Abstract
Na+/Ca2+ exchanger (NCX) is one of the major mechanisms for removing Ca2+ from the cytosol especially in cardiac myocytes and neurons, where their physiological activities are triggered by an influx of Ca2+. NCX contains a large intracellular loop (NCXIL) that is responsible for regulating NCX activity. Recent evidence has shown that proteins, including kinases and phosphatases, associate with NCX1IL to form a NCX1 macromolecular complex. To search for the molecules that interact with NCX1IL and regulate NCX1 activity, we used the yeast two-hybrid method to screen a human heart cDNA library and found that the C-terminal region of sarcomeric mitochondrial creatine kinase (sMiCK) interacted with NCX1IL. Moreover, both sMiCK and the muscle-type creatine kinase (CKM) coimmunoprecipitated with NCX1 using lysates of cardiacmyocytes and HEK293T cells that transiently expressed NCX1 and various creatine kinases. Both sMiCK and CKM were able to produce a recovery in the decreased NCX1 activity that was lost under energy-compromised conditions. This regulation is mediated through a putative PKC phosphorylation site of sMiCK and CKM. The autophosphorylation and the catalytic activity of sMiCK and CKM are not required for their regulation of NCX1 activity. Our results suggest a novel mechanism for the regulation of NCX1 activity.
Keywords: ATP, Calcium, Calcium Transport, Cardiac Hypertrophy, Creatine, Sodium Calcium Exchange, Creatine Kinase
Introduction
Calcium is known to be involved in many cellular activities, such as muscle contraction, hormone secretion, and neurotransmitter release (1). In cardiac myocytes, the increase of the cytosolic Ca2+ concentration ([Ca2+]i) is mediated by voltage-dependent Ca2+ channels in the plasma membrane and the ryanodine receptors in the endoplasmic reticulum. The Na+/Ca2+ exchanger (NCX)2 and the Ca2+-ATPase in the plasma membrane are responsible for exporting cytosolic Ca2+ out of cells and maintaining the necessary steep Ca2+ gradient across the membrane. It has been shown that the NCX transports 10 to 15 times more Ca2+ than the Ca2+-ATPase in the plasma membrane and therefore plays a major role in Ca2+ transport (2–4). Depending on the membrane potential and the Na+ and Ca2+ gradients across the plasma membrane, NCX can function in either the forward mode (extracellular Na+ in exchange for intracellular Ca2+), which serves well as a Ca2+ efflux mechanism under physiological conditions, or in the reverse mode (extracellular Ca2+ in exchange for intracellular Na+), which may be responsible for the damage that occurs in cardiac myocytes under pathological conditions such as during ischemia and heart failure (5–7).
Three mammalian NCX genes, NCX1, NCX2, and NCX3, have been identified (8–10). NCX1 is present in almost every tissue and is expressed at high levels in the heart, brain, and kidneys (11). NCX1 has 938 amino acids and has been modeled to contain 9 transmembrane segments (12). A large intracellular loop of NCX (NCXIL), which is located between the fifth and sixth transmembrane segments, is critical for the regulation of NCX activity by Na+, Ca2+, H+, ATP and phosphatidylinositol diphosphate (13). Several proteins, including protein kinase A (PKA), protein kinase C (PKC), muscle protein kinase A-anchoring protein, and the phosphatases PP1/PP2A, have been shown to exist within the NCX1 macromolecular complex (14).
Several lines of evidence suggest that the interaction of NCX with other molecules is closely associated with its physiological function. It has been shown that 14-3-3 protein interacts with the three NCX isoforms and inhibits NCX activity (15). In smooth muscle cells, NCX co-localizes with Na+/K+-ATPase in regions closed to the sarcoplasmic reticulum that store Ca2+, which suggests a connection between NCX with its associated Na+/K+-ATPase and Ca2+ release from the sarcoplasmic reticulum (16). Our previous results from a functional study of NCX1 in relation to the regulation of [Ca2+]i and catecholamine secretion in bovine chromaffin cells suggested that there is a close association between the exchanger, the Ca2+ channels, the Ca2+ pump in the endoplasmic reticulum, the mitochondria, and the exocytotic sites (17).
In this study, to further understand the regulation of the NCX1 activity, we used a yeast two-hybrid screening to search for molecules that interact with NCX1 and found that sarcomeric mitochondrial creatine kinase (sMiCK) interacts with NCX1IL. In addition to sMiCK, our results also showed that the cytoplasmic muscle-type CK (CKM) is able to interact with NCX1 in mammalian cells. In this study we provide evidence to support a novel mechanism for the regulation of NCX activity involving sMiCK and CKM. This is also a new and novel function for creatine kinase.
EXPERIMENTAL PROCEDURES
Animals and Reagents
Eight-week-old C57BL/6 mice were obtained from the animal facility of National Yang-Ming University. Handling of the animals was according to the University guidelines and was approved by the National Yang-Ming University Animal Care and Use Committee. The chemicals used in cell culture were purchased from GIBCO/BRL (Grand Island, NY). dATP, dCTP, dGTP, dTTP, and the protease inhibitor mixture were obtained from Roche (Mannheim, Germany). Vent® DNA polymerase and various restriction enzymes were purchased from New England Biolabs (Beverly, MA). The primers used for PCR were synthesized by Mission Biotech (Taipei, R.O.C.). All other chemicals, unless otherwise specified, were purchased from Sigma.
Vector Construction
To construct pBTM116- NCX1IL for the yeast two-hybrid analysis and pET30-NCX1IL for the GST pull-down assay, the intracellular loop fragment of bovine cardiac NCX1 (18–19) (amino acids 218–737) was amplified and cloned into the pBTM116 and pET30 vectors, respectively. In addition, various deletion mutants of NCX1IL were cloned into pBTM116. A cDNA encoding the full-length protein of sMiCK in pBluescript II SK was obtained from Dr. K. Peck (Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan, R.O.C.), and the sMiCK fragment was amplified by PCR to give pACT2-sMiCK for the yeast two-hybrid analysis and pGEX-4T1-sMiCK for the GST pull-down assay.
uMiCK and CKB were cloned from human brain poly(A)+ RNA (Clontech Laboratories, Mountain View, CA) and CKM was cloned from human heart poly(A)+ RNA (Clontech Laboratories) by reverse transcription- polymerase chain reaction (RT-PCR). The uMiCK(225–378), CKB(231–381), and CKM(231–381) were amplified with various appropriate primer pairs by PCR and were cloned into the pACT2 vector. To construct plasmids for transfection into HEK293T cells, wild-type and various CK mutants were cloned into the pEF1α-Myc/His vector, and NCX1 was cloned into the pEF1α-Flag vector, which was modified from pEF1α-Myc/His. All constructs were verified by sequencing.
