Abstract
Non-photochemical quenching (NPQ) of excess absorbed light energy is a fundamental process that regulates photosynthetic light harvesting in higher plants. Among several proposed NPQ mechanisms, aggregation-dependent quenching (ADQ) and charge transfer quenching have received the most attention. In vitro spectroscopic features of both mechanisms correlate with very similar signals detected in more intact systems and in vivo, where full NPQ can be observed. A major difference between the models is the proposed quenching site, which is predominantly the major trimeric light-harvesting complex II in ADQ and exclusively monomeric Lhcb proteins in charge transfer quenching. Here, we studied ADQ in both monomeric and trimeric Lhcb proteins, investigating the activities of each antenna subunit and their dependence on zeaxanthin, a major modulator of NPQ in vivo. We found that monomeric Lhcb proteins undergo stronger quenching than light-harvesting complex II during aggregation and that this is enhanced by binding to zeaxanthin, as occurs during NPQ in vivo. Finally, the analysis of Lhcb5 mutants showed that chlorophyll 612 and 613, in close contact with lutein bound at site L1, are important facilitators of ADQ.
Keywords: Carotenoid, Chloroplast, Fluorescence, Membrane Proteins, Photosynthesis, Photosynthetic Pigments, Plant, Reactive Oxygen Species (ROS), Non-photochemical Quenching
Introduction
Plants fix CO2 and synthesize sugars by absorbing light energy using two multiprotein complexes named photosystems I and II (PSI and PSII).2 Each complex has a core where charge transfer and electron transport occur and a light-harvesting antenna system composed of Lhc (light-harvesting complex) proteins. Lhc proteins belong to a superfamily with a highly conserved amino acid sequence, suggesting a common structure (1–4). Different members are associated with PSI (Lhca proteins) and PSII (Lhcb proteins), and their potential for aggregation also varies, such that they exist as trimers (LHCII is a trimer of Lhcb1 to -3), dimers (LHCI exists as Lhca1/4 and Lhca2/3 dimers), or monomers (Lhcb4 to -6) (5, 6).
Structural analysis of LHCII has shown that each subunit comprises three transmembrane helices (designated A, B, and C) and two amphipathic helices exposed on the thylakoid lumen surface, designated D and E (1). Each monomer coordinates four xanthophylls and 14 porphyrins, either Chl a or Chl b. Two carotenoid-binding sites, defined in Ref. 2 as L1 and L2, lie close to helices A and B and usually bind lutein (Lut), whereas a third site named N1, which specifically binds neoxanthin (Neo), lies near helix C (7). Finally, a peripheral and less stable binding site named V1 has been shown to accommodate violaxanthin (Vio) and/or zeaxanthin (Zea) (1, 8, 9), depending on the metabolic state of the chloroplast.
There are no structural data on monomeric Lhc proteins, so the structures are modeled on studies of LHCII trimers (1, 2) and PSI-LHCI (4, 5, 10). However, biochemical data indicate that Lhcb4 and Lhcb5 lack carotenoid-binding site V1 and have a different selectivity at site L2 (which binds Vio rather than Lut) and that Lhcb6 lacks site N1 (9, 11–13). These three proteins bind 8, 9, and 10 Chl molecules, respectively (11, 14–17). One important feature of monomeric Lhcb proteins is their ability to exchange the Vio ligand at site L2 with newly formed Zea, which is produced following lumen acidification under excess light conditions (8, 18–20), a process strictly related to photoprotection (21, 22). Excess light energy induces the accumulation of Chl singlet excited states, increasing the probability of Chl triplet formation. Such triplets can react with O2 to form reactive oxygen species resulting in photoinhibition (23). Lhcb proteins prevent photoinhibition by quenching Chl triplets (24, 25), by scavenging reactive oxygen species (23, 26, 27), and by preventing their formation by feedback de-excitation of singlet excited states (23), a mechanism known as non-photochemical quenching (NPQ). Lhcb subunits are known to play a key role in this process, as shown by mutants with a low Lhc protein content (28). Mechanisms proposed for singlet chlorophyll excited states quenching include charge transfer quenching (CTQ) and aggregation-dependent quenching (ADQ).
CTQ involves the formation of a carotenoid radical cation and a Chl radical anion upon excitation, which recombine at the ground state to dissipate the excitation energy (29–31). ADQ may occur in the trimeric LHCII, which is thought to undergo a conformational change to transfer energy from Chl a excited states to the short lived carotenoid S1 excited state, a conformational change that can be reproduced accurately in vitro during LHCII aggregation (32, 33). Recently, quenching has also been associated with the formation of Chl-Chl charge transfer states during the aggregation of LHCII trimers in vitro (34).
It is possible that ADQ occurs both in LHCII and in monomeric Lhcb proteins, possibly by different mechanisms. Indeed, two quenching sites have been identified by time-resolved fluorescence analysis in vivo, one located in the PSII core and the other in the peripheral antenna system (35). Electron microscopy coupled with reverse genetics has shown that the outer antenna, comprising LHCII trimers and Lhcb6, detaches from PSII supercomplexes, segregating into LHCII + Lhcb6-enriched domains (36, 37). Spectroscopic changes induced by the aggregation of LHCII trimers in vitro, notably changes in Raman resonance and low temperature fluorescence spectra, were also detected in leaves and chloroplasts under quenching conditions (32, 38, 39), suggesting similar conformational changes can be induced by in vitro aggregation and NPQ activation in vivo. Zea was also shown to increase fluorescence quenching in vitro in aggregated LHCII, Lhcb5, and Lhcb4 (40), although its role in LHCII trimers is currently unclear (8, 38, 40).
Although ADQ has been studied widely, there are no experimental data to show which chromophores are involved. Previous reports have suggested that ADQ occurs within the protein domain encompassing carotenoid-binding site L1 and Chl 610-611-612 through strong carotenoid/Chl coupling and energy transfer to the carotenoid excited state S1 (32, 38, 41, 42) or through Chl-Chl charge transfer (34). This putative quenching site is highly conserved in trimeric and monomeric Lhcb proteins (43).
Here, we report the localization and Zea dependence of ADQ using time-resolved and steady state spectroscopy on the different Lhcb proteins. In addition, targeted mutagenesis of Lhcb5 (44) showed that ADQ is strongly influenced by Chl molecules proximal to the Lut-binding site L1. This result complements previous studies of CTQ, which localized the quenching site to Chl 603, Chl 609, and Zea in carotenoid-binding site L2 (30), and suggests that different types of quenching may occur in different Lhc protein domains.
EXPERIMENTAL PROCEDURES
DNA Cloning, Mutations, and Isolation of Overexpressed Lhcb4 to -6 Apoproteins
The Arabidopsis thaliana lhcb5, lhcb4.1, and lhcb6 genes were cloned as previously reported (12, 43, 44). Mutations in lhcb5 were generated as previously described (44) using the QuikChangeTM site-directed mutagenesis kit (Stratagene). Wild-type Lhcb4, Lhcb6, and Lhcb5 and mutant Lhcb5 apoproteins were overexpressed in Escherichia coli strain SG13009 and reconstituted in vitro as described previously (15) with modifications (44, 45). Pure pigments were purchased from Sigma (Chl a and Chl b) or purified by HPLC (xanthophylls). When more than one carotenoid was present in the pigment mix, all species were added in equal amounts.
LHCII Isolation
In order to induce Zea accumulation in LHCII trimers, leaves were illuminated for 30 min at 1200 μmol m2 s−1. LHCII trimers were isolated from dark-adapted or illuminated A. thaliana leaves as described previously (8).
Pigment Analysis
HPLC analysis was performed as described (46). The Chl/carotenoid and Chl a/b ratios were determined independently by fitting the spectrum of acetone extracts to the spectra representing individual purified pigments (47).
Steady State Spectroscopy
Samples were diluted in 20 mm HEPES (pH 7.5), 0.2 m sucrose, 0.03% n-dodecyl-β-d-maltopyranoside (β-DM) to maintain proteins in a non-aggregated state or in 20 mm citrate (pH 5.5), 0.2 m sucrose, 0.003% β-DM for the induction of aggregation. Room temperature absorption spectra were recorded using a SLM-Aminco DK2000 spectrophotometer, with a sampling step wavelength of 0.4 nm. Fluorescence emission spectra were measured using a Horiba Jobin-Yvon Fluoromax-3 spectrofluorometer and corrected for the instrumental response. Low temperature (77 K) fluorescence was measured in a cryostat with 80% glycerol added to each mixture. Circular dichroism (CD) spectra were measured at 10 °C on a Jasco 600 spectropolarimeter using a R7400U-20 photomultiplier tube and samples dissolved in the same solvents used for absorption, with an OD of 1 at the maximum in the Qy transition. The measurements were performed in a 1-ml cuvette.
Time-resolved Fluorescence Analysis
LHCII trimers, Lhcb4 to -6 wild type proteins, and Lhcb5 mutants with or without bound Zea were diluted to the same chlorophyll concentration (80 ng/ml) in buffers promoting non-aggregation or aggregation, as described above. Mixtures of LHCII trimers and Lhcb6 monomers (3:1 molar ratio), each binding either Vio or Zea, were prepared in the same buffers. The level of aggregation in different samples was assessed by measuring the amplitude of light scattering at 750 nm. Time-resolved fluorescence spectroscopy was carried out at room temperature using the single-photon-timing method with a PicoQuant Fluotime 200. Kinetic analysis was performed with a PicoQuant FluoFit, at an excitation wavelength of 435 nm and detection wavelengths of 685 and 705 nm. Each sample was measured 10 times in two independent experiments. The fluorescence quantum yield (ϕ) was calculated from fluorescence decay lifetimes as τf/τ0f−, where τf is the average fluorescence lifetime and τ0f− is the time constant of Chl spontaneous emission in a constant protein environment (48), extrapolated from the LHCII fluorescence quantum yield (49).
