Abstract
Kv7 channels, especially Kv7.2 (KCNQ2) and Kv7.3 (KCNQ3), are key determinants for membrane excitability in the brain. Some chemical modulators of KCNQ channels are in development for use as anti-epileptic drugs, such as retigabine (D-23129, N-(2-amino-4-(4-fluorobenzylamino)-phenyl)), which was recently approved for clinical use. In addition, several other compounds were also reported to potentiate activity of the Kv7 channels. It is therefore of interest to investigate compound-channel interactions, so that more insights may be gained to aid future development of therapeutics. We have conducted a screen of 20,000 compounds for KCNQ2 potentiators using rubidium flux combined with atomic absorption spectrometry. Here, we report the characterization of a series of new structures that display isoform specificity and induce a marked reduction of deactivation distinct from that of retigabine. Furthermore, KCNQ2(W236L), a previously reported mutation that abolishes sensitivity to retigabine, remains fully sensitive to these compounds. This result, together with mutagenesis and other studies, suggests that the reported compounds confer a unique mode of action and involve new molecular determinants on the channel protein, consistent with the idea of recognizing a new site on channel protein.
Keywords: Drug Action, Ion Channels, Medicinal Chemistry, Potassium Channels, Protein-Drug Interactions, Chemical Biology
Introduction
Potassium channels represent a major group of drug targets critical for clinical treatment for a variety of diseases (1). Extensive biophysical studies of potassium channels in both native and recombinant systems have led to the appreciation that the close-open transition of a given channel may be influenced by both physical and chemical factors (2–4). For example, the large conductance, calcium-gated potassium channel (BK), which is normally activated by a combination of membrane depolarization and elevated intracellular calcium, may be activated by high concentrations of calcium alone, at the resting potential, without depolarization (5, 6). Among the Kv channels, voltage-gating appears to be “dominant,” and few channels could be gated by ligands in the absence of membrane depolarization.
Neuronal Kv7 (or KCNQ) channels are activated at subthreshold membrane potentials. Because activation of these channels could dampen membrane excitability, potentiation of these channels by synthetic chemical ligands is believed to be beneficial in treating diseases with hyperexcitibility, such as epilepsy and neuropathic pains (7, 8). Conversely, loss-of-function mutations in these channels lead to benign familial neonatal convulsion syndrome (9). Drug-induced potentiation of channel activity (i.e. increase of net current flow in a given time period) may be achieved by several mechanisms, including modulation of protein expression, channel gating, or single channel conductance. A number of compounds have been reported to display potentiating activities on KCNQ channels (e.g. see a recent review (10)). For example, potent anti-epileptic activities were reported for ICA-27243 in various rodent convulsant models (e.g. ED50 values were 1.5 and 2.2 mg/kg per oral for maximal electroshock- and pentylenetetrazole-induced seizure models in rats, respectively) (11–14). However, for most of these compounds, limited structure-activity relationship (SAR)6 data are available.
KCNQ channels are widely expressed in different tissues (15). There are five isoforms with more than 40% amino acid identity within the six-transmembrane segment core regions. Although the atomic structure of Kv7 channels is not yet available, structures of homologous Kv channels, such as Kv1.2, have been used to model the structures of Kv7 channels (16). Earlier evidence suggests that KCNQ2 to KCNQ5 are neuronal isoforms (17–20), whereas KCNQ1 is a non-neuronal isoform, found primarily in cardiac myocytes mediating IKs (slow delayed rectifies potassium current) current upon coassembly with non-conductive accessory KCNE1 subunits (17, 21, 22). Neuronal M currents are thought to be mediated by coassembly of KCNQ2 with KCNQ3 and to a lesser extent with KCNQ4 (17, 23). Recent evidence suggests that KCNQ2 homomultimers could be responsible for the M current in certain regions of the central nervous system (24). Thus, isoform-specific KCNQ activators would be important tools for exploring the composition and properties of M currents in native tissues.
Assays to monitor potassium channel activity can be broadly divided into three classes: conventional binding assays, flux-based assays, and electrophysiological recordings (25). Considering throughput and correlation with channel activity, flux assays have been more commonly used in high throughput screens (25, 26). Rubidium (Rb+) and thallium (Tl+) are two surrogate, heavy metal ions that permeate through most potassium channels. Because they are non-physiological ions, they allow improved signal-to-noise ratios in flux assays (27, 28).
In an effort to examine the chemical repertoire of KCNQ activators, we screened a collection of 20,000 compounds against KCNQ2 homomeric channels and identified multiple series of structures with distinct chemotypes. It is of importance both to characterize their regulatory attributes and to define active chemical groups. Here, we report an SAR study, which shows diverse effects of highly similar structures displaying profound differences in potentiating KCNQ homomultimeric channels. We also identify critical residues for the potentiating activities of these compounds, providing supportive evidence of a new binding site.
EXPERIMENTAL PROCEDURES
cDNAs and Mutagenesis
The KCNQ1–KCNQ5 cDNAs were gifts from Drs. T. Jentsch (Zentrum für Molekulare Neurobiologie, Hamburg), D. Makinnon (State University of New York, Stony Brook), M. Sanguinetti (University of Utah), M. Shapiro (University of Texas Health Science Center, San Antonio), and V. Vardanyan (Universität Hamburg), respectively. Point mutations were introduced by using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and verified by DNA sequencing.
Cell Culture and Transfection
Chinese hamster ovary (CHO) cells were grown in 50/50 DMEM/F-12 (Cellgro, Manassas, VA) with 10% fetal bovine serum (FBS), and 2 mm l-glutamine (Invitrogen). To express KCNQ channels and mutants, cells were split at 24 h before transfection, plated in 60-mm dishes, and transfected with Lipofectamine 2000TM reagent (Invitrogen), according to the manufacturer's instructions. At 24 h after transfection, cells were split and replated onto coverslips coated with poly-l-lysine (Sigma). A GFP cDNA (Amaxa, Gaithersburg, MD) was cotransfected to identify the transfected cells by fluorescence microscopy.
High Throughput Library Screening and Hits Validation using the Rb+ Flux Assay
A screen of the ChemBridge DiversetTM library of 20,000 compounds for KCNQ2 channel modulators was performed using a Rb+ flux assay reported earlier (29, 30). To validate the hits from the initial screen, compounds were cherry-picked from the original compound library and retested using the Rb+ efflux assay using wild type KCNQ2. To test whether the activator hits have similar effects on KCNQ2 W236L mutant channels, a Rb+ flux assay was performed using a CHO KCNQ2 W236L stable cell line with the same assay format. Hits were selected with the same criteria (increase or decrease of the Rb+ flux level by 3 times the S.D. compared with the DMSO control) for each assay format.
