Abstract
Accumulating evidence reveals that sole mutations in hENT3 cause a spectrum of human genetic disorders. Among these include H syndrome, characterized by scleroderma, hyperpigmentation, hypertrichosis, hepatomegaly, cardiac abnormalities and musculoskeletal deformities, pigmented hypertrichotic dermatosis with insulin-dependent diabetes syndrome, characterized by autoantibody-negative diabetes mellitus and skin deformities, familial Rosai-Dorfman disease, characterized by short stature, familial histiocytosis and sinus histiocytosis with massive lymphadenopathy (SHML), characterized by severe tissue infiltration of immune cells and swollen lymph nodes. hENT3 spectrum disorders share a common mutation and share overlapping clinical manifestations that display many intriguing resemblances to mitochondrial and lysosomal disorders. Although earlier studies identify hENT3 as a mitochondrial and a lysosomal nucleoside transporter, the precise connections between hENT3 and the pathophysiology of these disorders remain unresolved. In this study, we performed functional and biochemical characterization of these mutations in hENT3. We report severe reductions/losses of hENT3 nucleoside transport functions of hENT3 syndrome mutants. In addition to transport alterations, we provide evidence for possible loss of hENT3 functions in all H and pigmented hypertrichotic dermatosis with insulin-dependent diabetes syndromes due to either mistrafficking or altered stability of mutant hENT3 proteins.
Keywords: Genetic Diseases, Membrane Trafficking, Nucleoside, Skin, Transport Drugs
Introduction
H syndrome and pigmented hypertrichotic dermatosis with insulin-dependent diabetes (PHID)3 syndrome are recently described autosomal-recessive genetic disorders in humans (1, 2). H syndrome patients display symptoms such as cutaneous hyperpigmentation, hypertrichosis, heart anomalities, hearing loss, hepatomegaly hypogonadism, and etc. (1). Patients with PHID exhibit childhood onset of pigmented hypertrichotic skin lesions associated with insulin-dependent diabetes mellitus (2). H and PHID syndromes are allelic with each other and share common clinical features (e.g. hypertrichosis) (2). Two recent reports by Molho-Pessach et al. (1) and Cliffe et al. (2) reveal that both syndromes are caused solely by the mutations in the SLC29A3 gene. A new report by Morgan et al. (3) shows a common SLC29A3 mutation (hENT3-G437R) identified in H and PHID syndromes also is involved in familial Rosai-Dorfman disease and sinus histiocytosis with massive lymphadenopathy (SHML) disorders. Familial Rosai-Dorfman disease and SHML disorders exhibit massive tissue infiltration of histiocytes and plasma cells as well as enlarged lymph nodes. The overlapping symptoms shared by all hENT3 disorders (hypertrichosis, short stature, lymphadenopathy, etc.) demonstrate the involvement of SLC29A3 mutations in a spectrum of human genetic disorders.
The SLC29A3 gene encodes human equilibrative nucleoside transporter 3 (hENT3), a member of a largely conserved group of solute carrier (SLC) transporters called the ENT or SLC29 family (4, 5). These facilitative transporters mediate salvage of hydrophilic nucleosides as well as nucleoside analogs used in the treatment of cancers and viral diseases (4, 5). In comparison with the other human ENT members, hENT3 is unique in that it functions intracellularly with maximal activity at an acidic pH range of 5.5–6.5 (6, 7). Whereas Baldwin et al. (6) reported that hENT3 is localized partially in the late endosomes and lysosomes, our recent studies indicate that hENT3 is also localized in the mitochondria (7). Low pH transport properties and subcellular localization of hENT3 in lysosomes and in mitochondria suggest that hENT3 possibly transports nucleosides from the inside of the lysosomes to the cytoplasm (6) as well as across the inner mitochondrial membrane (7). Although these data indicate that hENT3 is likely to perform physiological functions relating to lysosomes and mitochondria, direct evidence linking hENT3 to these organelle functions has not yet been established.