Antibodies
pET30-NCX1IL and pGEX-4T1 -sMiCK were transformed into Escherichia coli, and the NCX1IL and GST-sMiCK proteins were purified and used to generate rabbit anti-NCX1IL and anti-GST-sMiCK antisera. The antibodies generated against NCXIL recognizes the full length NCX1 overexpressed in HEK293T cells, which have no detectable endogenous NCX, and endogenous mouse cardiac NCX1 (supplemental Fig. S1). Mouse anti-Myc (9E10, 1:10000) and anti-β-tubulin (E7, 1:10,000) was obtained from the Developmental Studies Hybridoma Bank (University of Iowa). Rabbit anti-CKM antiserum was purchased from Abgent (San Diego, CA). Rabbit anti-MnSOD antiserum was obtained from Upstate Biotechnology (Lake Placid, NY). Rabbit anti-Myc antiserum and other secondary antisera were purchased from Bethyl Laboratories (Montgomery, TX). The goat-anti-rabbit Alexa Fluor 555 was purchased from Molecular Probes (Invitrogen) and donkey anti-mouse IgG-FITC was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Yeast Two-hybrid Screening and Assay for β-Galactosidase
Yeast two-hybrid screening was employed to search for NCX1IL-interacting proteins (20). A human heart cDNA library constructed using pACT2 (Clontech Laboratories) was used as the prey and the plasmid pBTM116-NCX1IL was used as the bait. pBTM116-NCX1IL and the human heart cDNA library were sequentially transformed into yeast strain L40 (MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS:: (lexAop)4-HIS URA3:: (lexAop)8-lacZ GAL4 gal80) by a high efficiency method (21). Approximately 5 × 106 independent cDNA clones were screened, and positive clones were selected on synthetic complete medium lacking lysine, leucine, uracil, tryptophan, and histidine (-KLUTH) plates. Double transformants were grown in -KLUT medium, and the β-galactosidase activity was determined by measuring the cleavage of o-nitrophenyl-β-d-galactopyraniside (ONPG) solution (4 mg/ml ONPG in Z buffer). The A420 was measured to calculate the activity in Miller units (units = [A420 × 1000]/[V (ml) × T(min) × A600]). For the X-gal filter lift assay, yeast colonies were grown on the -KLUT plates and transferred from plates to filters to be overlaid with X-gal.
GST Pull-down Assay
Equal amounts of GST or GST-sMiCK were immobilized on glutathione- agarose beads, which were then incubated with NCX1IL in a GST pull-down buffer (100 mm NaCl, 50 mm MgCl2, 10% glycerol, 1% Nonidet P-40, 0.2% bovine serum albumin (w/v), protease inhibitors, and 25 mm HEPES, pH 7.5) on a rotator at 4 °C. After washing with GST pull-down buffer, the bound proteins were eluted by glutathione buffer (10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0) and analyzed by Western blotting using the indicated antibodies.
Cell Culture and Transfection
HEK293T cells were cultured at 37 °C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in a 5% CO2 incubator and then were transfected with the various constructs using Lipofectamine 2000 (Invitrogen) modified from the manufacturer's instructions. The medium was changed 6 h after transfection. For the co-immunoprecipitation experiments, HEK293T cells were cultured on 10-cm cell culture dishes (6 × 105 cells/dish) and co-transfected with 5 μg of pNCX1 and 5 μg of CK isozymes plasmids as indicated. For measuring the reverse-mode NCX1 activity, HEK293T cells were cultured on poly-l-lysine-coated 24 mm cover glasses (1 × 105 cells/cover glass) and were co-transfected with 0.1 μg of pGFP, 1.2 μg of pNCX1 and 0.4 μg of empty vector or 0.4 μg of the indicated CK isozymes plasmids. For immunocytochemical staining, HEK293T cells were grown on poly-l-lysine-coated 24-mm cover glasses (1 × 105 cells/cover glass) and co- transfected with 0.5 μg of pNCX1 and 0.5 μg of the indicated CK isozymes plasmids.
Protein Extraction, Immunoprecipitation, and Western Blot Analysis
One day after transfection, HEK293T cells were washed three times with phosphate-buffered saline (PBS) (136 mm NaCl, 1.76 mm KH2PO4, 2.68 mm KCl, and 8 mm Na2HPO4, pH 7.4) and resuspended in RIPA lysis buffer (150 mm NaCl, 1 mm EDTA, 0.1% Triton X-100, 50 mm NaF, 1 mm dithiothreitol, protease inhibitors, and 20 mm HEPES, pH 7.8). The cell lysates were cleared by centrifuging at 13,000 × g for 10 min at 4 °C, and then the supernatants were used for Western blotting analysis or immunoprecipitation. The protein concentration of each sample was determined using a BCA kit (Pierce).
Dissected C57BL/6 mouse ventricles were rinsed with ice-cold HBSS, pulverized in liquid nitrogen, and then homogenized in a glass homogenizer with extraction buffer (150 mm NaCl, 3 mm KCl, 5 mm EDTA, protease inhibitors, and 20 mm HEPES, pH 7.4). The lysate was centrifuged at 1,000 × g for 15 min at 4 °C. The supernatant was homogenized again on ice and then centrifuged at 5,000 × g for 10 min at 4 °C. The supernatant was then used for the immunoprecipitation experiments. To obtain the mitochondrial fraction and the membrane fraction of the cardiac myocytes, the pulverized ventricles were homogenized in the homogenization buffer (150 mm NaCl, 5 mm EDTA, 250 mm sucrose, protease inhibitors, and 20 mm HEPES, pH 7.8). The lysate was centrifuged at 10,000 × g for 15 min at 4 °C to obtain the mitochondrial fraction, and then the supernatant was centrifuged at 100,000 × g for 1 h at 4 °C to obtain the membrane fraction. The pellets were washed with homogenization buffer three times and then resuspended in RIPA lysis buffer.
Immunoprecipitation was performed by incubating the indicated primary antibodies with 30 μl protein A/G beads (Pierce) for 1 h at 4 °C, then the beads were washed with 1 ml of RIPA buffer at 4 °C three times. Detergent-extracted total protein (3 mg) in a reaction volume of 1 ml was then added and the mixture agitated on a rotor at 4 °C overnight. Next, the samples were spun down by low-speed centrifugation and washed five times with RIPA buffer. For Western blotting analysis, the tissue and cell lysate were subjected to SDS-PAGE and transferred to PVDF membranes. The membrane was blocked with PBS containing 0.3% skim milk and 0.5% bovine serum albumin for 1 h at room temperature and probed with the relevant primary antibodies at 4 °C overnight. After the membrane was washed and incubated with HRP-conjugated secondary antibodies at room temperature for 2 h, it was developed with an ECL kit (Amersham Biosciences, Buckinghamshire, England).
Immunocytostaining and Fluorescence Microscopy
One day after transfection, HEK293T cells grown on cover slips were rinsed three times with PBS and then fixed with 3.7% formaldehyde in PBS for 20 min at room temperature. The samples were stained with appropriate primary antibodies and then incubated with rhodamine-conjugated goat anti-rabbit antiserum or FITC-conjugated goat anti-mouse antiserum at 4 °C overnight. The cover slips were mounted with Vectashield (Vector Laboratories, Burlingame, CA) and viewed under an inverted confocal microscope (Zeiss Axiovert 200 m, Carl Zeiss, Jena, Germany) with a 63×/1.4 Plan-Apochromat objective.