High Throughput Library Screening Using the Tl+ Flux Assay
For screening the focused compound library, the Tl+ BTC flux assay was performed according to the published protocol (28) with modifications. Briefly, freshly cultured HEK293 cells stably expressing KCNQ2 channels were seeded onto poly-d-lysine-coated, 384-well Becton Dickinson plates by a Multidrop dispenser and incubated overnight at 37 °C in a 5% CO2 incubator. After manual removal of the medium, BTC-AM solution (2 μm, 50 μl) was added into the cell plates and incubated in the dark, at room temperature, for 60 min. Then the BTC-AM solution was manually removed, and cell plates were washed twice with chloride-free buffer and loaded with 25 μl/well of chloride-free buffer. The compound plates were prepared in chloride-free buffer with Tl+ stimulus solution; the final concentration of Tl+ in the assay wells was 7.5 mm, and the compound concentrations were 20 or 2 μm upon the addition of 25 μl/well from compound plates. Both compound plates and cell plates were mounted on the FDSS (Hamamatsu Photonics Ltd., Japan), and cell plates were preread with excitation at 480 nm and emission at 540 nm for 5 s, at a 1-Hz frequency, before the compounds were added into the cell plates. The fluorescence responses from cell plates were continuously monitored for 100 s at a 1-Hz frequency. The ratios of fluorescence (F/F0) were corrected for background autofluorescence and were used for quantification of KCNQ2 channel responses to the compound library. Retigabine (10 μm) and/or zinc pyrithione (ZnPy; 10 μm) served as positive controls. Negative controls were buffer solutions containing DMSO at the same concentrations used with compound application. The assay displayed a good Z′ factor (>0.5) and signal-to-noise ratio (>7).
Electrophysiological Recording in CHO Cells
Whole cell voltage clamp recording was carried out, using cultured CHO cells, at room temperature, with an Axopatch-200B amplifier (Molecular Devices, Sunnyvale, CA). The electrodes were pulled from borosilicate glass capillaries (World Precision Instruments, Sarasota, FL). When filled with the intracellular solution, the electrodes had resistances of 3–5 megaohms. Pipette solution contained 145 mm KCl, 1 mm MgCl2, 5 mm EGTA, 10 mm HEPES, 5 mm MgATP (pH 7.3 with KOH). During the recording, constant perfusion of extracellular solution was maintained using a BPS perfusion system (ALA Scientific Instruments, Westburg, NY). Extracellular solution contained 140 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1.5 mm MgCl2, 10 mm HEPES, 10 mm glucose (pH 7.4 with NaOH). Signals were filtered at 1 kHz and digitized using a DigiData 1322A, with pClamp 9.2 software (Molecular Devices, Sunnyvale, CA). Series resistance was compensated by 60–80%. In the present study, various voltage protocols were used. The holding potential was −120 mV in all voltage protocols, except where indicated. To elicit the currents, cells were stimulated by a series of 2,000-ms depolarizing steps, from −80 mV to +50 mV, in 10-mV increments.
Chemical Synthesis
Chemical synthesis was proceeded as shown in Reaction 1.
REACTION 1.
General Methods
Dicyclohexylcarbodiimide (DCC) was added and stirred for 30 min into a solution of carboxylic acid 1 (0.36 mmol) in tetrahydrofuran (THF) (2 ml) at 0 °C. Then amine 2 (0.3 mmol) was added to the mixture, followed by the addition of dimethyl aminopyridine (DMAP; 0.06 mmol). The reaction mixture was allowed to stir at room temperature overnight. After evaporating the volatiles, the mixture was partitioned by CH2Cl2 (2 × 20 ml) and saturated NaHCO3 solution (20 ml), dried over anhydrous Na2SO4, and concentrated in vacuo. The residue was purified by column chromatography to give the desired amide ztz series compounds as described (see supplemental material). The other analogues were synthesized in a similar manner.
ztz240 Preparation and Characterization
ztz240 was prepared using the general procedure in 85% yield. The characterization of the ztz240 series of compounds was examined by 1H and 13C NMR and high resolution mass spectrometry. These spectra show high purity (>95%) of these compounds. For ztz240, 1H NMR (CDCl3 + CD3OD, 500 MHz): δ = 8.42 (d, J = 2.5 Hz, 1H), 8.41–8.26 (m, 1H), 7.91–7.88 (m, 2H), 7.27 (d, J = 8.5 Hz, 1H), 7.11–7.07 (m, 2H); 13C NMR (CDCl3 + CD3OD, 125 MHz): δ = 166.1, 145.6, 141.4, 141.3, 134.9, 131.2, 130.3 × 2, 130.1, 124.4, 115.9 × 2. Detailed preparation procedure and purity information of other derivatives are available (see supplemental material).
Data and Statistical Analysis
Patch clamp data were processed using Clampfit 9.2 (Molecular Devices, Sunnyvale, CA) and then analyzed in GraphPad Prism 4 (GraphPad Software, San Diego, CA). Voltage-dependent activation curves were fitted with the Boltzmann equation, G = Gmin + (Gmax − Gmin)/(1 + exp(V − V½)/S)), where Gmax is the maximum conductance, Gmin is the minimum conductance, V½ is the voltage for reaching 50% of maximum conductance, and S is the slope factor. Dose-response curves were fitted with the Hill equation, E = Emax/ (1 + (EC50/C)P), where EC50 is the drug concentration producing half of the maximum response, and P is the Hill coefficient. The activation and deactivation traces were fitted with exponential equations containing one or two components using Clampfit 9.2. Data are presented as means ± S.E. Significance was estimated using paired two-tailed Student's t tests.
RESULTS
Identification of ztz240 as an Activator for KCNQ2
To identify compounds with potentiator activities for M currents, we generated stable cell lines for both KCNQ2 and KCNQ2/3. After a series of optimizations for several key high throughput screening parameters, a standard procedure of Rb+ flux assay was developed and used to screen a ChemBridge DiversetTM library of 20,000 compounds at a 10 μm final concentration (31). In this pilot screen against KCNQ2 homomeric channels, we selected 670 hits, including both potentiators and inhibitors. Among the 270 potentiator hits, 92 have been validated with more than 15% increase in signal intensity (3 × S.D., with Z′ = 0.62). To increase the probability of finding new and interesting structures with novel modes of action, we applied a triage criterion by examining the compound effects on KCNQ2(W236L), a mutation that abolishes sensitivity to retigabine, a well known KCNQ potentiator (32, 33). In addition to ZnPy, reported earlier (31), we have also found several other compound classes effective on KCNQ2(W236L)(Fig. 1A). We further tested 5337031, which potentiated KCNQ2 channel activities in a dose-dependent manner, with an EC50 value of 9.8 ± 0.2 μm.