Dynamic Light Scattering
The size of aggregates induced by detergent dilution was determined by dynamic light scattering using a ZETASIZER NANO S instrumentation as described in Ref. 50.
RESULTS
Time-resolved Fluorescence Analysis of Monomeric Lhcb4 to -6 Subunits and LHCII Trimers
In order to investigate ADQ and its dependence on Zea, monomeric and trimeric Lhcb proteins were refolded in vitro or isolated from thylakoids. Monomeric Lhcb proteins were reconstituted in vitro after adding pigment mixtures, as described previously (12, 15, 43–45, 51). All mixtures contained the same amounts of Chl a, Chl b, Neo, and Lut, but each mixture contained either Vio or Zea but never both, so that two batches of each Lhcb protein were prepared, consistently binding Lut and Neo but differing according to whether Vio or Zea was bound at site L2 (19, 20, 30, 52). Supplemental Table S1 shows the pigment composition of each refolded or native complex, and depending on the subunits, each complex contained different relative amounts of Lut, Neo, Vio, and Zea as previously reported and consistent with the composition of complexes purified from thylakoid membranes (15, 53, 54). All of the complexes were stable and properly folded, resulting in efficient energy transfer from Chl b and the xanthophylls to Chl a, as determined by fluorescence spectroscopy (not shown). Vio-binding LHCII trimers were purified from the thylakoid membranes of dark-adapted leaves, whereas Zea-binding LHCII trimers were obtained by illuminating the leaves (1200 μmol m−2 s−1) to induce partial substitution at the V1 site prior to isolation, with a final de-epoxidation index of 0.33 (8) (supplemental Table S1).
All complexes were analyzed in both concentrated detergent (0.03% β-DM) and in diluted detergent (0.003% β-DM), the latter falling below the critical micelle concentration of 0.01% for β-DM and thus promoting aggregation (55). Furthermore, the pH of the solution was adjusted to either 7.8 or 5.5, generating samples in unquenched or quenched states, respectively (56). Quenching was quantified by time-resolved fluorescence (38, 57, 58) with an emission wavelength of 685 nm.
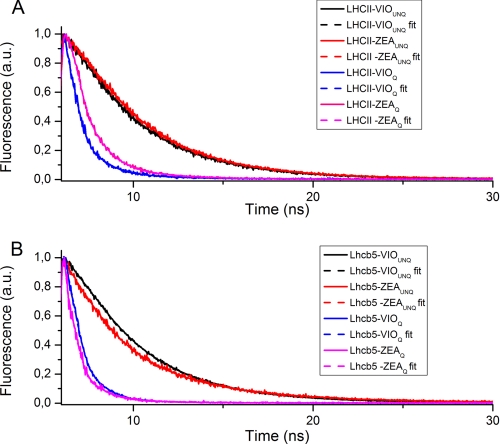
Fig. 1 shows the fluorescence decay traces and fitted curves for Lhcb5 and LHCII as examples, although the results for all complexes are provided in Table 1. Two exponential components were sufficient to fit all of the decay traces for unquenched samples, with decay components at 3.70–4.53 and 1.37–2.72 ns, respectively. The average fluorescence lifetime for Lhcb4 to -6 proteins was reduced when Zea rather than Vio was bound at site L2, in agreement with previous reports (12, 48, 59). In contrast, Zea binding at site V1 in trimeric LHCII had no effect on the fluorescence lifetime.
FIGURE 1.
Fluorescence decay of Lhb5 and LHCII under “quenching” and “unquenching” conditions. Shown are fluorescence decay traces (solid lines) of LHCII (A) and Lhcb5 (B) proteins recorded at 685 nm, under either quenching (Q) or unquenching (UNQ) conditions (see “Results” for details). Decay traces were then fitted using either two or three exponential functions for unquenching or quenching samples, respectively; resulting fitting curves (fit) are shown as dashed lines.
TABLE 1.
Time-resolved fluorescence analysis on Lhcb proteins in detergent or under conditions favoring aggregation
Shown are fitting results of fluorescence decay traces (emission detected at 685 nm) measured on recombinant Lhcb4 to -6 proteins, native LHCII trimers, and a mixture of Lhcb6 and LHCII trimers in a 3:1 molar ratio. Samples were reconstituted (Lhcb4 to -6) or purified (LHCII trimers) with violaxanthin (V) or zeaxanthin (Z). Different complexes were diluted in the presence of 0.03% β-DM and 20 mm HEPES (pH 7.8) or 0.003% β-DM and 20 mm citrate (pH 5.5), in order to induce unquenching (UNQ) or quenching (Q) conditions, respectively. Amp1–3, amplitude of the exponential components 1–3; τ1–3, decay time constants (ns) of the exponential curves 1–3 used to fit the fluorescence decay curves; τAV, average fluorescence decay lifetime (ns); φ: fluorescence quantum yield. Errors are less than 12% in each case.
| Protein | Amp1 | τ1 | Amp2 | τ2 | Amp3 | τ3 | τAV | χ2 | φ |
|---|---|---|---|---|---|---|---|---|---|
| % | ns | % | ns | % | ns | ns | ×10−2 | ||
| LHCII VUNQ | 21.41 | 1.42 | 78.59 | 4.01 | 3.46 | 1.10 | 8.90 | ||
| LHCII ZUNQ | 17.31 | 1.14 | 82.69 | 3.92 | 3.44 | 1.06 | 8.95 | ||
| Lhcb4 VUNQ | 40.72 | 2.42 | 59.28 | 4.56 | 3.69 | 1.05 | 9.49 | ||
| Lhcb4 ZUNQ | 57.94 | 1.76 | 42.06 | 4.18 | 2.78 | 1.14 | 7.15 | ||
| Lhcb5 VUNQ | 33.10 | 2.50 | 66.90 | 4.15 | 3.60 | 1.07 | 9.26 | ||
| Lhcb5 ZUNQ | 56.13 | 1.44 | 43.87 | 3.70 | 2.43 | 1.15 | 6.25 | ||
| Lhcb6 VUNQ | 49.58 | 1.82 | 50.42 | 4.22 | 3.030 | 1.19 | 7.79 | ||
| Lhcb6 ZUNQ | 69.78 | 1.52 | 30.22 | 4.33 | 2.369 | 1.13 | 6.09 | ||
| LHCII VUNQ + Lhcb6 VUNQ (3:1) | 25.96 | 1.352 | 74.04 | 3.969 | 3.28978 | 1.197 | 8.47 | ||
| LHCII ZUNQ + Lhcb6 ZUNQ (3:1) | 38.04 | 1.227 | 61.96 | 3.822 | 2.8349 | 1.135 | 7.30 | ||
| LHCII VQ | 66.85 | 0.11 | 31.46 | 0.50 | 1.69 | 3.09 | 0.28 | 1.03 | 0.72 |
| LHCII ZQ | 45.23 | 0.13 | 50.10 | 0.70 | 4.66 | 2.92 | 0.55 | 1.12 | 1.41 |
| Lhcb4 VQ | 67.18 | 0.09 | 31.40 | 0.49 | 1.42 | 2.56 | 0.25 | 1.22 | 0.64 |
| Lhcb4 ZQ | 70.42 | 0.08 | 28.38 | 0.48 | 1.20 | 2.33 | 0.22 | 1.09 | 0.57 |
| Lhcb5 VQ | 65.30 | 0.08 | 33.93 | 0.50 | 0.77 | 2.73 | 0.24 | 1.22 | 0.62 |
| Lhcb5 ZQ | 84.40 | 0.06 | 14.99 | 0.43 | 0.61 | 2.39 | 0.13 | 1.08 | 0.33 |
| Lhcb6 VQ | 87.05 | 0.07 | 12.05 | 0.39 | 0.90 | 3.52 | 0.14 | 0.98 | 0.36 |
| Lhcb6 ZQ | 84.31 | 0.05 | 15.03 | 0.37 | 0.66 | 2.51 | 0.11 | 1.01 | 0.26 |
| LHCII VQ + Lhcb6 VQ (3:1) | 87.36 | 0.11 | 12.04 | 0.50 | 0.60 | 2.95 | 0.17 | 1.32 | 0.44 |
| LHCII ZQ + Lhcb6 ZQ (3:1) | 83.55 | 0.12 | 16.00 | 0.45 | 0.44 | 3.70 | 0.19 | 1.31 | 0.49 |
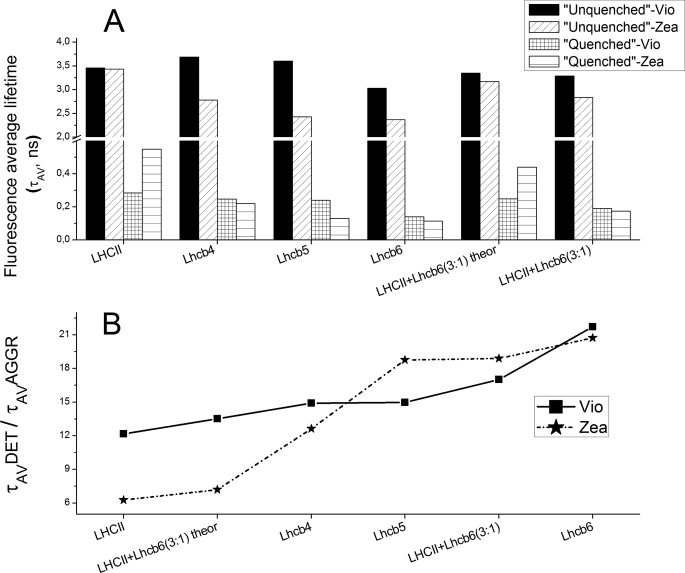
For the quenched samples, at least three exponential components were needed to fit the decay traces, including a short component at 40–130 ps, an intermediate component at 380–700 ps, and a long lasting component (in the nanosecond range) with a small amplitude. Average fluorescence lifetimes were significantly reduced in all aggregated samples compared with the corresponding non-aggregated complexes, ranging from 3–4 ns to 0.13–0.55 ns, depending on the sample. The strongest quenching induced by aggregation was observed for Lhcb6 in the presence of both Vio and Zea (Fig. 2). The effect of Zea differed according to which protein was analyzed. LHCII + Zea had longer lasting fluorescence (0.55 ns) than LHCII + Vio (0.28 ns), whereas the opposite trend was observed for monomeric Lhcb4 and Lhcb5. For Lhcb5, the average fluorescence lifetime was 240 ps in the presence of Vio and 130 ps in the presence of Zea due to the reduction of time constants associated with all three exponential components (Table 1). By plotting the ratio of the average fluorescence lifetimes in the unquenched and quenched samples, the effect of Zea and Vio on ADQ could be compared among the different Lhcb proteins (Fig. 2B). Lhcb6 showed the highest ratio in the presence of either Zea or Vio, indicating that fluorescence lifetime was highly dependent on aggregation, whereas trimeric LHCII showed the lowest ratio, indicating that fluorescence lifetime was only minimally dependent on aggregation, and in this context Zea appears to prevent quenching rather than stimulating it.