Measurement of Reverse Mode NCX1 Activity
One day after transfection, HEK293T cells were treated with 5 μg/ml oligomycin and 2 mm 2-deoxyglucose (2-DG) in a loading buffer (LB) (145 mm NaCl, 5 mm KCl, 1 mm MgCl2, 2 mm CaCl2, and 10 mm HEPES, pH 7.4) for 10 min at 37 °C to induce the energy-compromised conditions. After washing three times with LB, the cells were incubated with 5 μm Fura-2 AM (Molecular Probes) for 20 min at 37 °C. Fura-2-loaded cells were washed three times with LB and then loaded with Na+ by incubating in a Ca2+-free Na+ loading buffer (145 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm ouabain, 10 μm monensin, and 10 mm HEPES, pH 7.4) for 10 min at 37 °C. GFP-positive cells were selected for the measurement of the reverse-mode NCX1 activity by puffing them with Na+-free buffer (145 mm N-methylglucamine, 5 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 1 mm ouabain, and 10 mm HEPES, pH 7.4) for 30 s. The Fura-2 fluorescence ratio obtained by 340 nm and 380 nm excitation (F340/F380) was monitored with an inverted microscope (IX-70, Olympus Co., Tokyo, Japan) using a 40× oil immersion objective (UAPO 40× oil/340; N/A: 1.35; Olympus Corporation, Tokyo, Japan). [Ca2+]i was calculated as previously described (22–23). The amplitudes and the apparent initial rates of the [Ca2+]i rise upon puffing the cells with Na+-free solution for 30 s were then calculated and taken as a measure of the reverse-mode NCX activity.
ATP Assay
ATP levels were assayed with an ATP bioluminescent assay kit (Sigma). Luminescence units were then measured using a Perkin Elmer Victor 2 multilabel counter.
Statistical Analysis
Data are presented are mean ± S.E., and the results were analyzed by one-way ANOVA. Statistical significance is set at p < 0.05.
RESULTS
sMiCK Interacts with NCX1IL
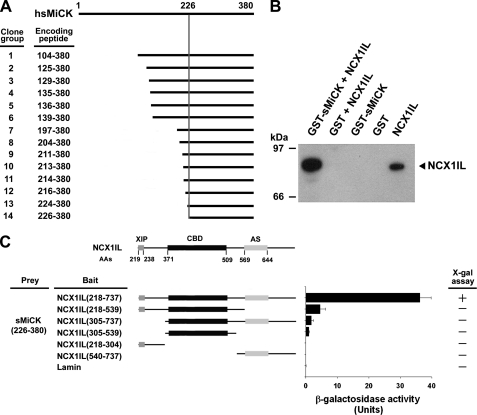
To search for proteins that interact with NCX1, yeast two-hybrid screening of a human heart cDNA library was carried out using NCX1IL (amino acids 218–737) as the bait. Approximately 5 × 106 transformants were screened. Among the 52 positive clones obtained from the yeast-two hybrid screening, 32 clones were identified to be sMiCK. The DNA sequences of these sMiCK clones were analyzed and separated into 14 different groups. The shortest region of sMiCK cDNA that contributed to the interaction between sMiCK and NCX1IL was the C terminus of sMiCK corresponding to amino acids 226–380 (Fig. 1A). The interaction between full-length sMiCK and NCX1IL was then examined by GST pull-down assay. Plasmids encoding NCX1IL and GST-sMiCK were transformed into E. coli and were incubated with glutathione-agarose beads. The results from Western blot analysis showed that NCX1IL was pulled down with GST-sMiCK (Fig. 1B).
FIGURE 1.
sMiCK is a candidate molecule that interacts with NCX1IL. A, schematic representation of the human sMiCK clones identified using yeast-two hybrid screening. The shortest region of sMiCK that is able to contribute to the interaction between sMiCK and NCX1IL is located at C terminus from amino acids 226–380. B, interaction of sMiCK with NCX1IL was analyzed by GST pull-down assay. NCX1IL was incubated with GST-sMiCK or GST immobilized on glutathione-agarose beads. The samples were then subjected to pull down and Western blot analysis was carried out with mouse anti-NCX1IL antibodies. C, interaction of various NCXIL deletion mutants with C terminus of sMiCK. Left panel: schematic diagram of the constructs of NCX1IL with various domains deleted, including the exchanger inhibitory peptide (XIP), Ca2+ binding domain (CBD) and alternative splicing region (AS). Right panel: results of β-galactosidase activity assay and X-gal filter lift assay (+, positive binding; −, negative binding).
NCX1IL was separated into the exchanger inhibitory peptide (XIP), Ca2+ binding domain and alternative splicing regions (24–26). To identify the regions in NCX1IL that may be responsible for interacting with sMiCK, various constructs containing these different regions of NCX1IL were constructed and transformed into yeast. The interactions between the different regions of NCX1IL and sMiCK were then examined by yeast two-hybrid assay, and the results show that the entire NCX1IL was required for the interaction (Fig. 1C).
Interaction between NCX1 and Various CK Isozymes
There are four creatine kinase (CK) isozymes in mammals, ubiquitous mitochondrial CK (uMiCK), sarcomeric mitochondrial CK (sMiCK), cytoplasmic brain type CK (CKB), and cytoplasmic muscle type CK (CKM); these isozymes are found in different tissues and have different subcellular localizations. The CK isozymes are important for the maintenance of cellular energy homeostasis; they catalyze the reversible transfer of a phosphate group from phosphocreatine to ADP to yield ATP and creatine. The human sMiCK shares 83% identity with uMiCK, and about 66% identity with the cytosolic CKB and CKM. sMiCK(226–380) shares 83% identity with uMiCK(225–378) and have about 68% identity with CKB(231–381) and CKM(231–381) (supplemental Fig. S2). To examine the interaction of NCX1 and the four CK isozymes in mammalian cells, full-length NCX1 and various CK isozymes were co-expressed in HEK293T cells and the co-immunoprecipitation experiments were performed. The results show that, in addition to sMiCK, CKM also immunoprecipitated with NCX1, apparently giving the highest amount of precipitate among the four isozymes (Fig. 2A). Furthermore, the interactions between NCX1 and sMiCK and CKM in the lysate from mouse ventricular myocytes were examined by co-immunoprecipitation with rabbit anti-NCX1 antiserum (Fig. 2B). The results show that both sMiCK and CKM in the mouse ventricular myocytes co-immunoprecipitated with NCX1.
FIGURE 2.
The interaction of NCX1 and CK isozymes in HEK293T cells and mouse cardiac myocytes. A, empty vector, puMiCK, psMiCK, pCKM, and pCKB (Myc-tagged) were individually co-transfected with pNCX1 into HEK293T cells. The interactions between NCX1 and the CK isozymes were analyzed by co-immunoprecipitation using mouse anti-Myc antibody. Western blot analysis was carried out with rabbit anti-NCX1IL antibodies and rabbit anti-Myc antibodies. B, interaction between NCX1 and sMiCK in mouse heart extract was analyzed by co-immunoprecipitation using rabbit anti-NCX1IL antibodies. Western blot analysis was carried out by rabbit anti-NCX1IL antibodies and rabbit anti-sMiCK antibodies. C, interaction of NCX1 and CKM in mouse heart extract was analyzed by co-immunoprecipitation using rabbit anti-NCX1 antibodies and then Western blotting using rabbit anti-NCX1IL antibodies and rabbit anti-CKM antibodies.
It is curious that in the above yeast two-hybrid assay only sMiCK and not CKM was found to interact with NCX1IL (Fig. 1A). To explore this, we then constructed plasmids encoding the C terminus of uMiCK(225–378), CKB(231–381), and CKM(231–381) and co-transform these together with NCX1IL into yeast. The results show that uMiCK(225–378), CKB(231–381), and CKM(231–381) do not interact with NCX1IL when examined by serial dilution on selective plates or by β-galactosidase activity assay (supplemental Fig. S3). The results suggest that despite the high sequence similarity among the various CK isoforms, the interaction between NCX and CK is highly specific for sMiCK in the yeast two-hybrid assay.