FIGURE 1.
Structures of primary screen hits and derivatives. A, a series of initial hits identified from the ChemBridge Diverset compound collection, with ChemBridge ID number as indicated. B, derivatives of nitrogen position and ring placements (X, Y = F, Cl, Br; n = 0, 1, 2). C, structures of ztz240 and ICA-27243 (ztz233; see Figs. 8 and 9).
We have pursued conventional chemical synthesis in an effort to both validate and evaluate efficacy and pharmacology of these compounds. Based on the inspection of several active structures from the primary screen (Fig. 1A), it appears that two aromatic moieties, linked via an amide bridge, play critical pharmacological roles in activation of KCNQ channels. These structures displayed considerable similarity to an ICAgen compound, ICA-27243 (Fig. 1C) (11–14). To explore this notion, we first examined a series of analogues with different nitrogen positions. Fig. 1B outlined the four classes of chemical scaffolds selected to incorporate additional modifications of positions 1 and 2, using a positional scanning approach. The number of compounds to be synthesized for testing is guided by the “Topliss scheme.” The Topliss scheme is an efficient strategy for identifying the optimum substitution on the benzene ring in an active lead compound for maximization of compound potency and minimizing the number of compounds to be synthesized by incorporation of hydrophobic, electronic, and steric effects (34, 35).
ztz240 (N-(6-chloro-pyridin-3-yl)-4-fluorobenzamide) was one of the initial 52 derivatives made (Fig. 1C and supplemental material). Fig. 2 shows KCNQ2 currents in transiently transfected cells elicited by depolarization steps from a holding potential of −80 mV, as described earlier (31). The depolarization steps slowly activated a series of non-inactivating outward potassium currents (Fig. 2A). In the absence of drug, the 800-ms duration of hyperpolarization step to −120 mV completely deactivated the channel, with a time constant of 20.6 ± 3.4 ms (n = 6). In the presence of 10 μm ztz240, we observed a significant increase in outward current amplitude, accompanied by considerable prolongation of inward current during the repolarization period at −120 mV, indicative of slowing in deactivation kinetics.
FIGURE 2.
ztz240 activation on KCNQ2 channels. A, the representative traces of KCNQ2, before and after application of 10 μm ztz240, elicited by the protocol as indicated. The holding potential is −80 mV, followed by a series of depolarization steps from −70 to +50 mV in 10-mV increments, followed by an 800-ms hypopolarization step to −120 mV to record the tail current. B, the representative traces of KCNQ2, with and without 10 μm ztz240, elicited by a modified protocol from A, as indicated. The holding potential was −120 mV. C, dose-response curve of ztz240 effects on outward current of KCNQ2 channels. The ratio of outward current amplitude in the presence of the compound (I) versus that in the absence of compound (I0) was plotted against compound concentration. The outward currents were elicited by a 2-s depolarization to +50 mV from the holding potential, −120 mV. D, voltage activation curves of KCNQ2, in the absence or presence of 10 μm ztz240. Error bars, S.E.
To examine the effect more closely, the holding potential was shifted to a more negative value of −120 mV that removes the initial, instantaneous current upon depolarization. With the modified protocol, the outward current potentiation was still observed. The channel closed slowly, taking ∼10 s at −120 mV. The voltage activation curve reveals that ztz240 dramatically left-shifted the V½ by more than 45 mV, from −11.4 ± 4.2 mV to −58.7 ± 7.7 mV (n = 4, p < 0.05) (Fig. 2D), consistent with the observation that ∼10% of the channels are in the open state, at −80 mV, in the presence of ztz240. The potentiation of outward current was dose-dependent, with a half-maximal value (EC50) of 5.8 ± 0.9 μm and a 2.2 ± 0.3-fold potentiation of current at maximal doses (n = 6) (Fig. 2C).
In comparison with effects by other activators, such as retigabine (32–33, 36) and ZnPy (31), ztz240 shows different effects, most noticeably on slowing channel deactivation. Normally, the deactivation of KCNQ2 channel could be fitted by a single exponential function, which was increased from 14.6 ± 1.3 to 27.2 ± 2.6 ms (n = 4) by retigabine or to 64.9 ± 12.6 ms by ZnPy (n = 5) (Fig. 3A) (31, 36). In contrast, ztz240 dramatically prolonged deactivation, which was better fitted by a biexponential function, with fast and slow time constants of 171.9 ± 62.5 ms and 3,230.5 ± 534.1 ms (n = 6), respectively (Fig. 3B).
FIGURE 3.
Effects on deactivation by ztz240 on KCNQ2. A, comparison of 10 μm ztz240, 10 μm ZnPy, and 10 μm retigabine (RTG) effects on KCNQ2, in the absence (black) and presence (gray) of drugs. The protocol is indicated at the bottom. B, histogram shows that ztz240 slows the deactivation of KCNQ2. The deactivation was fitted with a biexponential function with fast (τf) and slow (τs) components. C, the representative traces show deactivation phases of KCNQ2 channel in the absence (left) or presence of 10 μm ztz240 (right). The gray line indicates 0 pA (base line). The time scale and protocol used are as indicated. D, deactivation phase of KCNQ2 without drug was fitted by single exponential function, and the time constant (τ) was plotted against different voltages. E, deactivation of KCNQ2 after application of drug was fitted by biexponential function, and the time constants, both τf and τs, were plotted against different voltages, respectively. F, the normalized activation phases from the full traces (inset) in the control (black line) and after application of 10 μm ztz240 (gray line) are shown. The protocol in A was used. G, the activation phases in the absence or presence of ztz240 were fitted to a biexponential function, and the time constants (τf and τs) were plotted against different testing voltages. H, the dose-response relationship of ztz240 concentration to slow time constant of deactivation (τs). The protocol in A was used. Error bars, S.E.