FIGURE 2.
Average fluorescence lifetime ratios between samples in detergent and under conditions favoring aggregation. A, fluorescence average lifetimes of monomeric Lhcb4 to -6, trimeric LHCII proteins and of a mixture of Lhcb6 and LHCII trimers (3:1 molar ratio) in detergent (τAV UNQ) or under aggregation conditions (τAV Q). Time-resolved fluorescence analysis is reported in Table 1. Lhcb4 to -6 and LHCII trimer average fluorescence lifetimes were measured either in violaxanthin binding (Vio) or zeaxanthin binding (Zea) samples; calculated fluorescence average lifetimes of the LHCII and Lhcb6 mixture are reported (theor). B, average fluorescence lifetime ratios among samples measured in detergent (τAV UNQ) or in aggregation conditions (τAV Q), showing actual and calculated fluorescence average lifetimes of the LHCII and Lhc6 mixture under aggregation conditions.
Recently, NPQ was shown to be triggered by the dissociation of the LHCII-M·Lhcb6·Lhcb4 supercomplex catalyzed by protonation of PsbS (36) and enhanced by Zea synthesis. This event segregates two domains in grana membranes, one of which contains Lhcb6 together with LHCII-M and LHCII-L trimers. Two different quenching sites have been detected, one connected to the PSII core and the other unconnected (35, 60).
In order to mimic the formation of the LHCII-Lhcb6 domain, we analyzed a 3:1 molar ratio mixture of LHCII trimers and Lhcb6 monomers in the non-aggregated and aggregated states. Fluorescence lifetime data are shown in Table 1. Two exponential components were sufficient to fit the unquenched traces, but at least three were required to fit the quenched traces, with decay components similar to those observed in samples containing a single protein species. Fig. 2 compares the average fluorescence decay lifetimes of the mixtures containing Vio and Zea with the corresponding data from samples containing single subunits. Lifetimes predicted by computing linear combinations of individual samples are also shown to represent the absence of interactions between LHCII and Lhcb6. In the non-aggregated state, the observed and predicted lifetimes are the same, whereas in the aggregated state, the observed lifetimes are shorter than the predicted values (Fig. 2), indicating that the quenching of LHCII (the predominant species in the sample) is enhanced in the presence of Lhcb6. This effect is exacerbated in the presence of Zea. Similar results were observed when fluorescence emission was measured at 705 nm (supplemental Table S2), with a higher amplitude for the shortest component (<170 ps), implying that the quenching species has a greater far red emission, in agreement with previous in vivo data (38).
77 K Steady State Fluorescence Analysis of Lhcb4 to -6 and LHCII Trimers
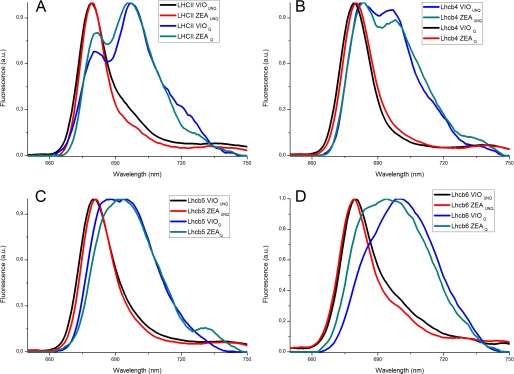
The formation of excitation traps by aggregation in LHCII is associated with the appearance of long wavelength-emitting forms, reflecting the mixed excitation/charge transfer state. These spectral forms have recently been associated with similar red-shifted forms detected in vivo in quenched leaves or chloroplasts (38, 39). In order to verify the appearance of these far red-emitting forms in Lhcb proteins and their dependence on Zea, we measured the 77 K emission fluorescence spectra of non-aggregated and aggregated Lhcb proteins (Fig. 3). The emission spectra of all unquenched samples had a characteristic single peak at ∼680 nm, whereas an additional signal at 697 nm appeared in the quenched samples, irrespective of which carotenoid was bound at the V1 site (38). A broadening of the high energy peaks was also observed. In the case of aggregated Lhcb4, emission spectra were characterized by two peaks at 681 and 698 nm, regardless of which carotenoid was present. Lhcb6 and Lhcb5 also showed significant spectral changes, although in both cases aggregation produced a single broad peak encompassing both the 685–695 and 684–699 nm ranges rather than the two discrete peaks observed for LHCII and Lhcb4. Nevertheless, the broad emission spectrum represents the convolution of two bands, as is more evident in the case of Lhcb5. Carotenoid selectivity at the L2 site therefore contributed only small differences to the spectra, and it is interesting to note that Lhcb6 showed the most significant red sift following aggregation, regardless of whether Zea or Vio was present.
FIGURE 3.
77 K emission spectra of Lhcb proteins in detergent or under conditions favoring aggregation. Fluorescence emission spectra (recorded at 77 K) of Lhcb proteins binding either Vio or Zea. Different samples were diluted with detergent (UNQ) or under aggregation conditions (Q) as described under “Results.” A, LHCII trimers; B, Lhcb4 complexes; C, Lhcb5 complexes; D, Lhcb6 complexes. Black traces, violaxanthin-binding complexes in detergent; red traces, zeaxanthin-binding complexes in detergent; blue traces, violaxanthin-binding complexes under aggregation conditions; cyan traces, zeaxanthin-binding complexes under aggregation conditions.
In Vitro Refolding of Lhcb5 Mutants Lacking Chlorophyll-binding Residues
The sites of energy dissipation in Lhc proteins are thought to be the Chl molecules with the lowest transition energies, namely Chl 612-610-611 (1, 61–63), and Lut bound at the L1 site (32, 38, 39). To verify this model and/or identify other chromophores involved in quenching, we analyzed a series of Lhcb5 mutants in which the Chl-binding residues were disrupted by targeted mutagenesis (44). Lhcb5 was chosen for a series of reasons: (i) it undergoes ADQ even more efficiently than LHCII (Figs. 1 and 2 and Table 1) (64, 65); (ii) most Chl-binding sites can be specifically targeted in Lhcb5, whereas LHCII and Lhcb6 have sites without a ligand that could be experimentally modified; (iii) quenching in Lhcb5 is enhanced by Zea just like NPQ in vivo, whereas LHCII and Lhcb4 show little dependence on this xanthophyll (Fig. 2); and (iv) the occupancy of each Chl-binding site in Lhcb5 has been well characterized (44). On this basis, we prepared two samples for each Lhcb5 mutant (containing either Vio or Zea in addition to the standard Chl a, Chl b, Lut, and Neo). Our nomenclature for the mutants reflects the targeted Chl-binding site, as described previously (2), with a suffix indicating whether Vio or Zea are present. For example, mutant A2VIO is mutant N179F described in Ref. 44, which has a mutated Chl-binding site A2 (2) coordinating Chl 612 (1) (Table 2). All of the pigment·protein complexes folded correctly in vitro in the presence of either Zea or Vio, resulting in efficient energy transfer from Chl b and xanthophylls to Chl a as reported previously (44). Supplemental Table S2 summarizes the results of pigment analysis for each of the mutants as compared with wild-type Lhcb5. In samples containing Vio, our results are consistent with previous studies, with most mutants losing one Chl but mutants A4VIO and A5VIO each losing two (Chl 611 and 609, respectively) in addition to Chl 602 and 603 (44). The carotenoid content was conserved in all mutants, although a marginal loss of Lut and Neo was observed in A4VIO, A5VIO, B5VIO, and B6VIO (44). Samples containing Zea and Vio had similar pigment contents, although A3ZEA and B5ZEA presumably lose more than one Chl because the loss of a single chlorophyll would imply that only Chl a is bound at these sites, a situation not found in Lhcb5.Vio or any other Lhc protein analyzed thus far (12, 44, 66–69). We suggest that A3ZEA and B5ZEA lose Chl 614 and 603, respectively, similar to Lhca1 (69). It is worth noting that, as previously reported (44), it was not possible to produce a Lhcb5 complex with a mutation in the A1 Chl-binding site because the refolded complex was extremely unstable in the presence of both Vio and Zea.
TABLE 2.