Effect of CK on NCX1 Activity under Energy-compromised Conditions
We next studied the physiological significance of the interactions between NCX1 and CK. CK is known to be important for the maintenance of cellular energy homeostasis. It has been shown that NCX activity is stimulated by ATP (27) and is inhibited under an ATP-depleted condition in rat cardiomyocytes (28) and in Chinese hamster ovary cells that stably expressed bovine cardiac NCX (29). The effects of CK on the NCX1 activity were therefore examined under energy-compromised conditions, which were induced by treating the cells with oligomycin and 2-DG. The cellular ATP concentration in the HEK293T cells was decreased by at least 60% compared with that under control conditions (supplemental Table S1).
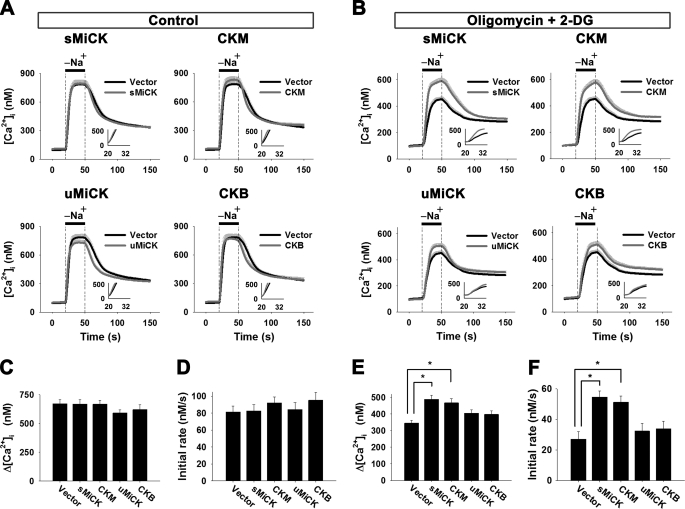
Both NCX1 and CK were overexpressed in HEK293T cells and then the reverse-mode NCX1 activity was measured by preloading the cells with Na+ in the absence of extracellular Ca2+, and the amplitude and the apparent initial rates of the [Ca2+]i rise upon puffing Na+-free solution were taken as a measure of the activity. Note that HEK293T cells do not express NCX1 protein and have no detectable endogenous NCX activity (supplemental Figs. S1B and S4). Under control conditions, co-expression of the four CK isozymes with NCX1 did not affect NCX1 activity (Fig. 3, A, C, D). Under the energy-compromised conditions, NCX1 activity was significantly decreased; the amplitude and the initial rate were decreased to 50 and 35% of the control cells, respectively (Fig. 3, A versus B). Interestingly, there was a significant increase in the NCX1 activity in the presence of sMiCK or CKM, but not in the presence of uMiCK or CKB, using co-transfected cells (Fig. 3, B, E, and F). The amplitude and the initial rate were recovered to 73 and 67% of the control cells in the sMiCK co-transfected cells and to 70 and 63% when co-transfected CKM cells were examined. The results show that the two CK isozymes, sMiCK and CKM, which interact with NCX, can produce a part recovery of the decreased reverse-mode NCX1 activity that occurs under energy-compromised conditions.
FIGURE 3.
sMiCK and CKM maintain NCX1 activity under energy-compromised conditions. HEK293T cells were co-transfected with pGFP, pNCX1, and plasmids encoding various CK isozymes as indicated. Cells with GFP fluorescence were selected to measure the reverse-mode NCX activity by puffing with a Na+-free buffer for 30 s as indicated. The NCX activity was estimated by the amplitude (Δ[Ca2+]i = peak [Ca2+]i − basal [Ca2+]i) and the apparent initial rate (insets) of the [Ca2+]i changes upon puffing of Na+-free buffer. A, C, D, reverse-mode NCX activity under control conditions (empty vector, n = 25; uMiCK, n = 24; sMiCK, n = 26; CKB, n = 28 and CKM n = 28 cells). B, E, F, reverse-mode NCX activity under energy compromised conditions induced by treatment of oligomycin and 2-DG (empty vector, n = 25; uMiCK, n = 31; sMiCK, n = 32; CKB, n = 28; and CKM, n = 25 cells). Data are mean ± S.E. from three individual experiments. *, p < 0.05.
The C Terminus of sMiCK and CKM Is Required for the Regulation of NCX1 Activity
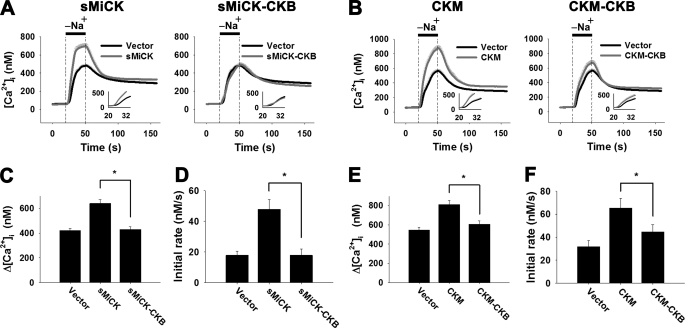
To further study whether the C termini of sMiCK and CKM are responsible for regulation of NCX1 activity, the C terminus of sMiCK and CKM were replaced by the corresponding region of CKB, amino acids 231–381. The chimeric CK, sMiCK-CKB and CKM-CKB, were co-expressed with NCX1 in the HEK293T cells. The NCX1 activity in the sMiCK-CKB or CKM-CKB co-expressing cells was similar to that in the control empty vector transfected cells under energy-compromised conditions (Fig. 4, A and B); this is shown by the similar amplitudes and the similar apparent initial rates for the [Ca2+]i changes upon puffing the cells with Na+-free solution (Fig. 4, C–F). The results demonstrate that the C termini of sMiCK and CKM are critical for the recovery of decreased NCX1 activity under the energy-compromised conditions.
FIGURE 4.
Chimeric sMiCK-CKB and CKM-CKB have no effects on the NCX1 activity under energy-compromised conditions. HEK293T cells were co-transfected with pGFP, pNCX1, and empty vector, psMiCK, psMiCK-CKB, pCKM or pCKM-CKB as indicated. After treatment with oligomycin and 2-DG, cells with GFP fluorescence were selected for measuring the reverse-mode NCX activity by puffing with a Na+-free buffer for 30 s, as indicated. The NCX activity was estimated by the amplitude (Δ[Ca2+]i = peak [Ca2+]i − basal [Ca2+]i) and the apparent initial rate (insets) of the [Ca2+]i changes upon puffing of Na+-free buffer. Experiments using cells overexpressing wild-type and chimeric CKs were performed in parallel. A, C, D, NCX activity in cells overexpressed wild-type sMiCK or sMiCK-CKB (empty vector, n = 28; sMiCK, n = 27; sMiCK-CKB, n = 24 cells). B, E, F, NCX activity in cells overexpressed wild-type CKM or CKM-CKB (empty vector, n = 37; CKM, n = 27; CKM-CKB, n = 33 cells). Data are mean ± S.E. from three individual experiments. *, p < 0.05.