We then studied the effects of ztz240 on deactivation at different voltages. Currents were elicited by depolarization to +50 mV, followed by hyperpolarization to different testing voltages from −100 to −140 mV (Fig. 3C). The deactivation rate was plotted against voltages in the absence (Fig. 3D) or presence (Fig. 3E) of ztz240. The slow time constant (τs) at −120 mV was plotted against the ztz240 concentration (Fig. 3H) and revealed an EC50 value of 5.9 ± 0.5 μm (n = 6), which is essentially identical to the EC50 obtained from the increased conductance (Fig. 2C). The activation phase could be fitted by a biexponential function, and ztz240 displayed only modest effects (Fig. 3, F and G). In contrast, retigabine greatly accelerated the KCNQ activation rate (31, 36)(Fig. 3A).
Molecular Specificity and Critical Residues for ztz240 Modulation
The KCNQ family includes five members, termed as KCNQ1–KCNQ5, that share high sequence homology. The subtype sensitivity to activators can be used to distinguish different type activators and may be related to molecular determinants that confer the molecular interaction. For example, retigabine potentiates the homomultimers of all KCNQs except KCNQ1, whereas ZnPy, in contrast, potentiates all but KCNQ3. Application of 10 μm ztz240 potentiates KCNQ4 and KCNQ5 more potently than it does KCNQ2, based on the increase in current amplitude. Neither KCNQ1 nor KCNQ3 displays detectable sensitivity to ztz240 (Fig. 4, A and B and Table 1). ztz240 potentiates KCNQ4 and KCNQ5 more potently and with a different profile compared with KCNQ2. First, the left shift of activation V½ by 10 μm ztz240 was less for KCNQ4 and KCNQ5 channels than for KCNQ2 (KCNQ2 from −11.4 ± 4.2 to −58.7 ± 7.7 mV; KCNQ4 from −10.9 ± 1.3 to −27.9 ± 4.4 mV; and KCNQ5 from −19.9 ± 2.6 to −31.0 ± 2.3 mV) (Table 1). Second, the two-phase deactivation seen for KCNQ2 was not observed in KCNQ4 and KCNQ5 channels in the presence of ztz240. Considering the different subtype specificity of ztz240 from that of retigabine or ZnPy, these data are consistent with the notion that ztz240 activates KCNQ channels by a mechanism different from retigabine and ZnPy.
FIGURE 4.
The subtype specificity of ztz240 on KCNQ channels. A, whole cell currents of CHO cells transfected individually with the indicated cDNAs were recorded in the absence (left) and presence (right) of 10 μm ztz240. B, histogram shows 10 μm ztz240 potentiation on KCNQ1 to KCNQ5. The normalized current amplitude is shown. C, the representative traces of mutants W236L, L245A, L279A, and L245A/L279A in the absence (black) and presence (gray) of 10 μm ztz240 (left) and corresponding activation curves (right). The representative traces of the mutants were elicited by depolarization to +50 mV from the holding potential at −120 mV. Error bars, S.E.
TABLE 1.
ztz240 effects on different KCNQ subtypes
I/I0 indicates the effects of the compounds on the amplitude of outward current of KCNQ channels. I0 is the amplitude of outward current in the absence of the compound. I is the amplitude of outward current in the presence of the compound. All currents were elicited by 2-s depolarization from −80 to +50 mV with 10-mV increments from a holding potential of −120 mV. To calculate I/I0, the depolarization was +50 mV and was measured at the end of depolarization. To calculate V½, the tail current elicited following −120 mV was measured. All values are mean ± S.E. (n = 3–6).
| I/I0 |
V½ |
τ deactivation |
|||
|---|---|---|---|---|---|
| Control | 10 μm ztz240 | Control | 10 μm ztz240 | ||
| mV | ms | ||||
| KCNQ1 | 0.9 ± 0.0 | −13.5 ± 1.8 | −16.6 ± 2.9 | 32.9 ± 5.2 | 51.8 ± 5.6 |
| KCNQ2 | 1.9 ± 0.2 | −11.4 ± 4.2 | −58.7 ± 7.7 | 20.6 ± 3.4 | τs = 3230.5 ± 534.1 |
| τf = 171.9 ± 62.5 | |||||
| KCNQ3 | 1.0 ± 0.2 | −39.8 ± 4.1 | −43.1 ± 5.4 | 13.4 ± 2.7 | 26.0 ± 2.6 |
| KCNQ4 | 2.4 ± 0.4 | −10.9 ± 1.3 | −27.9 ± 4.4 | 12.2 ± 2.9 | 15.9 ± 0.9 |
| KCNQ5 | 3.5 ± 0.4 | −19.9 ± 2.6 | −31.0 ± 2.3 | 7.7 ± 1.1 | 13.6 ± 3.4a |
a p < 0.05.
Efforts to identify key residue(s) necessary for retigabine sensitivity (i.e. molecular determinants) have revealed one conserved tryptophan in KCNQ2–KCNQ5 that is critical for retigabine sensitivity. In the case of KCNQ2, the mutant, W236L, is functional but no longer sensitive to retigabine (32, 33). However, KCNQ2(W236L) remains sensitive to ZnPy. Our recent work revealed that two leucine residues in S5 (Leu249) and S5-S6 linker (Leu275) of KCNQ2 are key determinants for the ZnPy-mediated voltage activation curve shift (31). Therefore, we first tested ztz240 effects on W236L and found that, in the presence of 10 μm ztz240, W236L was potentiated to 1.7 ± 0.2-fold, similar to that observed with wild-type KCNQ2, 1.9 ± 0.2 (n = 3, p > 0.05). ztz240 also left-shifted the voltage activation curve of W236L significantly, from −21.9 ± 1.2 to −38.6 ± 7.6 mV (n = 4, p < 0.05). Furthermore, its effects on the deactivation persisted and could be better fitted by a biexponential function, which is also consistent with wild-type KCNQ2 (Fig. 4 and Table 2). Similarly, we tested ztz240 effects on L249A, L275A, and double mutation L249A/L275A, the key determinants for ZnPy sensitivity. As shown in Fig. 4, C and D, and Table 2, all of the three mutants are sensitive to ztz240. Noticeably, all of the three mutants show significant left shifting of the voltage activation curve after application of ztz240. Taken together, mutants of the reported residues critical for conferring the sensitivity to either retigabine or ZnPy remain sensitive to ztz240, providing further evidence that ztz240 acts on or mediates its effects via different molecular determinants.
TABLE 2.