Time-resolved fluorescence analysis on Lhcb5 chlorophyll-binding site mutants in detergent or under conditions favoring aggregation
Shown are fitting results of fluorescence decay traces (emission detected at 685 nm) measured on recombinant Lhcb5 complexes mutated at different chlorophyll-binding sites, reconstituted in the presence of chlorophyll a, chlorophyll b, lutein, neoxanthin, and Vio or Zea. Samples were diluted in the presence of 0.03% β-DM and 20 mm HEPES (pH 7.8) or 0.003% β-DM and 20 mm citrate (pH 5.5), in order to induce unquenching (UNQ) or quenching (Q) conditions, respectively. Fluorescence emission was recorded at 685 nm. Amp1–3, amplitude of the exponential components 1–3; τ1–3, decay time constants (ns) of the exponential curves 1–3 used to fit the fluorescence decay curves; τAV, average fluorescence decay lifetime (ns). φ, fluorescence quantum yield. Mutations and chlorophylls coordinated by mutated residues are indicated. Errors are less than 12% in each case.
| Sample | Mutated residues | Chl coordinated | Amp1 | τ1 | Amp2 | τ2 | Amp3 | τ3 | τAV | c2 | φ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| % | ns | % | ns | % | ns | ns | ×10−2 | ||||
| WT VioUNQ | 33.10 | 2.50 | 66.90 | 4.15 | 3.60 | 1.07 | 9.26 | ||||
| A2 VioUNQ | N179F | Chl 612 | 29.99 | 2.18 | 70.01 | 4.17 | 3.57 | 1.01 | 9.18 | ||
| A3 VioUNQ | Q193L | Chl 613 | 65.06 | 2.77 | 34.94 | 4.87 | 3.50 | 1.03 | 9.00 | ||
| A4 VioUNQ | E65V/R181L | Chl 602 | 73.98 | 3.16 | 26.02 | 5.25 | 3.71 | 1.16 | 9.54 | ||
| A5 VioUNQ | H68F | Chl 603 | 61.08 | 3.07 | 38.92 | 4.98 | 3.81 | 1.14 | 9.80 | ||
| B3 VioUNQ | H208L | Chl 614 | 37.20 | 2.52 | 62.80 | 4.19 | 3.57 | 1.09 | 9.18 | ||
| B5 VioUNQ | E137V | Chl 609 | 41.29 | 2.54 | 58.71 | 4.07 | 3.44 | 1.10 | 8.85 | ||
| B6 VioUNQ | E129V | Chl 606 | 47.70 | 2.38 | 52.30 | 4.74 | 3.62 | 1.09 | 9.31 | ||
| WT ZeaUNQ | 56.13 | 1.44 | 43.87 | 3.70 | 2.43 | 1.15 | 6.25 | ||||
| A2 ZeaUNQ | N179F | Chl 612 | 49.29 | 1.75 | 50.70 | 4.07 | 2.93 | 1.08 | 7.54 | ||
| A3 ZeaUNQ | Q193L | Chl 613 | 52.05 | 1.71 | 47.95 | 4.69 | 3.14 | 1.06 | 8.08 | ||
| A4 ZeaUNQ | E65V/R181L | Chl 602 | 59.20 | 1.94 | 40.80 | 4.57 | 3.01 | 1.13 | 7.74 | ||
| A5 ZeaUNQ | H68F | Chl 603 | 59.94 | 2.37 | 40.06 | 4.70 | 3.30 | 1.08 | 8.49 | ||
| B3 ZeaUNQ | H208L | Chl 614 | 64.29 | 1.80 | 35.71 | 4.51 | 2.77 | 1.19 | 7.13 | ||
| B5 ZeaUNQ | E137V | Chl 609 | 50.68 | 1.69 | 49.32 | 3.88 | 2.77 | 1.04 | 7.13 | ||
| B6 ZeaUNQ | E129V | Chl 606 | 60.94 | 1.79 | 39.06 | 4.73 | 2.94 | 1.06 | 7.56 | ||
| WT VioQ | 65.30 | 0.08 | 33.93 | 0.50 | 0.77 | 2.73 | 0.24 | 1.22 | 0.62 | ||
| A2 VioQ | N179F | Chl 612 | 50.26 | 0.11 | 49.74 | 0.73 | 3.13 | 3.08 | 0.52 | 1.20 | 1.34 |
| A3 VioQ | Q193L | Chl 613 | 60.82 | 0.08 | 34.74 | 0.71 | 4.44 | 2.98 | 0.43 | 1.17 | 1.11 |
| A4 VioQ | E65V/R181L | Chl 602 | 74.91 | 0.06 | 23.17 | 0.57 | 1.92 | 2.93 | 0.23 | 1.07 | 0.59 |
| A5 VioQ | H68F | Chl 603 | 81.74 | 0.19 | 16.53 | 0.60 | 1.73 | 3.17 | 0.31 | 1.20 | 0.80 |
| B3 VioQ | H208L | Chl 614 | 80.22 | 0.03 | 18.15 | 0.69 | 1.63 | 3.11 | 0.20 | 1.12 | 0.51 |
| B5 VioQ | E137V | Chl 609 | 73.14 | 0.05 | 25.24 | 0.58 | 1.62 | 2.78 | 0.22 | 1.28 | 0.57 |
| B6 VioQ | E129V | Chl 606 | 80.83 | 0.04 | 18.18 | 0.47 | 1.00 | 3.22 | 0.15 | 1.26 | 0.39 |
| WT ZeaQ | 84.40 | 0.06 | 14.99 | 0.43 | 0.61 | 2.39 | 0.13 | 1.08 | 0.33 | ||
| A2 ZeaQ | N179F | Chl 612 | 73.45 | 0.09 | 23.16 | 0.49 | 3.40 | 2.13 | 0.25 | 1.00 | 0.64 |
| A3 ZeaQ | Q193L | Chl 613 | 70.54 | 0.24 | 22.47 | 1.12 | 6.99 | 3.06 | 0.63 | 1.16 | 1.62 |
| A4 ZeaQ | E65V/R181L | Chl 602 | 78.94 | 0.06 | 18.60 | 0.46 | 2.46 | 2.60 | 0.20 | 0.96 | 0.51 |
| A5 ZeaQ | H68F | Chl 603 | 83.23 | 0.07 | 16.15 | 0.41 | 0.62 | 2.71 | 0.14 | 1.05 | 0.36 |
| B3 ZeaQ | H208L | Chl 614 | 79.11 | 0.05 | 20.42 | 0.35 | 0.48 | 2.34 | 0.12 | 0.99 | 0.31 |
| B5 ZeaQ | E137V | Chl 609 | 65.05 | 0.09 | 32.86 | 0.47 | 2.09 | 2.33 | 0.26 | 1.02 | 0.67 |
| B6 ZeaQ | E129V | Chl 606 | 87.93 | 0.04 | 11.69 | 0.29 | 0.39 | 2.18 | 0.08 | 0.95 | 0.21 |
Time-resolved Fluorescence Analysis of Lhcb5 Mutants
Each mutant Lhcb5 protein was prepared in the detergent solutions described above to yield non-aggregated and aggregated samples, and each was prepared in the presence of either Vio or Zea. Time-resolved fluorescence analysis (Table 2) revealed that the fluorescence decay traces of unquenched samples could be fitted to curves using two exponential functions, with time constants in the nanosecond range similar to the wild-type Lhcb5 protein. All complexes containing Vio had average fluorescence lifetimes similar to that of the wild type protein (∼3.6 ns), except mutant A5, which showed a slight delay in both decay components and thus an average fluorescence lifetime of 3.8 ns. In samples containing Zea, the average fluorescence lifetimes were reduced to 2.2–2.8 ns, with A3ZEA and A5ZEA mutants having the longest lifetimes (Table 2). Interestingly, all of the mutants had longer fluorescence lifetimes than the wild-type protein. This suggests that delocalized quenching promoted by Zea is not specifically dependent on one particular Chl and instead is likely to reflect multiple weak interactions.
The fluorescence decay traces of mutants in the aggregated state in the presence of either Vio or Zea required three exponential curves, characterized by two short components (<100 ps and 300–730 ps) and a longer component in the nanosecond range with small amplitude, similar to that of the wild type. All samples showed at least 5-fold more quenching after aggregation, irrespective of which Chl site was lost. The decay kinetics and average fluorescence lifetimes of Vio-binding mutants A4-A5VIO, B3VIO, and B5VIO were similar to those of the wild type in the presence of Vio (0.24–0.30 ns), whereas B6VIO showed even faster decay (0.15 ns). In contrast, A2VIO and A3VIO mutants had a longer fluorescence lifetime (0.52 and 0.43 ns, respectively) due to the prevalence of the intermediate component from 500 to 700 ps. A similar but smaller effect was observed for B3VIO, which lacks Chl 614 (the closest chlorophyll to Chl 613). Almost all of the quenched Zea-binding mutant complexes were characterized by decay components similar to those of wild type complexes in the presence of Zea, with minor differences reflecting small changes in the amplitude of the longest component. Clear differences in the decay time constants were detected in A3ZEA, with the two short components delayed to 240 and 1.12 ns, implying that the loss of Chl 613 reduces the efficiency of ADQ. The florescence decay was more rapid in B6ZEA compared with the wild type under quenching conditions, and similar results were obtained when fluorescence emission was detected at 705 nm, although the amplitude of the faster component (<100 ps) was higher.