Subcellular Localization of the NCX1 and CK Isozymes
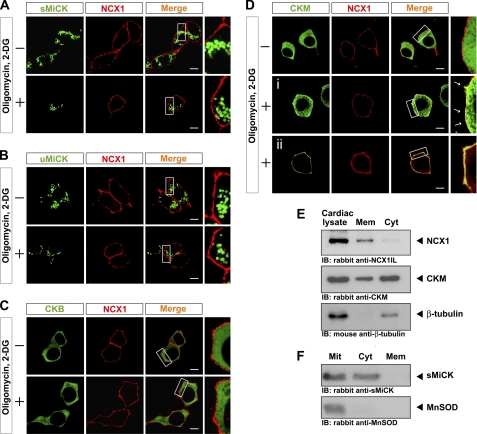
Because mitochondrial sMiCK and cytosolic CKM can recover reverse-mode NCX1 activity under energy-compromised conditions, the subcellular localizations of NCX1 and CKs were then examined. The subcellular localization of NCX1 and the four CK isozymes were as expected; NCX1 was localized to the plasma membrane, uMiCK and sMiCK were localized to the mitochondria (Fig. 5, A and B) based on the fact that they co-localized with the mitochondrial marker MitoTracker orange (data not shown), and CKM and CKB were distributed in the cytosol (Fig. 5, C and D).
FIGURE 5.
Subcellular localization of NCX1 and the CK isozymes in HEK293T cells under control and energy-compromised conditions. NCX1 and the four CK isozymes were transiently expressed in HEK293T cells, which were treated with (+) or without (−) oligomycin and 2-DG. Immunocytochemistry was performed with rabbit anti-NCX1IL antibodies and mouse anti-Myc antibody. The subcellular localization of NCX1 (red) and the four CK isozymes (green), (A) uMiCK, (B) sMiCK, (C) CKB, and (D) CKM, were examined by confocal microscopy. D, i and ii, two different patterns of CKM distribution under the energy-compromised conditions. Scale bars are 5 μm in all images. E, presence of NCX1 and CKM in membrane fraction (Mem) and cytosolic fraction (Cyt) of ventricular cardiac myocytes. F, presence of sMiCK and MnSOD in mitochondrial fraction (Mit), cytosolic fraction (Cyt), and membrane fraction (Mem) isolated from ventricular cardiac myocytes.
Under energy-compromised conditions, the subcellular localizations of NCX1, uMiCK, sMiCK, and CKB show a similar pattern to that found under the control conditions. Interestingly, under the energy-compromised conditions, CKM was recruited to the plasma membrane and co-localized with NCX1 (Fig. 5D). Most cells showed preferential localization of CKM to the plasma membrane (Fig. 5D, i), but a few cells (∼10%) showed predominant localization of CKM to the plasma membrane (Fig. 5D, ii).
The subcellular localizations of NCX1, sMiCK, and CKM were further examined in the membrane and cytosolic fractions isolated from mouse cardiac tissues. NCX1 was detected only in the membrane fraction and not in the cytosolic fraction, while CKM was present in both membrane and cytosolic fractions (Fig. 5E). The results are compatible with the above immunostaining results (Fig. 5D). Our results were also in accord with previous studies showing that some CKM is found in the plasma membrane of cardiac muscle cells (30–31).
Surprisingly, although not detected in the immunostaining images, sMiCK was detected not only in mitochondrial fraction with the mitochondrial marker MnSOD, but, in addition, the protein was also present in the cytosolic fraction (Fig. 5F). The presence of sMiCK in the cytosol suggests that some sMiCK may be directly interacting with NCX in the plasma membrane.
A Putative PKC Phosphorylation Site on sMiCK and CKM Is Required for the Regulation of NCX1 Activity
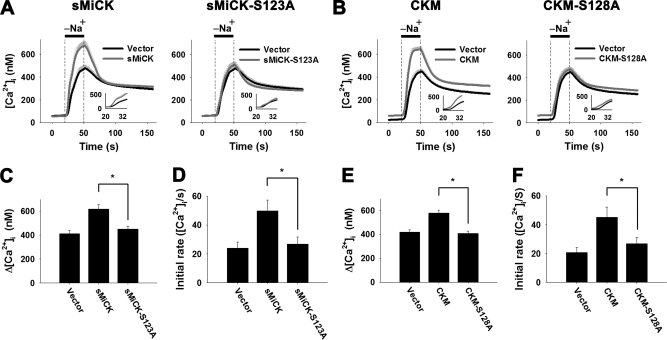
It has been shown that CKM is phosphorylated by PKC, possibly at Ser-128 (32). To examine the role of this serine residue in CK mediated recovery of the decreased NCX1 activity, plasmids encoding the Ser-128 equivalent mutants of sMiCK and CKM, namely sMiCK-S123A and CKM-S128A, were constructed and co-transfected with NCX1 into HEK293T cells. The results show that sMiCK-S123A and CKM-S128A failed to produce a recovery in the decreased NCX1 activity under energy-compromised conditions (Fig. 6, A and B); there was no significant difference in the amplitude and the apparent initial rates of the [Ca2+]i rise between sMiCK-S123A or CKM-S128A transfected cells and the empty vector-transfected cells (Fig. 6, C–F). Results show that the putative PKC phosphorylation site of sMiCK and CKM is critical to their regulation of NCX1 activity.
FIGURE 6.
The putative PKC phosphorylation site of sMiCK and CKM is crucial for the effect of CK on the NCX1 activity. HEK293T cells were co-transfected with pGFP, pNCX1, and plasmids encoding sMiCK-S123A and CKM-S128A, putative PKC phosphorylation site mutants of sMiCK and CKM, respectively. After treatment with oligomycin and 2-DG, cells with GFP fluorescence were selected for measuring the reverse-mode NCX activity by puffing with a Na+-free buffer for 30 s, as indicated. The NCX activity was estimated by the amplitude (Δ[Ca2+]i = peak [Ca2+]i − basal [Ca2+]i) and the apparent initial rate (insets) of the [Ca2+]i changes upon puffing of Na+-free buffer. A, C, D, NCX activity in cells overexpressing wild-type sMiCK or sMiCK-S123A (empty vector, n = 29; sMiCK, n = 35; sMiCK-S123A, n = 29 cells). B, E, F, NCX activity in cells overexpressing wild-type CKM or CKM-S128A (empty vector, n = 27; CKM, n = 27; CKM-S128A, n = 31 cells). Data are mean ± S.E. from three individual experiments. *, p < 0.05.
Creatine Kinase Enzyme Activity Is Not Required for the Regulation of NCX1 Activity
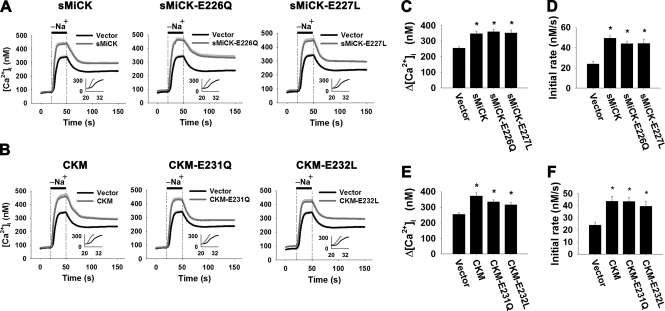
A conserved negatively charged amino acid cluster, EED, located in the active site of CK isozymes, has been shown to be essential for CK enzyme activity (33). To study whether CK enzyme activity is required for the recovery of NCX1 activity under energy-compromised conditions, catalytic site mutants of sMiCK and CKM, namely sMiCK-E226Q, sMiCK-E227L, CKM-E231Q, or CKM-E232L were transiently expressed with NCX1 in HEK293T cells. These catalytic site mutants showed no detectable CK activity (data not shown). All mutants of sMiCK or CKM showed the same ability as the wild-type sMiCK and CKM for the recovery of decreased NCX1 activity under energy- compromised conditions (Fig. 7). It appears that CK catalytic activity is not required for the regulation of NCX1 activity.