The molecular determinants of ztz240 on KCNQ2 channels
The protocols used are same as indicated in the legend to Table 1. All mutants are KCNQ2 background. I, I0, and V½ are the same as those referred to in the legend to Table 1.
| I/I0 |
V½ |
τ deactivation |
|||
|---|---|---|---|---|---|
| Control | 10 μm ztz240 | Control | 10 μm ztz240 | ||
| mV | ms | ||||
| KCNQ2 | 1.9 ± 0.2 | −12.9 ± 1.2 | −55.3 ± 1.1 | 20.6 ± 3.4 | τs = 3,230.5 ± 534.1 |
| τf = 171.9 ± 62.5 | |||||
| W236L | 1.7 ± 0.2 | −21.9 ± 1.1 | −38.6 ± 7.6 | 18.9 ± 1.7 | τs = 835.9 ± 83.3 |
| τf = 36.2 ± 3.4 | |||||
| L249A | 1.8 ± 0.3 | −7.5 ± 1.5 | −41.8 ± 2.5 | 8.6 ± 1.6 | τs = 1103.3 ± 283.4 |
| τf = 38.4 ± 5.4 | |||||
| L275A | 1.8 ± 0.1 | −3.6 ± 5.4 | −28.3 ± 3.4 | 12.2 ± 0.4 | τs = 586.5 ± 60.4 |
| τf = 26.9 ± 2.5 | |||||
| L249A/L275A | 2.3 ± 0.2 | 7.9 ± 0.9 | −9.1 ± 1.4 | 10.5 ± 2.5 | τs = 406.1 ± 14.8 |
| τf = 14.4 ± 1.8 | |||||
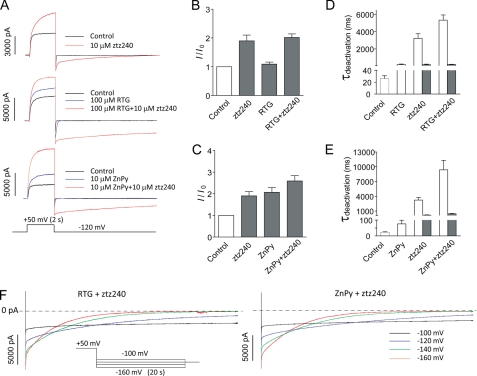
To gain further insights into ztz240 effects, simultaneous application of two different potentiators to KCNQ2 was tested. The rationale is that if both compounds could simultaneously bind to the same channel complex, the net effects are likely to be non-additive and display unique characteristics. If, however, one compound has a “dominant” effect by, for example, occupying the site and preventing binding of the second compound, the resultant current should resemble that when the compound is applied alone. We first measured the effects of ztz240 in the presence of 100 μm retigabine (saturated concentration). As shown in Fig. 5A (middle), 100 μm retigabine (RTG) increased outward current (1.1 ± 0.2-fold) and slowed deactivation to 180.6 ± 66.7 ms from 25.8 ± 5.2 ms (n = 5). Supplement of 10 μm ztz240 further potentiated the outward current (2.0 ± 0.1-fold), and, more dramatically, it slowed the deactivation (Fig. 5A, middle). The deactivation can only be fitted with two time constants (Fig. 5D). Different from ztz240 alone, the tail current persists, displaying a much longer duration up to 40 s (Fig. 5, A, D, and F). Similarly, combinatorial application of 10 μm ZnPy and ztz240 also showed similar effects (Fig. 5, A–C). A saturated dose of 30 μm ZnPy was also used in this experiment. However, it has caused significant instability of recording, due to its toxicity (data not shown). The non-additive effects and pronounced prolongation of tail currents observed only in the presence of the mixed compounds, distinctive from the effects by individual compounds, indicate that ztz240 can modulate the same channel complex in combination with either retigabine or ZnPy (Fig. 5, D and F).
FIGURE 5.
Non-additive effects on KCNQ2 channel using mixed active compounds. A, representative traces in the absence or presence of drugs are shown as indicated and color-coded. B and C, histograms represent increase in outward current amplitude (I/I0) in the absence or presence of drugs as indicated. D and E, histograms represent time constants of deactivation in the absence or presence of drugs as indicated. For controls, RTG and ZnPy, the unfilled bars indicate the time constant revealed by single exponential equation fitting. For ztz240 alone or ztz240 mixed with RTG or ZnPy, the deactivation could optimally be fitted with an exponential equation of two components: fast (filled bars) and slow (unfilled bars) time constants (also see Fig. 3E). F, representative traces in the presence of RTG + ztz240 (left) and ZnPy + ztz240 (right) elicited by the protocol as indicated. The voltage steps are color-coded as indicated. The dotted lines represent the base line of no current. Error bars, S.E.
Perhaps the most striking effect of ztz240 on KCNQ channels is the change of deactivation rate. We next sought to identify residues critical for ztz240 effects. The S6 segment is thought to be involved in gating modulation of channels. Previous studies of RL-3 (an activator for KCNQ1), retigabine, and ZnPy have identified residues in the S6 segment critical for their effects (31–33, 37). In addition to the highly conserved gating hinge glycine (for KCNQ2, this is Gly301) in voltage-gated potassium channels, a PVP (proline-valine-proline) or PAG (for KCNQ2 Pro308-Ala309-Gly310) bend was proposed as the “second hinge” of Kv channels (38–41). We tested whether changing either proline or alanine of the second hinge would cause alteration of ztz240 effects. The proline 308 mutant resulted in a non-functional channel. Interestingly, Ala309 mutants to cysteine, valine, and glycine abolish the current potentiation (Fig. 6, A and B). These changes also significantly and differentially affected the deactivation (Fig. 6C). The slow deactivation time constants decreased to 1793.4 ± 319.3 ms for A309C (n = 5, p < 0.05), 1678.9 ± 291.8 ms for A309V (n = 7, p < 0.05), and 4135.6 ± 251.9 ms for A309G (n = 5). In contrast, this series of mutants preserved similar sensitivity to retigabine and ZnPy (Fig. 6A). Taken together, these data revealed one critical residue that selectively changes ztz240 effects on KCNQ2 channels and are consistent with the notion that ztz240 potentiates KCNQ channels by acting on a new drug-channel binding site.
FIGURE 6.
Point mutations revealed the critical residue for ztz240 activity. A, the currents of wild type KCNQ2 and the indicated mutants in the absence or presence of 10 μm ztz240, 10 μm RTG, and 10 μm ZnPy, respectively. The S6 sequence of KCNQ2 is shown in the inset, and two hinges are highlighted in boldface type. B, histogram comparing outward current potentiation of A309C, A309V, and A309G with ztz240. C, histogram comparing deactivation of A309C, A309V, and A309G. Error bars, S.E.