77 K Steady State Fluorescence Analysis of Lhcb5 Mutants
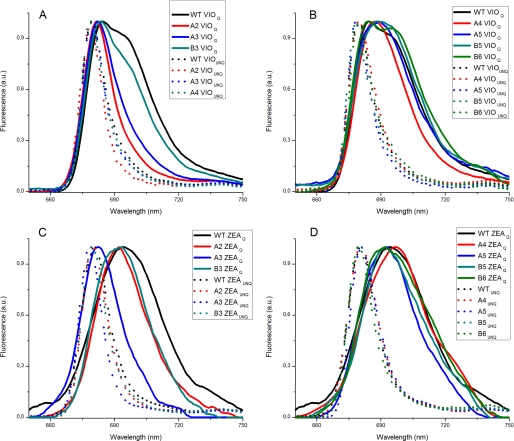
We recorded 77 K emission fluorescence spectra for Lhcb5 mutants in the aggregated state in order to identify the Chl molecules responsible for the red-shifted fluorescence emission (i.e. those involved in the formation of mixed excitation/charge transfer states). Fig. 4 compares the emission spectra for Lhcb5 mutants in the unquenched and quenched states, showing that all unquenched mutants have fluorescence emission spectra similar to that of the wild type, although A2VIO and A3ZEA have 77 K fluorescence emission peaks that are blue-shifted by 2 nm compared with the wild type. Under conditions promoting aggregation, mutants A4-5VIO, B5VIO, and B6VIO still present emission spectra similar to that of the wild type protein in the presence of Vio (Fig. 4B), with two main peaks at 685 and 700 nm. In contrast, the amplitude of the 700 nm broad component is strongly reduced in the A2VIO and A3VIO mutants, and the 685 nm peak is blue-shifted to 680 nm, possibly due to the missing low energy band (Fig. 4A). Mutant B3VIO is intermediate, showing a slight reduction in the far red fluorescence emission tail. With Zea bound to the L2 binding site, only B6ZEA and A4ZEA show emission spectra similar to that of the wild type in the presence of Zea, with a broad peak at 698 nm, representing the sum of the red and far red components (Fig. 4, C and D). The amplitude of the far red tail was reduced in the spectra from mutants A2ZEA, B3ZEA, B5ZEA, and A5ZEA, particularly in the case of A3ZEA, where the single peak showed a significant blue shift, and the far red component was completely absent.
FIGURE 4.
77 K emission spectra of Lhcb5 WT and mutants in detergent or under conditions favoring aggregation. 77 K fluorescence emission spectra for Lhcb5 wild type and chlorophyll-binding mutants. Different samples, binding either Vio or Zea, were diluted with detergent (UNQ, dashed traces) or under aggregation conditions (Q, solid lines) as described under “Results.” A, wild type (black traces), A2 (red traces), A3 (blue traces), and B3 (cyan traces) violaxanthin-binding complexes; B, wild type (black traces), A4 (red traces), A5 (blue traces), B5 (cyan traces), and B6 (green traces) violaxanthin-binding complexes; C, wild type (black traces), A2 (red traces), A3 (blue traces), and B3 (cyan traces) zeaxanthin-binding complexes; D, wild type (black traces), A4 (red traces), A5 (blue traces), B5 (cyan traces), and B6 (green traces) zeaxanthin-binding complexes.
Conformational Changes Induced by Aggregation in Lhcb5
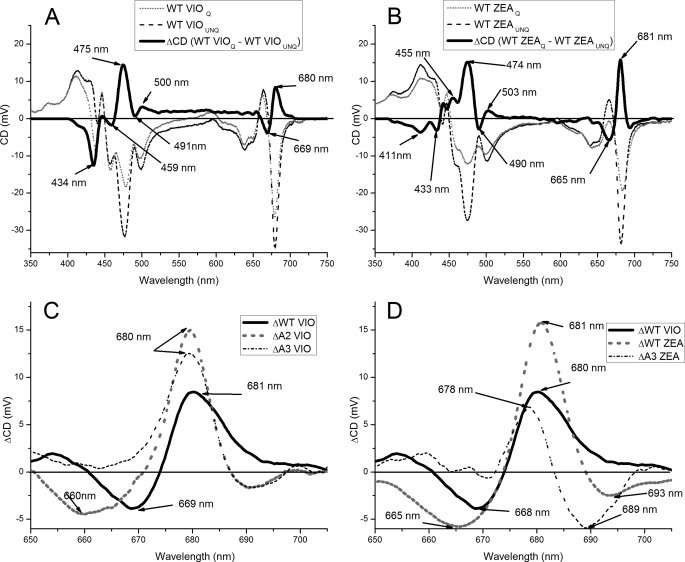
Conformational changes induced by aggregation in LHCII correlate with specific changes in CD spectra (70). In order to confirm the involvement of specific Chl-binding sites in ADQ, we used CD spectroscopy to investigate the occurrence of conformational changes induced by aggregation in wild type Lhcb5 and mutants, with Vio or Zea bound at site L2. As shown in Fig. 5, when the unquenched spectrum of Lbcb5 + Vio was subtracted from the equivalent quenched spectrum, characteristic components were observed at 680 (+), 669 (−), 500 (−), 491 (+), 475 (+), 459 (−), and 434 (−) nm. In Lbcb5 + Zea, these components were shifted to 681 (+), 665 (−), 503 (+), 490 (-), 474 (+), 455 (+), and 433 (−) nm, and two additional signals appeared at 411 nm (−) and 693 (−) nm. These results demonstrate that conformational changes induced by the aggregation of Lhcb5 are partially dependent on which carotenoid binds at the L2 site. The CD spectra of Lhcb5 mutants + Vio showed similar modulations upon aggregation (data not shown), with the exception of A2VIO and A3VIO. In these cases, the differences resulted in a sharper signal at 680 (+) nm, with the red-most contribution eliminated. The CD spectra of Lhcb5 mutants + Zea were also similar to that of the wild type, with the exception of A3ZEA where differences in the red region of the spectrum were blue-shifted, with negative components at 689 and 665 nm and a positive peak at 678 nm (Fig. 5).
FIGURE 5.
CD spectra of Lhcb5 WT and mutants. A, CD spectra of Vio binding Lhcb5 wild type in detergent (UNQ, dashed black line) and under aggregation conditions (Q, solid black line) and the difference between them (gray dashed traces). Difference spectrum peaks are indicated. B, CD spectra of Zea binding Lhcb5 wild type in detergent (UNQ, dashed black line) and under aggregation conditions (Q, solid black line) and the difference between them (gray dashed traces). Difference spectrum peaks are indicated. C, CD difference spectra in the 650–750 nm region of Vio binding Lhcb5 wild type (solid black line), A2 (dashed black line), and A3 (dashed gray line) mutants, calculated by subtracting the spectrum measured in detergent (UNQ) from the spectrum measured under aggregation conditions (Q). D, CD difference spectra in the 650–750 nm region of Vio binding Lhcb5 wild type (solid black line), zeaxanthin binding Lhcb5 wild type (dashed black line), and the zeaxanthin binding mutant A3 (dashed gray line), calculated by subtracting the spectrum measured in detergent (UNQ) from the spectrum measured under aggregation conditions (Q).
Aggregation States of Different Lhcb Proteins
The results presented above show that different Lhcb proteins undergo similar conformational changes upon aggregation, and these conformational changes are associated with fluorescence quenching. To determine whether the efficiency of quenching relates quantitatively to the degree of aggregation, we measured the distribution of particle sizes by dynamic light scattering under the same conditions used for fluorescence lifetime measurements. We observed particles of two different sizes, 100 nm and 1 μm, under conditions promoting aggregation (supplemental Fig. S1). Lhcb6 and LHCII formed 1-μm particles. It should be noticed that by aggregating LHCII monomers or trimers, the same aggregated size is obtained, implying a determinant role of the protein monomers in this process. Lhcb4 formed 100-nm particles, and Lhcb5 switched from 100-nm to 1-μm particles, depending on whether it bound Viola or Zea (supplemental Fig. S1). Single point mutations in Lhcb5 did not influence particle size distribution (supplemental Fig. S1).
DISCUSSION
We have analyzed the behavior of different Lhcb proteins undergoing ADQ, in light of multiple reports over the last 20 years suggesting a correlation between fluorescence quenching induced by in vitro aggregation of LHCII trimers and the induction of NPQ in vivo (17, 32, 38–40, 55, 57, 58, 64, 71, 72). In particular, changes in the Raman resonance spectrum of LHCII trimers during aggregation correlate with similar signals detected in leaves when NPQ is induced, suggesting that Lhc proteins undergo the same conformational change during aggregation in vitro and NPQ activation in vivo (32). Similarly, far red fluorescence in quenched leaves is associated with red-shifted emission forms in LHCII oligomers in vitro (38, 39). However, these reports focused primarily on LHCII, in either its monomeric or trimeric aggregation state, whereas earlier reports suggested that Lhcb4 and Lhcb5 were even more efficient facilitators of ADQ than LHCII (17, 64, 65). Moreover, although Zea is a strong inducer of NPQ in vivo (73), its role in ADQ has never been clarified. Finally, the precise location of the quenching sites (i.e. the identity of the chromophores involved in NPQ) has yet to be determined.