FIGURE 7.
The effect of catalytic site mutants of sMiCK and CKM on NCX1 activity under energy-compromised conditions. HEK293T cells were co-transfected with pGFP, pNCX1, and plasmid encoding catalytic site mutants of sMiCK or CKM as indicated. After treatment with oligomycin and 2-DG, cells with GFP fluorescence were selected for measuring the reverse-mode NCX activity by puffing with a Na+-free buffer for 30 s, as indicated. The NCX activity was estimated by the amplitude (Δ[Ca2+]i = peak [Ca2+]i − basal [Ca2+]i) and the apparent initial rate (insets) of the [Ca2+]i changes upon puffing of Na+-free buffer. A, C, D, NCX activity in cells overexpressing wild-type sMiCK, sMiCK-E226Q, or sMiCK-E227L (empty vector, n = 31; sMiCK, n = 27; sMiCK-E226Q, n = 25; sMiCK-E227L, n = 30 cells). B, E, F, NCX activity in cells overexpressing wild-type CKM, CKM-E231Q, or CKM-E232L (empty vector, n = 31; CKM, n = 27; CKM-E231Q, n = 27; CKM-E232L, n = 26 cells). Data are mean ± S.E. from three individual experiments. *, p < 0.05.
Autophosphorylation of sMiCK and CKM Is Not Required for the Regulation of NCX1 Activity
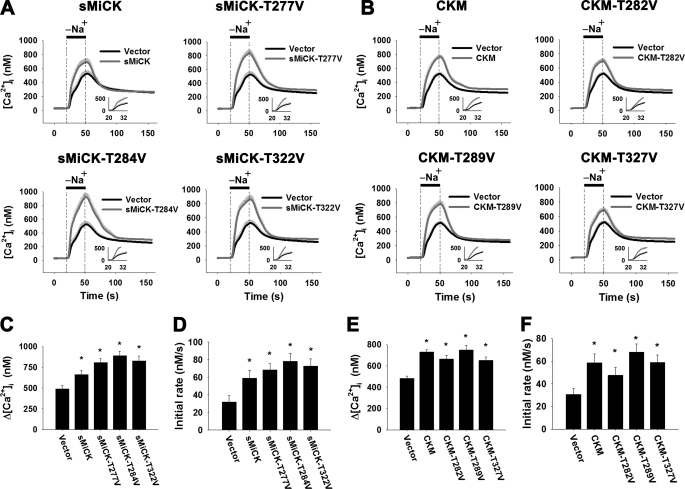
Several threonine residues of CKM have been identified as the autophosphorylation sites (34). In this study, we generated autophosphorylation site mutants of sMiCK and CKM, namely sMiCK-T277V, sMiCK-T284V, sMiCK-T322V, CKM-T282V, CKM-T289V, and CKM-T327V; these mutant proteins were then individually co-expressed with NCX1 in HEK293T cells. These mutants of sMiCK or CKM show similar ability to that of wild-type sMiCK and CKM in terms of the recovery of NCX1 activity (Fig. 8). The results suggest that autophosphorylation of sMiCK and CKM is not involved in the effect of CK on the recovery of NCX1 activity.
FIGURE 8.
The effect of expression of autophosphorylation site mutants of sMiCK and CKM on NCX1 activity under energy-compromised conditions. HEK293T cells were co-transfected with pGFP, pNCX1, and plasmid encoding autophosphorylation site mutants of sMiCK or CKM as indicated. After treatment with oligomycin and 2-DG, cells with GFP fluorescence were selected for measuring the reverse-mode NCX activity by puffing with a Na+-free buffer for 30 s, as indicated. The NCX activity was estimated by the amplitude (Δ[Ca2+]i = peak [Ca2+]i − basal [Ca2+]i) and the apparent initial rate (insets) of the [Ca2+]i changes upon puffing of Na+-free buffer. A, C, D, NCX activity in cells overexpressing wild-type sMiCK, sMiCK-T277V, sMiCK-T284V, or sMiCK-T322V (empty vector, n = 29; sMiCK, n = 29; sMiCK-T277V, n = 37; sMiCK-T284V, n = 33; sMiCK-T322V, n = 33 cells). B, E, F, NCX activity in cells overexpressing wild-type CKM, CKM-T282V, CKM-T289V, or CKM-T327V (empty vector, n = 32; CKM, n = 26; CKM-T282V, n = 33; CKM-T289V, n = 29; CKM-T327V, n = 29 cells). Data are mean ± S.E. from three individual experiments. *, p < 0.05.
DISCUSSION
In this study, we discovered a novel way by which NCX1 activity is regulated that involves two CK isozymes, sMiCK and CKM, which are able to recover the decreased NCX1 activity under energy-compromised conditions. The C-terminal region and the putative PKC phosphorylation Ser residue of sMiCK and CKM are critical for the effect of these CKs on NCX1 activity but catalytic activity and autophosphorylation of sMiCK and CKM are not required.
The C-terminal regions of sMiCK and CKM were found to be responsible for their interaction with NCX1 and the effect on the NCX1 activity. The results are in accord with the three-dimensional structures of CKs, which show that the C-terminal region is located on the outside of the octameric mitochondrial and dimeric cytosolic CK isozymes and that the N-terminal regions of CK isozymes are known to be critical for CK dimerization (human uMiCK: PDB code 1qk1 (35), chicken sMiCK: PDB code 1crk (36), chicken CKB: PDB code 1qh4 (37), and human CKM: PDB code 1i0e (38)). Interestingly, the putative PKC phosphorylation serine residue, which is required for the effects of CK on NCX activity, is located adjacent to the C terminus based on the three-dimensional structure. It is possible that phosphorylation of the serine residue is critical for maintaining the C terminus in a proper position for maintaining NCX activity. Although the C-terminal regions of the four CK isozymes share a high degree of amino acid sequence homology, the effect on the recovery of NCX1 activity is specific for CKM and sMiCK. When their C termini were replaced by that of CKB, the effect on NCX activity was abolished.