Chemical Structures Critical for Slowing the Deactivation
To examine the SAR of ztz240, we designed and synthesized a series of analogues with different and combinatorial replacements on the two aromatic rings, for a total of 52 compounds, whose activities were first determined by a Tl+-based assay using a kinetic fluorescence reader (see “Experimental Procedures”) (28). Among them, six compounds (ztz222, ztz233, ztz239, ztz245, ztz250, and zls1726; Fig. 7A) were selected. They showed fluorescence signal increases equivalent to the positive controls: 10 μm retigabine and 10 μm ZnPy (Fig. 7B) displayed concentration-correlated signal increases (Fig. 7C). The whole cell patch clamp testing confirmed that the six compounds did indeed potentiate the KCNQ2 current (Fig. 7D), with potentiation ranging from 1.6- to 2.7-fold.
FIGURE 7.
The derivatives of ztz240 activate KCNQ2 channels. A, chemical structure of derivatives of ztz240 and six other derivatives. B, heat map of Tl+ flux assay (F/F0) for the derivatives in duplicate. The Tl+ flux assay is detailed under “Experimental Procedures.” Columns 1 and 2 show controls (buffer, Tl+ stimulus solutions, retigabine, ZnPy, and 5337031) for KCNQ2-HEK293 cells, and columns 23 and 24 show controls (buffer, Tl+ stimulus solutions, retigabine, ZnPy, and 5337031) for wild-type HEK293 cells. C, scatter plot Tl+ flux assay for the derivatives in two concentrations (2 and 20 μm). Black, not active in both concentrations; green, active only in high concentration (20 μm); blue, active in both concentrations. F0 and F, fluorescence before and after compound addition, respectively. D, histograms of whole cell patch clamp currents in the presence of the indicated compounds at 10 μm. Error bars, S.E.
Examination of the Tl+ assay results reveals that the amide linker between the phenyl ring and the pyridine ring is not tolerant of change. The nitrogen position in the heterocyclic ring is also critical for activity. Only those with nitrogen located at the 3-position exhibit pronounced activity. Moreover, the substitution pattern of fluorine(s) in the phenyl ring plays an important role. The fluorine(s) at position 4′ and/or 3′ is essential for strong activation, whereas the 2-position deteriorates the effect (see supplemental material).
The unique mode of action conferred by ztz240 is characterized by significant, prolonged deactivation and left shifting of activation V½; therefore, we used both the time constant of slow deactivation phase and ΔV½ as parameters to examine the SAR of the ztz240 analogues. As shown in Fig. 8, switching the fluorine from 4′ (ztz240) to 3′ (ztz239) resulted in a significant decrease in the slow time constant, from 3,230.5 ± 534.1 to 1,075.8 ± 44.8 ms (n = 3), and of ΔV½ from −42.4 ± 1.2 to −24.9 ± 3.8 mV (n = 3). After adding back the 4′-fluorine (ztz233), the slow time constant was restored to 3,074.2 ± 959.5 ms (n = 3). These studies demonstrate a more important role of the 4′-fluorine position relative to that of the 3′-fluorine position, and the 3′-fluorine position did not incur a negative effect on the activity. A similar trend is observed for compounds ztz250, ztz245, and ztz222, which bear bromine at position 4′ instead. These outcomes support the conclusion that fluorine substitution of the phenyl ring contributes to the slow deactivation. In contrast, it seems that either chlorine or bromine substitutions (e.g. ztz240 and ztz250) in the pyridine ring are acceptable for conferring the changes in deactivation and left shifting of V½. The phenomena are also observed with ztz239 versus ztz245 and with ztz233 versus ztz222, respectively.
FIGURE 8.
The roles of fluorine, chlorine, or bromine substitutions in KCNQ activation and deactivation. The chemical structure of the compounds, as indicated, and the corresponding representative traces in the absence (black) and presence (gray) of corresponding compounds are shown. The slow time constant (τs) (numbers in italic type) and shifting of V½ (numbers in normal type) of each compound are as indicated in the inset. For the recording protocol, see Fig. 3A.
Conversely, we compared two series of compounds by fixing the position of either 4′-fluorine or 3′- and 4′-fluorine on the phenyl ring. Compound ztz252, without the chlorine or bromine substituent, gave rise to only a slight left shift of V½ (−5.7 ± 2.3 mV) and did not exhibit slow deactivation (Fig. 9, left); the overall potentiation of outward current was minimal. A similar trend is observed with zls1726, which bears 3′- and 4′-fluorine but lacks chlorine or bromine. These results support the notion that the chlorine or bromine moiety plays an important role in mediating slow deactivation. Hence, the two modes of potentiation, hypopolarizing shift of V½ and slowing of deactivation, appear to be contributed by different groups on the compounds.
FIGURE 9.
The diagram shows the chlorine or bromine substitution is necessary for ztz240 activity on activation and deactivation of KCNQ2. The chemical structure of the compounds, as indicated, and the corresponding representative traces in the absence (black) and presence (gray) are shown beside the compound structure. The slow deactivation time constant (τs) (numbers in italic type) and shifting of V½ (numbers in normal type) of each compound are indicated in each inset. NA, not applicable (the compound did not cause two-phase deactivation). For the recording protocol, see Fig. 3A.
ztz233 is, in fact, identical to the structure of ICA-27243, which has been reported for its potent effects on both recombinant KCNQ2/3 channels and anti-epileptic activities (12, 14). The EC50 values for KCNQ2/3 were 5.7 ± 0.6 μm for ztz233 and 6.1 ± 1.2 μm for ztz240. This compares well with the reported EC50 of 4.8 ± 1.6 μm for ICA-27243 (14). Their effects on KCNQ4, under our experimental protocols, however, were comparable with EC50 values (9.4 ± 4.4 μm for ztz233 and 12.2 ± 1.0 μm for ztz240), similar to that in the earlier report (14) (Fig. 10). The slowing of KCNQ2/3 deactivation by ztz240 is markedly reduced (994.7 ± 178.0 and 71.1 ± 30.1 ms, respectively), consistent with the lack of effects on KCNQ3 subunits (Figs. 4 and 10).
FIGURE 10.