ADQ in Lhcb Proteins
Comparing the average fluorescence lifetime of different Lhcb proteins before and after ADQ (Table 1) revealed that monomeric Lhcb4 to -6 and trimeric LHCII are strongly quenched following aggregation (Fig. 2), with a 10–30-fold reduction in fluorescence quantum yield (Table 1). The effect appears to be greater in monomeric antennas, with Lhcb6 showing the shortest fluorescence lifetime and the lowest quantum yield (Fig. 2B). The amplitude of quenching appears unrelated to particle size in solutions containing 0.003% β-DM because Lhcb proteins that differ in the amplitude of quenching and its dependence on Zea (e.g. Lhcb6 and LHCII) have the same particle size. Instead, the size of the aggregates appears to depend on the properties of individual Lhcb monomers because both monomeric and trimeric LHCII have similar sized aggregates (supplemental Fig. S1). Furthermore, wild-type Lhcb5 and Lhcb5 Chl-binding mutants with different fluorescence lifetimes exhibit the same aggregation behavior and undergo a switch between small and large aggregates when Zea binds at the L2 site, consistent with a previously reported change in conformation (74). We conclude that the observed changes in aggregate particle size are unrelated either to changes in the fluorescence properties of the complexes under aggregating conditions or to the conformational changes detected by CD spectroscopy. This is consistent with previous reports showing that strong CD spectra and fluorescence lifetime changes are observed when comparing proteins dissolved in buffers containing detergent concentration above and below the critical micellar concentration, whereas a further reduction in detergent concentration, although increasing particle size, does not significantly affect either the CD spectrum or the fluorescence lifetime (75). We cannot completely exclude the possibility that changes in the fluorescence lifetime of Lhcb5 induced by Zea might in part reflect changes in aggregation size.
The formation of a quenched state in aggregated LHCII was previously correlated with the presence of red-shifted forms due to the formation of mixed excitation/charge transfer states (38, 76). Here, we observed a similar red shift and broadening of emission spectra in all Lhcb complexes upon aggregation. LHCII and Lhcb4 show two peaks with amplitudes at 680 and 697–698 nm, whereas red shifting of the emission peaks is even more pronounced in Lhcb5 and Lhcb6. Another spectroscopic signature of LHCII in the quenched state induced by aggregation is a conformational change that can be monitored by CD spectroscopy (70). We observed similar changes in the CD spectra for Lhcb5 (Fig. 5), which suggests that all Lhcb proteins have the ability to undergo similar conformational changes in response to aggregation. In addition to these common features, we also observed unique properties, such as the red-most emission forms and the lowest aggregation-induced fluorescence yield in Lhcb6. These results imply that, whenever processes similar to ADQ occur in plants during the induction of NPQ, monomeric Lhcb proteins can be involved along with LHCII and show higher efficiency excitation energy quenching. In this context, it is interesting to note that the addition of Lhcb6 monomers to LHCII trimers enhances the intensity of quenching compared with LHCII alone. Moreover, Zea binding enhances this effect even further (Table 1 and Fig. 2). The simplest explanation is that, upon aggregation, excitation energy is delocalized among the LHCII/Lhcb6 aggregates and is quenched in the most efficient manner, namely by Lhcb6 (Table 1 and supplemental Table S2). The formation of specific domains within grana membranes enriched in LHCII and Lhcb6 oligomers has been recently demonstrated in vivo following NPQ (35–37, 60). Although it is rather unlikely that membrane proteins can aggregate in the lipid phase of the thylakoid membrane due to the inaccessibility of the hydrophobic membrane surfaces, multiple protein-protein interactions between antenna proteins have been shown to induce quenching in liposomes, probably by inducing conformational change(s) by their cooperative effect (48).
Effect of Zeaxanthin on ADQ in Lhcb Proteins
All Lhcb4 to -6 complexes undergo a reduction in fluorescence yield in solution when Zea binds to the L2 site, as reported previously (40, 74, 77). Interestingly, Zea present in the LHCII purified from plants exposed to excess light does not affect the complex fluorescence yield in solution or upon aggregation (8). Previously, Zea has been shown to have a positive effect on ADQ in both monomeric Lhcb proteins (72) and LHCII trimers isolated from A. thaliana npq2 mutants (38). This appears to conflict with our results, but the apparent discrepancy can be explained by the different pigment composition of LHCII trimers from npq2 mutants and wild-type plants exposed to intense light. The npq2 mutant lacks the enzyme zeaxanthin epoxidase and is therefore unable to synthesize Neo or Vio. LHCII proteins must therefore fold in the presence of Lut and Zea as the only xanthophylls, and these are incorporated into the inner L1 and L2 sites and the external V1 site, respectively, leaving the N1 site empty (24, 38). In wild-type plants, LHCII is synthesized in the presence of Vio and Neo, whereas Zea is only produced under excess light stress and binds to the only accessible external V1 site (8). The absence of Neo, in particular, may have a strong influence on ADQ, as discussed below.
We used LHCII isolated from plants exposed to excess light and with the N1 site therefore correctly occupied by Neo. Hence, we found that Zea, solely occupying the V1 site, has no effect in ADQ, in agreement with previous results (8). The LHCII.Zea preparation used in our investigation has a de-epoxiation index of 0.33 (i.e. it is not highly enriched in Zea, suggesting that this might be the reason for the poor quenching effect). However, the absence of a Zea-quenching effect in LHCII has been reported before (8, 40), and we observed an increase in the average fluorescence lifetime compared with LHCII.Vio (Table 1), supporting the view that Zea accumulation is ineffective in promoting quenching in LHCII.
In contrast to LHCII, the monomeric Lhcb4 to -6 proteins all bind Zea at the L2 site in vivo (9, 18–20). The same site is occupied in our recombinant proteins, making them representative of the native proteins when plants are exposed to excess light in vivo (12, 44, 78). Lhcb5.Zea undergoes ADQ much more efficiently than Lhcb5.Vio, whereas the differential for Lhcb6 is smaller, possibly due to the fact that this protein is already quenched in the presence of Vio (Fig. 2). These results imply that the capacity of individual Lhcb proteins to undergo enhanced ADQ upon Zea binding is specifically dependent on the properties of the individual gene product.
ADQ Is Regulated by Chlorophylls Located near Lut Bound at the L1 Site
Site-specific mutation allows the functions of different Chl-binding sites to be determined (12, 30, 44, 49), and this has been used to investigate the architecture of the CT quenching site (30) and the origin of the red-shifted fluorescence emission forms in LHCI (79). We used this approach with Lhcb5 to identify the chromophores involved in ADQ. First, we showed that mutations do not significantly affect the fluorescence lifetimes of Lhcb5.Vio in solution, implying that quenching is only triggered upon aggregation. The only exception was mutant A5, whose fluorescence lifetime exceeded that of wild-type Lhcb5.Vio, reflecting its involvement in a different type of quenching mechanism localized in the L2 domain (12, 30, 44). The strongest effects on fluorescence decay were observed in the A2VIO and A3VIO mutants, and 77 K fluorescence emission analysis showed that the same mutants were unable to produce a red-shifted emission band, a signature representing the formation of quenching sites (30, 38, 39, 79). This result is consistent with the absence of features induced by aggregation in the CD spectra of the same mutants (Fig. 5). It is important to note that this effect is specific because other mutants with intact Chl 612 and 613 show similar spectral properties to the wild type protein. In mutant B3VIO, small differences in fluorescence and CD spectra were observed, consistent with the proximity of the mutation to Chl 613 (1). On this basis, we conclude that Chl 612 and 613 are both involved in the conformational changes induced by ADQ. Chl 614, even if not indispensable, might be involved in modulating the energy level of the Chl 613 S1 transition through its ability to establish excitonic interactions (1, 44).
When Zea is bound to Lhcb5, multiple Chl-binding sites regulate the singlet chlorophyll excited state in the unquenched and non-aggregated protein because average fluorescence lifetimes are in all cases longer than those of the wild type protein + Zea. However, the A5ZEA mutant has a longer fluorescence lifetime than any other mutant (Table 1), suggesting a preferential role for Chl 603. Upon aggregation, the florescence lifetime of the A3ZEA mutant is far longer than that of the wild type or any other mutant, the most red-shifted 77 K fluorescence emission is absent (Fig. 4), and aggregation-specific CD signals are lost (Fig. 5). On this basis, we conclude that Chl 613 becomes or participates in the formation of the major quenching site in Zea-binding Lhcb5. It has been suggested that efficient energy quenching by aggregation requires the S1 transition energy level obtained through the establishment Chl-Chl excitonic interactions to be repressed, probably creating a mix of charge transfer state and exciton delocalization, responsible for 77 K fluorescence emission red shift (Figs. 3 and 4) (38, 39). Excitonic interactions are therefore essential for efficient ADQ, whereas a lone Chl cannot achieve the quenching effect. Indeed, Chl 613 is known to share a strong excitonic interaction with Chl 614, whereas Chl 612 forms excitonic interactions with Chl 611 and Chl 610. All of these chlorophylls are located close to Lut in the L1 site, and our results therefore support the hypothesis that the domain containing L1 is preferentially involved in ADQ (1, 80). Chl 612 and 613 play a prominent role, especially the latter when Zea is bound to the L1 site. It is worth noting that, although some mutants are affected more than others, the average fluorescence lifetime of all of them is reduced at least 5-fold following aggregation, suggesting that Lhcb proteins in this state undergo conformational changes to multiple dissipative states with the contribution of all Chl molecules bound to the protein. Interestingly, mutant B6 in both its Vio- and Zea-binding forms shows increased fluorescence quenching compared with the wild type. Because this mutant has lost Chl 606 and, partially, the Neo bound at site N1, we conclude that, although Chl 606 and Neo are not needed for ADQ, Neo somehow prevents the complete transition to the dissipative conformation. This, in turn, is consistent with the fact that a larger ADQ effect is observed in Lhcb6 (Fig. 2), the only PSII antenna protein that cannot bind Neo (11). We hypothesize that the conformational change induced by aggregation involves helix C twisting with respect to helices A and B, and this is prevented when Neo binds in the groove in between these two domains (1, 7).