The fact that NCX1 is predominantly expressed in cardiac myocytes, where sMiCK and CKM are also present, supports the physiological implications of the interaction between CK and NCX1. NCX is known to play a critical role in the maintenance of Ca2+ homeostasis in cardiomyocytes (39–40) and thus in the modulation of cardiac contractility. It has been shown that NCX activity is altered during cardiac remodeling in the hypertrophic heart and during heart failure (7, 41). The hearts from CKM and sMiCK double knock-out mice have been demonstrated to show increased sensitivity to ischemia-reperfusion injury and show a greater increase in diastolic Ca2+ concentration during ischemia (42); therefore the interaction between NCX1 and CKs may be important to prolonging normal heart functioning when a pathological condition such as ischemia starts to develop. In sMiCK knock-out mice, there are no obvious change in the morphology and functioning of the muscles (43). Nonetheless, a significant change in muscle force generation and cell morphology is found in CKM and sMiCK double knock-out mice (44). Therefore, this supports the hypothesis that there is a specific and direct coupling between the cytosolic and mitochondrial CKs that helps to maintain the normal physiology and functioning of the heart muscles.
sMiCK interacts with NCX1 in yeast, HEK293T cells, and cardiac myocytes and the expression of sMiCK increases the reverse-mode activity of NCX1 under energy compromised conditions; however, it is puzzling that colocalization of sMiCK and NCX1 could not be observed in this study. One possibility is that the amount of sMiCK that interacts with NCX1 is too small to be detected and that sMiCK is highly concentrated in the mitochondria, which renders the detection of cytosolic sMiCK more difficult. It has previously been shown that sMiCK is present in a large proteolipid complex composed of adenine translocator, voltage-dependent anion channels, and cytochrome c in the intermembrane space of mitochondria (45). It is also possible that sMiCK is only released into the cytosol under pathological conditions and that is when it interacts with NCX1.
For CKM, the immunoprecipitation results showed that CKM interacted with NCX1 in HEK293T cells and cardiac myocytes. Moreover, under energy-compromised conditions CKM translocated from the cytosol to the plasma membrane, where it co-localized with NCX1. Moreover, CKM mutants without the catalytic enzyme activity were also able, after energy depletion, to recover the reduced reverse-mode NCX1 activity to the same degree as wild- type CKM. Thus, a direct physical interaction between CKM and NCX1 is able to account for the recovery of the decreased reverse-mode NCX1 activity under the energy-compromised conditions.
CKM has been shown to have a physical interaction with the cardiac ATP-sensitive K+ channels and regulates K+ channels activity by controlling the ATP/ADP ratio near these channels. This finding is based on the results obtained by adding CKM substrates that change K+ channels activity in an excised membrane patch from guinea pig cardiomyocytes (46). The cytosolic brain-type creatine kinase, CKB, has been shown to interact with the K+-Cl− co-transporter, which is important in neurons for the regulation of the resting intracellular Cl− levels (47). Results obtained using a dominant negative CKB mutant and a CK inhibitor support the hypothesis that CKB regulates K+-Cl− co-transporter activity by changing the ATP concentration in the vicinity of the transporter (48). Our results clearly show that the activity of CK is not required for the effect of CK on NCX activity and the evidence supports the idea that phosphorylation of sMiCK at serine 123 and CKM at serine 128, probably by PKC, is critical for the effect of these CKs on NCX. It has been shown that NCX1IL is phosphorylated by PKC (49); however, the regulation of NCX activity by PKC remains controversial. PKC up-regulates NCX1 activity in rat aortic smooth muscle cells (50), neonatal cardiomyocytes (49), and hepatocytes (51). In contrast, PKC down-regulates NCX activity in other cell types such as bovine chromaffin cells (52). It is possible that some as yet unknown factors participate in the regulation of the NCX activity in these different tissues and that this contribute to these contradictory results. In this context, the interaction of NCX1 with CK represents an indirect regulation of NCX activity by PKC. It is possible that CK interacts with NCX via its C terminus under control conditions (Fig. 2), and under energy-compromised conditions CK is phosphorylated by PKC or other kinases, which further induces the C terminus of CK to take up a position for maintaining NCX activity.
In this study, we have made a novel finding that the ATP depletion induced decrease of NCX1 activity could be recovered by co-expressing this exchanger with CKM or sMiCK. Furthermore, the subcellular localization of CKM was changed by ATP depletion, which appears to correlate with the recovery in NCX activity. Our results show not only a novel mechanism for regulation of NCX1 activity by CK, but also a novel function for CK. This regulation is mediated via a putative PKC phosphorylation site of CKM and sMiCK and is independent of the catalytic activity and autophosphorylation of CK. Taken together, our results provide new insights into and an in-depth understanding of the regulation of cardiac physiology.
Supplementary Material
Acknowledgments
We thank Dr. S.-M. Shih (Institute of Biomedical Sciences, Academia Sinica) for help in establishing the yeast two-hybrid screening. We thank Dr. Mei-Yu Chen (National Yang-Ming University, Taipei) for providing the human brain cDNA. We thank Dr. Chien-Yuan Pan and Dr. De-Ming Yang for excellent technical assistance. We thank Dr. Mei-Lin Wu for valuable help and Dr. Ralph Kirby for critical reading of this manuscript.
This study was supported by grants from the National Science Council (NSC94-2320-B010-032, NSC95-2320-B010-026-MY3) and the Ministry of Education, Aim for the Top University Plan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- NCX
- sodium-calcium exchanger
- NCXIL
- large intracellular loop of sodium-calcium exchanger
- uMiCK
- ubiquitous creatine kinase
- sMiCK
- sarcomeric creatine kinase
- CKB
- brain-type creatine kinase
- CKM
- muscle-type creatine kinase
- XIP
- exchanger inhibitory peptide
- 2-DG
- 2-deoxyglucose.
REFERENCES
- 1.Clapham D. E. (2007) Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 2.Bers D. M. (2000) Circ. Res. 87, 275–281 [DOI] [PubMed] [Google Scholar]
- 3.Bers D. M. (2002) Nature 415, 198–205 [DOI] [PubMed] [Google Scholar]