Comparison of ztz233 (ICA-27243) and ztz240 effects on KCNQ2/3 and KCNQ4. A and B, representative traces of ztz233 (ICA-27243) and ztz240 effects on KCNQ2/3 heteromultimers, under the same voltage steps as shown in Fig. 3A. C and D, G-V curve of ztz233 (ICA-27243) and ztz240 on KCNQ2/3 (n = 3–6). The same protocol for wild type KCNQ2 was used in Fig. 2B. E–H, dose-response curves of ztz233 (ICA-27243) and ztz240 on KCNQ2/3 and KCNQ4 as indicated (n = 4–10). Error bars, S.E.
DISCUSSION
The physiological roles of KCNQ channels have provided a clear rationale for both identification and pharmacological characterization of modulatory chemicals, either as a lead for drug development or as probes to understand channel gating mechanisms or functional roles. Through a screen of 20,000 compounds by Rb+ flux, we have isolated a number of compounds of distinct chemotypes. Both the present report and our earlier work have demonstrated that zinc pyrithione and ztz240 have distinct isoform specificity. Comparing specific mutant KCNQ2 channels with loss of sensitivity to zinc pyrithione or retigabine, our results support the notion that these compounds are not only representatives of different chemotypes but also that they recognize potentially different sites and act by a unique mechanism of action to cause potentiation.
Our screen utilized two different tracer ions, rubidium and thallium; both are permeable to most potassium channels. The level of validation from the primary screen is around 30%, depending on the triage criteria and hit selection threshold. Among those compounds that failed to validate, some may have interesting pharmacology that would justify further characterization. For example, one would predict that a “potassium” ionophore could also enable the readout signal, under conditions applied in our screen. But these compounds would probably fail to show channel specificity. The selection of chemotypes for further characterization was motivated by several factors. Our lead compound 5337031 and its related structures (Fig. 1A) resulting from our primary screen with a simple structure have provided a good starting point for its structural modification to seek more potent KCNQ activators, despite the fact that its structure is similar to that of ICA-27243. Initially, we decided to replace the “CH” moiety with an “N” in one of the phenyl rings. The conventional wisdom is that incorporation of a nitrogen atom can increase the chance of improving binding affinity to targeted ion channels, due to its positive charge characteristics. This is seen in several KCNQ modulators, including, for example, XE991, flupirtine, and retigabine (10). Accordingly, we positioned screening of the nitrogen at 2′, 3′, and 4′ to determine the SAR. Moreover, we incorporated halogens into the phenyl rings, guided by the Topliss scheme (34, 35) because halogens can provide quick SAR information about electronic and steric effects on bioactivities. It is noteworthy that nowadays, it has become general practice in drug design to incorporate fluorine atom(s) into bioactive molecules to modulate their biological properties (42). It has been demonstrated that in many cases, the substitution of hydrogen atom(s) with fluorine(s) in a biologically active molecule improves its properties, including potency and/or bioavailability. It is believed that these effects result from its distinctive characteristics as the most electronegative element, including a highly strong C–F bond and a size similar to that of the hydrogen atom. Such effects have been observed again in current studies of ztz222, -233, -239, -240, -245, -250, and -252. Finally, put together, the studies afford a nice example and an effective strategy for design and development of potent and selective ligand-targeting ion channels.
ztz240 and several related structures display rather pronounced effects to slow deactivation. Although both zinc pyrithione and retigabine have been known for their effects of slowing deactivation, the effect of ztz compounds on deactivation is far significant. The experimental results of the pronounced prolongation of tail currents in Fig. 5 were only observed by mixed compounds, either ztz240 with retigabine or ztz240 with zinc pyrithione. This provides compelling evidence suggesting that both compounds act on the channel to cause this non-additive effect. Mutagenesis studies reveal that Ala309 appears to preferentially affect ztz240 effects (Fig. 6). In addition, retigabine potentiates all but KCNQ1, whereas zinc pyrithione potentiates all KCNQ but KCNQ3. In contrast, ztz240 has subunit specificity for KCNQ2, KCNQ4, and KCNQ5 (Fig. 4). Therefore, the molecular mechanism of the ztz240 effect is unique, mediated by specific molecular determinants, and hence consistent with recognizing a new site. However, further experiments are needed to reach a firm conclusion.
The effect has been seen over a wide range of testing voltages (Fig. 3), arguing that the in vivo activity of ICA-27243 reported earlier is due to its effects on KCNQ channels (12). The effects of these compounds on receptor mediated suppression remain unknown and require further investigation. Potentiation compounds with minimal effects on voltage-mediated activation but profound effects on deactivation could be of great interest. One would predict that these compounds would act under the voltages that physiologically gate the channels of interest, reminiscent of allosteric modulators for receptors, which function only after a native ligand engages with the receptor. Thus, a deliberate search for channel potentiators with no or little effect on gating voltage is of special benefit. Indeed, the high throughput screening feasibility during the primary screen to specifically look for compounds affecting deactivation remains a major obstacle. Nonetheless, one could imagine innovative approaches by taking advantage of the wealth of structure function studies, which harbor abundant mutants and data that may be used to profile hits or even to conduct primary screens.
Supplementary Material
Acknowledgments
We thank Drs. M. Sanguinetti and T. McDonald for gifts of cDNAs. We thank Dr. Owen McManus and members of the Li laboratory for valuable discussions and comments on the manuscript. We thank Shunyou Long for automation support and Xiaofang Huang, Amy Steven, and Kaiping Xu for technical assistance.
Addendum
While this manuscript was under review, Padilla et al. (43) reported that ICA-27243 (ztz233 referenced in this report) interacts with a site in the voltage-sensitive domain.
This work was supported, in whole or in part, by National Institutes of Health Grants GM078579 and MH084691 (to M. L.). This work was also supported by a postdoctoral fellowship award from the American Heart Association (to Z. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental material, including compound structures and descriptions.
- SAR
- structure-activity relationship
- RTG
- retigabine
- ZnPy
- zinc pyrithione.