The identification of the chlorophylls and carotenoids primarily responsible for ADQ does not provide a complete explanation of the molecular mechanisms facilitating energy dissipation following aggregation, which has previously been attributed to energy transfer to carotenoid excited state S1 or the formation of a Chl-Chl charge transfer state (32, 38). Our results cannot exclude either of these mechanisms. Strong Chl-Chl excitonic interactions are detectable in our quenched samples by examining CD and 77 K fluorescence spectra, as reported previously (38). Although our data suggest that Chl-Chl excitonic interactions may be involved in ADQ, possibly through their ability to reduce the energy level of S1 transition(s), the final quenching site is likely to be the Lut molecule bound to L1. Furthermore, the induction of NPQ in vivo has been positively correlated with the S1 carotenoid excited state (81).
ADQ Versus CTQ in Vitro
The physico-chemical nature of the NPQ mechanism has long been debated, due to the importance of this process for photosynthesis and plant life (82–84). Full induction of NPQ requires a trans-thylakoid pH differential, the protonation of PsbS, the accumulation of Zea or Lut, and the presence of Lhcb proteins (73, 85–91). Evidence supporting both ADQ and CTQ can be generated in vitro using native or recombinant Lhcb proteins (30–32, 55, 55). We have previously shown that CTQ is localized in monomeric Lhcb4 to -6 subunits and does not occur in LHCII (30, 31, 92). Zea bound at site L2 has a dual role in CTQ, directly participating in the reaction and also playing an allosteric role, allowing the formation of a Lut radical cation in the L1 site of Lhcb5 (30, 31, 92). Here, we have shown that all Lhcb proteins can undergo ADQ, but the process is much more efficient in Lhcb5 and -6 and much less efficient in LHCII (Fig. 2). Zea is not indispensable for ADQ but enhances the process in Lhcb5 and Lhcb6. Although CTQ involves Chl 603 and 609 and strongly depends on carotenoids occupying site L2 (30), ADQ mainly involves chlorophylls proximal to Lut occupying site L1.
Relevance of CTQ and ADQ for NPQ in Vivo
The mechanism responsible for NPQ in vivo is difficult to determine because it is impossible to perform detailed spectroscopic analysis in optically challenging materials, such as leaves and unicellular algae. Nevertheless, CTQ has been demonstrated in isolated thylakoids (29, 85), and, although the aggregation of proteins within a lipid membrane might appear unlikely, the induction of NPQ in leaves gives rise to spectral signatures similar to those of Lhc proteins aggregating in vitro (32, 38, 93). It has also been difficult to establish whether NPQ occurs in the monomeric Lhcb proteins (30, 31, 37, 94) or in the major LHCII (32, 57). The recent discovery that the PSII supercomplex dissociates during NPQ and that its components segregate into different membrane domains (36), whereas two distinct quenching sites are activated (35), suggests that NPQ may involve multiple mechanisms. Our results indicate that wherever ADQ occurs in plants during the induction of NPQ, both monomeric Lhcb proteins and trimeric LHCII can be involved, but the former contribute more efficiently to excitation energy quenching.
Supplementary Material
Acknowledgments
We thank Dr. Barbara Cellini and Prof. Carla Voltattorni (Dipartimento di Scienze Morfologico-Biomediche, University of Verona) for dynamic light scattering measurements.
This work was supported by Italian Ministry of Research Special Fund for Basic Research Grant PRIN 20073YHRLE and by EU project 238017 HARVEST.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1 and S2.
- PSI and PSII
- photosystem I and II, respectively
- Chl
- chlorophyll
- LHCI and LHCII
- major light-harvesting complex of photosystem I and II, respectively
- NPQ
- non-photochemical quenching
- Lut
- lutein
- Vio
- violaxanthin
- Neo
- neoxanthin
- Zea
- zeaxanthin
- CTQ
- charge transfer quenching
- ADQ
- aggregation-dependent quenching
- β-DM
- n-dodecyl-β-d-maltopyranoside.
REFERENCES
- 1.Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004) Nature 428, 287–292 [DOI] [PubMed] [Google Scholar]
- 2.Kühlbrandt W., Wang D. N., Fujiyoshi Y. (1994) Nature 367, 614–621 [DOI] [PubMed] [Google Scholar]
- 3.Jansson S. (1999) Trends Plant Sci. 4, 236–240 [DOI] [PubMed] [Google Scholar]
- 4.Amunts A., Drory O., Nelson N. (2007) Nature 447, 58–63 [DOI] [PubMed] [Google Scholar]
- 5.Ben-Shem A., Frolow F., Nelson N. (2003) Nature 426, 630–635 [DOI] [PubMed] [Google Scholar]
- 6.Bassi R., Høyer-Hansen G., Barbato R., Giacometti G. M., Simpson D. J. (1987) J. Biol. Chem. 262, 13333–13341 [PubMed] [Google Scholar]
- 7.Croce R., Remelli R., Varotto C., Breton J., Bassi R. (1999) FEBS Lett. 456, 1–6 [DOI] [PubMed] [Google Scholar]
- 8.Caffarri S., Croce R., Breton J., Bassi R. (2001) J. Biol. Chem. 276, 35924–35933 [DOI] [PubMed] [Google Scholar]
- 9.Ruban A. V., Lee P. J., Wentworth M., Young A. J., Horton P. (1999) J. Biol. Chem. 274, 10458–10465 [DOI] [PubMed] [Google Scholar]
- 10.Amunts A., Toporik H., Borovikova A., Nelson N. (2010) J. Biol. Chem. 285, 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caffarri S., Passarini F., Bassi R., Croce R. (2007) FEBS Lett. 581, 4704–4710 [DOI] [PubMed] [Google Scholar]
- 12.Passarini F., Wientjes E., Hienerwadel R., Croce R. (2009) J. Biol. Chem. 284, 29536–29546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid V. H. (2008) Cell. Mol. Life Sci. 65, 3619–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dainese P., Bassi R. (1991) J. Biol. Chem. 266, 8136–8142 [PubMed] [Google Scholar]
- 15.Giuffra E., Cugini D., Croce R., Bassi R. (1996) Eur. J. Biochem. 238, 112–120 [DOI] [PubMed] [Google Scholar]
- 16.Pagano A., Cinque G., Bassi R. (1998) J. Biol. Chem. 273, 17154–17165 [DOI] [PubMed] [Google Scholar]
- 17.Ruban A. V., Young A. J., Horton P. (1996) Biochemistry 35, 674–678 [DOI] [PubMed] [Google Scholar]
- 18.Jahns P., Wehner A., Paulsen H., Hobe S. (2001) J. Biol. Chem. 276, 22154–22159 [DOI] [PubMed] [Google Scholar]
- 19.Wehner A., Grasses T., Jahns P. (2006) J. Biol. Chem. 281, 21924–21933 [DOI] [PubMed] [Google Scholar]
- 20.Morosinotto T., Baronio R., Bassi R. (2002) J. Biol. Chem. 277, 36913–36920 [DOI] [PubMed] [Google Scholar]
- 21.Alboresi A., Ballottari M., Hienerwadel R., Giacometti G. M., Morosinotto T. (2009) BMC Plant Biol. 9, 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton P., Ruban A. V. (1992) Photosynth. Res. 34, 375–385 [DOI] [PubMed] [Google Scholar]
- 23.Niyogi K. K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359 [DOI] [PubMed] [Google Scholar]
- 24.Mozzo M., Dall'Osto L., Hienerwadel R., Bassi R., Croce R. (2008) J. Biol. Chem. 283, 6184–6192 [DOI] [PubMed] [Google Scholar]
- 25.Dall'Osto L., Lico C., Alric J., Giuliano G., Havaux M., Bassi R. (2006) BMC Plant Biol. 6, 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dall'Osto L., Cazzaniga S., North H., Marion-Poll A., Bassi R. (2007) Plant Cell 19, 1048–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dall'Osto L., Cazzaniga S., Havaux M., Bassi R. (2010) Mol. Plant. 3, 576–593 [DOI] [PubMed] [Google Scholar]
- 28.Havaux M., Dall'osto L., Bassi R. (2007) Plant Physiol. 145, 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt N. E., Zigmantas D., Valkunas L., Li X. P., Niyogi K. K., Fleming G. R. (2005) Science 307, 433–436 [DOI] [PubMed] [Google Scholar]
- 30.Ahn T. K., Avenson T. J., Ballottari M., Cheng Y. C., Niyogi K. K., Bassi R., Fleming G. R. (2008) Science 320, 794–797 [DOI] [PubMed] [Google Scholar]
- 31.Avenson T. J., Ahn T. K., Zigmantas D., Niyogi K. K., Li Z., Ballottari M., Bassi R., Fleming G. R. (2008) J. Biol. Chem. 283, 3550–3558 [DOI] [PubMed] [Google Scholar]
- 32.Ruban A. V., Berera R., Ilioaia C., van Stokkum I. H., Kennis J. T., Pascal A. A., van Amerongen H., Robert B., Horton P., van Grondelle R. (2007) Nature 450, 575–578 [DOI] [PubMed] [Google Scholar]
