Loop T, Kammermeier L, Fan Y, Radimerski T, Strahm MC, Certa U, Reichert H. Gliogenesis in Drosophila: genome-wide analysis of downstream genes of glial cells missing in the embryonic nervous system. Development. 2002;129:3295–3309. doi: 10.1242/dev.129.14.3295. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton K. A., Jr., Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Hiromi Y, Gehring WJ, Goodman CS. Expression and function of the segmentation gene fushi tarazu during Drosophila neurogenesis. Science. 1988;239:170–175. doi: 10.1126/science.2892267. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Venkatesh TR, Teplow DB, Benzer S. Neuronal development in the Drosophila retina: monoclonal antibodies as molecular probes. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]
- Klambt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-W. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Jr., Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- Ma Y, Certel K, Gao Y, Niemitz E, Mosher J, Mukherjee A, Mutsuddi M, Huseinovic N, Crews ST, Johnson WA, Nambu JR. Functional interactions between Drosophila bHLH/PAS, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. J Neurosci. 2000;20:4596–4605. doi: 10.1523/JNEUROSCI.20-12-04596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkeley Drosophila Genome Project http://www.fruitfly.org/
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, WoodageT. Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld MJ, Jacobs JR. Mesectodermal cell fate analysis in Drosophila midline mutants. Mech Dev. 1994;46:3–13. doi: 10.1016/0925-4773(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Soriano NS, Russell S. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- Zhao G, Skeath JB. The Sox-domain containing gene Dichaete/fish-hook acts in concert with vnd and ind to regulate cell fate in the Drosophila neuroectoderm. Development. 2002;129:1165–1174. doi: 10.1242/dev.129.5.1165. [DOI] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Morel V, Schweisguth F. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 2000;14:377–388. [PMC free article] [PubMed] [Google Scholar]
- King V, Tower J. Aging-specific expression of Drosophila hsp22. Dev Biol. 1999;207:107–118. doi: 10.1006/dbio.1998.9147. [DOI] [PubMed] [Google Scholar]
- Kurapati R, Passananti HB, Rose MR, Tower J. Increased hsp22 RNA levels in Drosophila lines genetically selected for increased longevity. J Gerontol A Biol Sci Med Sci. 2000;55:B552–9. doi: 10.1093/gerona/55.11.b552. [DOI] [PubMed] [Google Scholar]
- Karunanithi S, Barclay JW, Brown IR, Robertson RM, Atwood HL. Enhancement of presynaptic performance in transgenic Drosophila overexpressing heat shock protein Hsp70. Synapse. 2002;44:8–14. doi: 10.1002/syn.10048. [DOI] [PubMed] [Google Scholar]
- Karunanithi S, Barclay JW, Robertson RM, Brown IR, Atwood HL. Neuroprotection at Drosophila synapses conferred by prior heat shock. J Neurosci. 1999;19:4360–4369. doi: 10.1523/JNEUROSCI.19-11-04360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong KH, Ogashiwa T, Matsuo T, Fuyama Y, Aigaki T. Application of the gene search system to screen for longevity genes in Drosophila. Biogerontology. 2001;2:209–217. doi: 10.1023/A:1011517325711. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM. heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 1996;10:3003–3017. doi: 10.1101/gad.10.23.3003. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Nicole LM, Tanguay RM. On the specificity of antisense RNA to arrest in vitro translation of mRNA coding for Drosophila hsp 23. Biosci Rep. 1987;7:239–246. doi: 10.1007/BF01124795. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Steller H, Pirrotta V. P transposons controlled by the heat shock promoter. Mol Cell Biol. 1986;6:1640–1649. doi: 10.1128/mcb.6.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Kornberg RD. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975;72:2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyrovska N, Frydman J, Hohfeld J, Hartl FU. Directionality of polypeptide transfer in the mitochondrial pathway of chaperone-mediated protein folding. Biol Chem. 1998;379:301–309. doi: 10.1515/bchm.1998.379.3.301. [DOI] [PubMed] [Google Scholar]