REFERENCES
- 1.Ashcroft F. M. (2006) Nature 440, 440–447 [DOI] [PubMed] [Google Scholar]
- 2.Doyle D. A. (2004) Trends Neurosci. 27, 298–302 [DOI] [PubMed] [Google Scholar]
- 3.Sigworth F. J. (1994) Q. Rev. Biophys. 27, 1–40 [DOI] [PubMed] [Google Scholar]
- 4.Bezanilla F., Stefani E. (1994) Annu. Rev. Biophys. Biomol. Struct. 23, 819–846 [DOI] [PubMed] [Google Scholar]
- 5.Horrigan F. T., Aldrich R. W. (2002) J. Gen. Physiol. 120, 267–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rae J. L., Dewey J., Rae J. S., Cooper K. (1990) Curr. Eye Res. 9, 847–861 [DOI] [PubMed] [Google Scholar]
- 7.Lawson K., McKay N. G. (2006) Curr. Pharm. Des. 12, 459–470 [DOI] [PubMed] [Google Scholar]
- 8.Surti T. S., Jan L. Y. (2005) Curr. Opin. Investig. Drugs 6, 704–711 [PubMed] [Google Scholar]
- 9.Singh N. A., Westenskow P., Charlier C., Pappas C., Leslie J., Dillon J., Anderson V. E., Sanguinetti M. C., Leppert M. F. (2003) Brain 126, 2726–2737 [DOI] [PubMed] [Google Scholar]
- 10.Xiong Q., Gao Z., Wang W., Li M. (2008) Trends Pharmacol. Sci. 29, 99–107 [DOI] [PubMed] [Google Scholar]
- 11.Redrobe J. P., Nielsen A. N. (2009) Behav. Brain Res. 198, 481–485 [DOI] [PubMed] [Google Scholar]
- 12.Roeloffs R., Wickenden A. D., Crean C., Werness S., McNaughton-Smith G., Stables J., McNamara J. O., Ghodadra N., Rigdon G. C. (2008) J. Pharmacol. Exp. Ther. 326, 818–828 [DOI] [PubMed] [Google Scholar]
- 13.Rogawski M. A. (2000) Trends Neurosci. 23, 393–398 [DOI] [PubMed] [Google Scholar]
- 14.Wickenden A. D., Krajewski J. L., London B., Wagoner P. K., Wilson W. A., Clark S., Roeloffs R., McNaughton-Smith G., Rigdon G. C. (2008) Mol. Pharmacol. 73, 977–986 [DOI] [PubMed] [Google Scholar]
- 15.Brown D. A. (2008) J. Physiol. 586, 1781–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange W., Geissendörfer J., Schenzer A., Grötzinger J., Seebohm G., Friedrich T., Schwake M. (2009) Mol. Pharmacol. 75, 272–280 [DOI] [PubMed] [Google Scholar]
- 17.Kubisch C., Schroeder B. C., Friedrich T., Lütjohann B., El-Amraoui A., Marlin S., Petit C., Jentsch T. J. (1999) Cell 96, 437–446 [DOI] [PubMed] [Google Scholar]
- 18.Schroeder B. C., Hechenberger M., Weinreich F., Kubisch C., Jentsch T. J. (2000) J. Biol. Chem. 275, 24089–24095 [DOI] [PubMed] [Google Scholar]
- 19.Shapiro M. S., Roche J. P., Kaftan E. J., Cruzblanca H., Mackie K., Hille B. (2000) J. Neurosci. 20, 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H. S., Brown B. S., McKinnon D., Cohen I. S. (2000) Mol. Pharmacol. 57, 1218–1223 [PubMed] [Google Scholar]
- 21.Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. (1996) Nature 384, 78–80 [DOI] [PubMed] [Google Scholar]
- 22.Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Nature 384, 80–83 [DOI] [PubMed] [Google Scholar]
- 23.Wang H. S., Pan Z., Shi W., Brown B. S., Wymore R. S., Cohen I. S., Dixon J. E., McKinnon D. (1998) Science 282, 1890–1893 [DOI] [PubMed] [Google Scholar]
- 24.Schwarz J. R., Glassmeier G., Cooper E. C., Kao T. C., Nodera H., Tabuena D., Kaji R., Bostock H. (2006) J. Physiol. 573, 17–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunlop J., Bowlby M., Peri R., Vasilyev D., Arias R. (2008) Nat. Rev. Drug Discov. 7, 358–368 [DOI] [PubMed] [Google Scholar]
- 26.Xu J., Wang X., Ensign B., Li M., Wu L., Guia A., Xu J. (2001) Drug Discov. Today 6, 1278–1287 [DOI] [PubMed] [Google Scholar]
- 27.Terstappen G. C. (1999) Anal. Biochem. 272, 149–155 [DOI] [PubMed] [Google Scholar]
- 28.Weaver C. D., Harden D., Dworetzky S. I., Robertson B., Knox R. J. (2004) J. Biomol. Screen 9, 671–677 [DOI] [PubMed] [Google Scholar]
- 29.Sun H., Liu X., Xiong Q., Shikano S., Li M. (2006) J. Biol. Chem. 281, 5877–5884 [DOI] [PubMed] [Google Scholar]
- 30.Sun H., Shikano S., Xiong Q., Li M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16964–16969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong Q., Sun H., Li M. (2007) Nat. Chem. Biol. 3, 287–296 [DOI] [PubMed] [Google Scholar]
- 32.Wuttke T. V., Seebohm G., Bail S., Maljevic S., Lerche H. (2005) Mol. Pharmacol. 67, 1009–1017 [DOI] [PubMed] [Google Scholar]
- 33.Schenzer A., Friedrich T., Pusch M., Saftig P., Jentsch T. J., Grötzinger J., Schwake M. (2005) J. Neurosci. 25, 5051–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Topliss J. G. (1972) J. Med. Chem. 15, 1006–1011 [DOI] [PubMed] [Google Scholar]
- 35.Topliss J. G. (1977) J. Med. Chem. 20, 463–469 [DOI] [PubMed] [Google Scholar]
- 36.Tatulian L., Delmas P., Abogadie F. C., Brown D. A. (2001) J. Neurosci. 21, 5535–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seebohm G., Pusch M., Chen J., Sanguinetti M. C. (2003) Circ. Res. 93, 941–947 [DOI] [PubMed] [Google Scholar]
- 38.Seebohm G., Strutz-Seebohm N., Ureche O. N., Baltaev R., Lampert A., Kornichuk G., Kamiya K., Wuttke T. V., Lerche H., Sanguinetti M. C., Lang F. (2006) Biophys. J. 90, 2235–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webster S. M., Del Camino D., Dekker J. P., Yellen G. (2004) Nature 428, 864–868 [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. (2002) Nature 417, 515–522 [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. (2002) Nature 417, 523–526 [DOI] [PubMed] [Google Scholar]
- 42.Müller K., Faeh C., Diederich F. (2007) Science 317, 1881–1886 [DOI] [PubMed] [Google Scholar]
- 43.Padilla K., Wickenden A. D., Gerlach A. C., McCormack K. (2009) Neurosci. Lett. 465, 138–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.