- 33.Berera R., Herrero C., van Stokkum I. H., Vengris M., Kodis G., Palacios R. E., van Amerongen H., van Grondelle R., Gust D., Moore T. A., Moore A. L., Kennis J. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5343–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller M. G., Lambrev P., Reus M., Wientjes E., Croce R., Holzwarth A. R. (2010) Chemphyschem 11, 1289–1296 [DOI] [PubMed] [Google Scholar]
- 35.Holzwarth A. R., Miloslavina Y., Nilkens M., Jahns P. (2009) Chem. Phys. Lett. 483, 262–267 [Google Scholar]
- 36.Betterle N., Ballottari M., Zorzan S., de Bianchi S., Cazzaniga S., Dall'osto L., Morosinotto T., Bassi R. (2009) J. Biol. Chem. 284, 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bianchi S., Dall'Osto L., Tognon G., Morosinotto T., Bassi R. (2008) Plant Cell 20, 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miloslavina Y., Wehner A., Lambrev P. H., Wientjes E., Reus M., Garab G., Croce R., Holzwarth A. R. (2008) FEBS Lett. 582, 3625–3631 [DOI] [PubMed] [Google Scholar]
- 39.Johnson M. P., Ruban A. V. (2009) J. Biol. Chem. 284, 23592–23601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wentworth M., Ruban A. V., Horton P. (2000) FEBS Lett. 471, 71–74 [DOI] [PubMed] [Google Scholar]
- 41.Rogl H., Kühlbrandt W. (1999) Biochemistry 38, 16214–16222 [DOI] [PubMed] [Google Scholar]
- 42.Pascal A. A., Liu Z., Broess K., van Oort B., van Amerongen H., Wang C., Horton P., Robert B., Chang W., Ruban A. (2005) Nature 436, 134–137 [DOI] [PubMed] [Google Scholar]
- 43.Mozzo M., Passarini F., Bassi R., van Amerongen H., Croce R. (2008) Biochim. Biophys. Acta 1777, 1263–1267 [DOI] [PubMed] [Google Scholar]
- 44.Ballottari M., Mozzo M., Croce R., Morosinotto T., Bassi R. (2009) J. Biol. Chem. 284, 8103–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bassi R., Croce R., Cugini D., Sandonà D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10056–10061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilmore A. M., Yamamoto H. Y. (1991) Plant Physiol. 96, 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Croce R., Canino G., Ros F., Bassi R. (2002) Biochemistry 41, 7334–7343 [DOI] [PubMed] [Google Scholar]
- 48.Moya I., Silvestri M., Vallon O., Cinque G., Bassi R. (2001) Biochemistry 40, 12552–12561 [DOI] [PubMed] [Google Scholar]
- 49.Formaggio E., Cinque G., Bassi R. (2001) J. Mol. Biol. 314, 1157–1166 [DOI] [PubMed] [Google Scholar]
- 50.Cellini B., Bertoldi M., Montioli R., Laurents D. V., Paiardini A., Voltattorni C. B. (2006) Biochemistry 45, 14140–14154 [DOI] [PubMed] [Google Scholar]
- 51.Plumley F. G., Schmidt G. W. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connelly J. P., Müller M. G., Bassi R., Croce R., Holzwarth A. R. (1997) Biochemistry 36, 281–287 [DOI] [PubMed] [Google Scholar]
- 53.Bassi R., Pineau B., Dainese P., Marquardt J. (1993) Eur. J. Biochem. 212, 297–303 [DOI] [PubMed] [Google Scholar]
- 54.Dainese P., Marquardt J., Pineau B., Bassi R. (1992) in Research in Photosynthesis (Murata N. ed) Vol. I, pp. 13–20, Kluwer Academic Publishers, Dordrecht [Google Scholar]
- 55.Horton P., Ruban A. V., Rees D., Pascal A. A., Noctor G., Young A. J. (1991) FEBS Lett. 292, 1–4 [DOI] [PubMed] [Google Scholar]
- 56.Ruban A. V., Horton P. (1994) Photosynth Res. 40, 181–190 [DOI] [PubMed] [Google Scholar]
- 57.van Oort B., van Hoek A., Ruban A. V., van Amerongen H. (2007) FEBS Lett. 581, 3528–3532 [DOI] [PubMed] [Google Scholar]
- 58.Mullineaux C. W., Pascal A. A., Horton P., Holzwarth A. R. (1993) Biochim. Biophys. Acta 1141, 23–28 [Google Scholar]
- 59.Betterle N., Ballottari M., Hienerwadel R., Dall′Osto L., Bassi R. (2010) Arch. Biochem. Biophys., in press [DOI] [PubMed] [Google Scholar]
- 60.van Oort B., Alberts M., de Bianchi S., Dall'Osto L., Bassi R., Trinkunas G., Croce R., van Amerongen H. (2010) Biophys. J. 98, 922–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Novoderezhkin V. I., Palacios M. A., van Amerongen H., van Grondelle R. (2005) J. Phys. Chem. B 109, 10493–10504 [DOI] [PubMed] [Google Scholar]
- 62.Calhoun T. R., Ginsberg N. S., Schlau-Cohen G. S., Cheng Y. C., Ballottari M., Bassi R., Fleming G. R. (2009) J. Phys. Chem. B 113, 16291–16295 [DOI] [PubMed] [Google Scholar]
- 63.van Grondelle R., Novoderezhkin V. I. (2006) Phys. Chem. Chem. Phys. 8, 793–807 [DOI] [PubMed] [Google Scholar]
- 64.Wentworth M., Ruban A. V., Horton P. (2001) Biochemistry 40, 9902–9908 [DOI] [PubMed] [Google Scholar]
- 65.Wentworth M., Ruban A. V., Horton P. (2004) Biochemistry 43, 501–509 [DOI] [PubMed] [Google Scholar]
- 66.Remelli R., Varotto C., Sandonà D., Croce R., Bassi R. (1999) J. Biol. Chem. 274, 33510–33521 [DOI] [PubMed] [Google Scholar]
- 67.Morosinotto T., Mozzo M., Bassi R., Croce R. (2005) J. Biol. Chem. 280, 20612–20619 [DOI] [PubMed] [Google Scholar]
- 68.Mozzo M., Morosinotto T., Bassi R., Croce R. (2006) Biochim. Biophys. Acta 1757, 1607–1613 [DOI] [PubMed] [Google Scholar]
- 69.Morosinotto T., Castelletti S., Breton J., Bassi R., Croce R. (2002) J. Biol. Chem. 277, 36253–36261 [DOI] [PubMed] [Google Scholar]
- 70.Ruban A. V., Calkoen F., Kwa S. L. S., van Grondelle R., Horton P., Dekker J. P. (1997) Biochim. Biophys. Acta 1321, 61–70 [Google Scholar]
- 71.Barzda V., Mustárdy L., Garab G. (1994) Biochemistry 33, 10837–10841 [DOI] [PubMed] [Google Scholar]
- 72.Ruban A. V., Phillip D., Young A. J., Horton P. (1997) Biochemistry 36, 7855–7859 [DOI] [PubMed] [Google Scholar]
- 73.Demmig-Adams B. (1990) Biochim. Biophys. Acta 1020, 1–24 [Google Scholar]
- 74.Dall'Osto L., Caffarri S., Bassi R. (2005) Plant Cell 17, 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bassi R., Silvestri M., Dainese P., Moya I., Giacometti G. M. (1991) J. Photochem. Photobiol. B 9, 335–354 [Google Scholar]
- 76.Ruban A. V., Rees D., Noctor G. D., Young A., Horton P. (1991) Biochim. Biophys. Acta 1059, 355–360 [Google Scholar]
- 77.Crimi M., Dorra D., Bösinger C. S., Giuffra E., Holzwarth A. R., Bassi R. (2001) Eur. J. Biochem. 268, 260–267 [DOI] [PubMed] [Google Scholar]
- 78.Gastaldelli M., Canino G., Croce R., Bassi R. (2003) J. Biol. Chem. 278, 19190–19198 [DOI] [PubMed] [Google Scholar]
- 79.Morosinotto T., Breton J., Bassi R., Croce R. (2003) J. Biol. Chem. 278, 49223–49229 [DOI] [PubMed] [Google Scholar]
- 80.Kühlbrandt W., Wang D. N. (1991) Nature 350, 130–134 [DOI] [PubMed] [Google Scholar]
- 81.Bode S., Quentmeier C. C., Liao P. N., Hafi N., Barros T., Wilk L., Bittner F., Walla P. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12311–12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noctor G., Rees D., Young A., Horton P. (1991) Biochim. Biophys. Acta 1057, 320–330 [Google Scholar]
- 83.Horton P., Hague A. (1988) Biochim. Biophys. Acta 932, 107–115 [Google Scholar]
- 84.DemmigAdams B., Adams W. W., Barker D. H., Logan B. A., Bowling D. R., Verhoeven A. S. (1996) Physiol. Plant 98, 253–264 [Google Scholar]
- 85.Li Z., Ahn T. K., Avenson T. J., Ballottari M., Cruz J. A., Kramer D. M., Bassi R., Fleming G. R., Keasling J. D., Niyogi K. K. (2009) Plant Cell 21, 1798–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Havaux M., Dall'Osto L., Cuiné S., Giuliano G., Bassi R. (2004) J. Biol. Chem. 279, 13878–13888 [DOI] [PubMed] [Google Scholar]
- 87.Niyogi K. K., Shih C., Soon Chow W., Pogson B. J., Dellapenna D., Björkman O. (2001) Photosynth. Res. 67, 139–145 [DOI] [PubMed] [Google Scholar]
- 88.Niyogi K. K., Grossman A. R., Björkman O. (1998) Plant Cell 10, 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li X. P., Muller-Moule P., Gilmore A. M., Niyogi K. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li X. P., Björkman O., Shih C., Grossman A. R., Rosenquist M., Jansson S., Niyogi K. K. (2000) Nature 403, 391–395 [DOI] [PubMed] [Google Scholar]
- 91.Pogson B. J., Niyogi K. K., Björkman O., DellaPenna D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Avenson T. J., Ahn T. K., Niyogi K. K., Ballottari M., Bassi R., Fleming G. R. (2009) J. Biol. Chem. 284, 2830–2835 [DOI] [PubMed] [Google Scholar]
- 93.Ruban A. V., Johnson M. P. (2009) Photosynth. Res. 99, 173–183 [DOI] [PubMed] [Google Scholar]
- 94.Kovács L., Damkjaer J., Kereïche S., Ilioaia C., Ruban A. V., Boekema E. J., Jansson S., Horton P. (2006) Plant Cell 18, 3106–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.