- 4.Oceandy D., Stanley P. J., Cartwright E. J., Neyses L. (2007) Biochem. Soc. Trans. 35, 927–930 [DOI] [PubMed] [Google Scholar]
- 5.Imahashi K., Pott C., Goldhaber J. I., Steenbergen C., Philipson K. D., Murphy E. (2005) Circ. Res. 97, 916–921 [DOI] [PubMed] [Google Scholar]
- 6.Kusuoka H., Camilion de Hurtado M. C., Marban E. (1993) J. Am. Coll. Cardiol. 21, 240–248 [DOI] [PubMed] [Google Scholar]
- 7.Mattiello J. A., Margulies K. B., Jeevanandam V., Houser S. R. (1998) Cardiovasc. Res. 37, 424–431 [DOI] [PubMed] [Google Scholar]
- 8.Komuro I., Wenninger K. E., Philipson K. D., Izumo S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4769–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Matsuoka S., Hryshko L. V., Nicoll D. A., Bersohn M. M., Burke E. P., Lifton R. P., Philipson K. D. (1994) J. Biol. Chem. 269, 17434–17439 [PubMed] [Google Scholar]
- 10.Nicoll D. A., Quednau B. D., Qui Z., Xia Y. R., Lusis A. J., Philipson K. D. (1996) J. Biol. Chem. 271, 24914–24921 [DOI] [PubMed] [Google Scholar]
- 11.Quednau B. D., Nicoll D. A., Philipson K. D. (1997) Am. J. Physiol. 272, C1250–C1261 [DOI] [PubMed] [Google Scholar]
- 12.Ren X., Nicoll D. A., Philipson K. D. (2006) J. Biol. Chem. 281, 22808–22814 [DOI] [PubMed] [Google Scholar]
- 13.DiPolo R., Beaugé L. (2006) Physiol. Rev. 86, 155–203 [DOI] [PubMed] [Google Scholar]
- 14.Schulze D. H., Muqhal M., Lederer W. J., Ruknudin A. M. (2003) J. Biol. Chem. 278, 28849–28855 [DOI] [PubMed] [Google Scholar]
- 15.Pulina M. V., Rizzuto R., Brini M., Carafoli E. (2006) J. Biol. Chem. 281, 19645–19654 [DOI] [PubMed] [Google Scholar]
- 16.Moore E. D., Etter E. F., Philipson K. D., Carrington W. A., Fogarty K. E., Lifshitz L. M., Fay F. S. (1993) Nature 365, 657–660 [DOI] [PubMed] [Google Scholar]
- 17.Pan C. Y., Kao L. S. (1997) J. Neurochem. 69, 1085–1092 [DOI] [PubMed] [Google Scholar]
- 18.Aceto J. F., Condrescu M., Kroupis C., Nelson H., Nelson N., Nicoll D., Philipson K. D., Reeves J. P. (1992) Arch. Biochem. Biophys. 298, 553–560 [DOI] [PubMed] [Google Scholar]
- 19.Pan C. Y., Chu Y. S., Kao L. S. (1998) Biochem. J. 336, 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordentoft I., Jørgensen P. (2003) Biochem. J. 374, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. (1995) Yeast 11, 355–360 [DOI] [PubMed] [Google Scholar]
- 22.Grynkiewicz G., Poenie M., Tsien R. Y. (1985) J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 23.Pan C. Y., Tsai L. L., Jiang J. H., Chen L. W., Kao L. S. (2008) J. Neurochem. 107, 658–667 [DOI] [PubMed] [Google Scholar]
- 24.Kofuji P., Lederer W. J., Schulze D. H. (1994) J. Biol. Chem. 269, 5145–5149 [PubMed] [Google Scholar]
- 25.Levitsky D. O., Nicoll D. A., Philipson K. D. (1994) J. Biol. Chem. 269, 22847–22852 [PubMed] [Google Scholar]
- 26.Xu W., Denison H., Hale C. C., Gatto C., Milanick M. A. (1997) Arch. Biochem. Biophys. 341, 273–279 [DOI] [PubMed] [Google Scholar]
- 27.Collins A., Somlyo A. V., Hilgemann D. W. (1992) J. Physiol. 454, 27–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haworth R. A., Biggs A. V. (1997) J. Mol. Cell Cardiol. 29, 503–514 [DOI] [PubMed] [Google Scholar]
- 29.Condrescu M., Gardner J. P., Chernaya G., Aceto J. F., Kroupis C., Reeves J. P. (1995) J. Biol. Chem. 270, 9137–9146 [DOI] [PubMed] [Google Scholar]
- 30.Saks V. A., Lipina N. V., Sharov V. G., Smirnov V. N., Chazov E., Grosse R. (1977) Biochim. Biophys. Acta 465, 550–558 [DOI] [PubMed] [Google Scholar]
- 31.Grosse R., Spitzer E., Kupriyanov V. V., Saks V. A., Repke K. R. (1980) Biochim. Biophys. Acta 603, 142–156 [DOI] [PubMed] [Google Scholar]
- 32.Lin G., Liu Y., MacLeod K. M. (2009) Cell Mol. Life Sci. 66, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eder M., Stolz M., Wallimann T., Schlattner U. (2000) J. Biol. Chem. 275, 27094–27099 [DOI] [PubMed] [Google Scholar]
- 34.Stolz M., Hornemann T., Schlattner U., Wallimann T. (2002) Biochem. J. 363, 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eder M., Fritz-Wolf K., Kabsch W., Wallimann T., Schlattner U. (2000) Proteins 39, 216–225 [DOI] [PubMed] [Google Scholar]
- 36.Fritz-Wolf K., Schnyder T., Wallimann T., Kabsch W. (1996) Nature 381, 341–345 [DOI] [PubMed] [Google Scholar]
- 37.Eder M., Schlattner U., Becker A., Wallimann T., Kabsch W., Fritz-Wolf K. (1999) Protein Sci. 8, 2258–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Y. Q., Tang L., Zhou H. M., Lin Z. J. (2001) Acta Crystallogr. D. Biol. Crystallogr. 57, 1196–1200 [DOI] [PubMed] [Google Scholar]
- 39.Reppel M., Fleischmann B. K., Reuter H., Sasse P., Schunkert H., Hescheler J. (2007) Cardiovasc. Res. 75, 99–108 [DOI] [PubMed] [Google Scholar]
- 40.Wakimoto K., Kobayashi K., Kuro O. M., Yao A., Iwamoto T., Yanaka N., Kita S., Nishida A., Azuma S., Toyoda Y., Omori K., Imahie H., Oka T., Kudoh S., Kohmoto O., Yazaki Y., Shigekawa M., Imai Y., Nabeshima Y., Komuro I. (2000) J. Biol. Chem. 275, 36991–36998 [DOI] [PubMed] [Google Scholar]
- 41.Weber C. R., Piacentino V., 3rd, Houser S. R., Bers D. M. (2003) Circulation 108, 2224–2229 [DOI] [PubMed] [Google Scholar]
- 42.Spindler M., Meyer K., Strömer H., Leupold A., Boehm E., Wagner H., Neubauer S. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H1039–H1045 [DOI] [PubMed] [Google Scholar]
- 43.Steeghs K., Heerschap A., de Haan A., Ruitenbeek W., Oerlemans F., van Deursen J., Perryman B., Pette D., Brückwilder M., Koudijs J., Jap P., Wieringa B. (1997) J. Neurosci. Methods 71, 29–41 [DOI] [PubMed] [Google Scholar]
- 44.Steeghs K., Benders A., Oerlemans F., de Haan A., Heerschap A., Ruitenbeek W., Jost C., van Deursen J., Perryman B., Pette D., Brückwilder M., Koudijs J., Jap P., Veerkamp J., Wieringa B. (1997) Cell 89, 93–103 [DOI] [PubMed] [Google Scholar]
- 45.Schlattner U., Tokarska-Schlattner M., Wallimann T. (2006) Biochim. Biophys. Acta 1762, 164–180 [DOI] [PubMed] [Google Scholar]
- 46.Crawford R. M., Ranki H. J., Botting C. H., Budas G. R., Jovanovic A. (2002) FASEB J. 16, 102–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue K., Ueno S., Fukuda A. (2004) FEBS Lett. 564, 131–135 [DOI] [PubMed] [Google Scholar]
- 48.Inoue K., Yamada J., Ueno S., Fukuda A. (2006) J. Neurochem. 96, 598–608 [DOI] [PubMed] [Google Scholar]
- 49.Iwamoto T., Pan Y., Wakabayashi S., Imagawa T., Yamanaka H. I., Shigekawa M. (1996) J. Biol. Chem. 271, 13609–13615 [DOI] [PubMed] [Google Scholar]
- 50.Iwamoto T., Wakabayashi S., Shigekawa M. (1995) J. Biol. Chem. 270, 8996–9001 [DOI] [PubMed] [Google Scholar]
- 51.Ikari A., Sakai H., Takeguchi N. (1998) Eur. J. Pharmacol. 360, 91–98 [DOI] [PubMed] [Google Scholar]
- 52.Tokumura A., Okuno M., Fukuzawa K., Houchi H., Tsuchiya K., Oka M. (1998) Biochim. Biophys. Acta 1389, 67–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.