In a study involving 10 families affected with H syndrome, the following mutations and their consequences were documented (1). Two missense mutations (1279G→A, 1309G → A) involve substitution of Gly427 by Ser (G427S; 1279G→A) and Gly437 by Arg (G437R; 1309G→A), and one deletion mutation (1045delC) leads to a frameshift from amino acid position 345 (345FS) and early C-terminal truncation of the protein at residue 404 (1). PHID syndrome is caused by five different mutations: 940delT, 1330G→T, 347T→G, 1309G→A, and 1346C→G (2). Three mutations involve single amino acid changes, namely, substitutions of Met116 by Arg (M116R; 347T→G), Gly437 by Arg (G437R; 1309G→A), and Thr449 by Arg (T449R; 1346C→G). The deletion mutation 940delT leads to a frameshift from amino acid position 314 (314FS) and truncation at residue 444, and the nonsense mutation 1330G→T leads to truncation at residue 444 (E444X) (2). Although mutations in hENT3 are known to cause H and PHID syndromes, the mechanistic basis of development and pathogenesis of both of these syndromes are completely unknown. To elucidate the mechanistic basis of these syndromes, we functionally characterized hENT3 mutations.
EXPERIMENTAL PROCEDURES
Materials
NIH 3T3 fibroblasts (PA317) cells were purchased from ATCC (Manassas, VA). [3H]adenosine was obtained from Moravek Radiochemicals (Brea, CA) and [35S]methionine was obtained from MP Biomedicals (Solon, OH). A polyclonal antibody against hENT3 was described earlier (7), and goat polyclonal antibodies against C and N termini were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa 488- and 594-conjugated donkey anti-goat secondary antibody was obtained from Invitrogen. Uridine, DAPI, hexadimethrine bromide, MG132, leupeptin, and other chemicals were from Sigma. Collagenase A was from Roche Applied Science. BCA protein assay reagent was from Pierce. Fetal calf serum and horse serum were from HyClone Laboratories (Logan, UT). Fluorescent antifade mounting reagent and penicillin-streptomycin were obtained from Molecular Probes, Invitrogen.
Site-directed Mutagenesis
pOX-Δ36hENT3 and pLNCX2-hENT3HA constructs were described previously (7). Point mutations identified in H and PHID syndromes were introduced into pOX-Δ36hENT3 and LNCX2-Δ36hENT3 constructs using QuikChange® site-directed mutagenesis kit from Stratagene (La Jolla, CA) per the manufacturer's protocols. Mutagenic primers were synthesized and purified in HPLC grade (Integrated DNA Technologies) for mutagenesis. The following primer sets were used: 1045delC (forward, 5′- CCAAGTTTTTCATCCCCTCACTACCTTCCTCC-3′ and reverse, 5′-GGAGGAAGGTAGTGAGGGGATGAAAAACTTGG-3′); 1279G→A (forward, 5′-GGCTCAGCAACAGCTACCTCAGCACC-3′ and reverse, 5′-GGTGCTGAGGTAGCTGTTGCTGAGCC-3′); 1309G→A (forward, 5′-CCCTCCTCTACAGGCCTAAGATTGTG-3′ and reverse, 5′-CACAATCTTAGGCCTGTAGAGGAGGG-3′); 347T→G (forward, 5′-CCCTCCTCTACAGGCCTAAGATTGTG-3′ and reverse, 5′-GAAGTTGGCCACCAGGCACAGCCTGGAGGGCACGGTG-3′); 940delT (forward, 5′-GCTTCTGTGTCACC(del)ACGTCTTCTTCATCAC-3′ and reverse, 5′-GTGATGAAGAAGACGT(del)GGTGACACAGAAGC-3′); 1330G→T, (forward, 5′-CTAAGATTGTGCCCAGGTAGCTGGCTGAG-3′ and reverse, 5′-CTCAGCCAGCTACCTGGGCACAATCTTAG-3′); and 1346C→G (forward, 5′-GAGCTGGCTGAGGCCAGGGGAGTGGTGATGTCCTTTTATG-3′ and reverse, 5′-CATAAAAGGACATCACCACTCCCCTGGCCTCAGCCAGCTC-3′). The mutated plasmids were isolated using E.Z.N.A.TM plasmid mini prep kit I (Omega Bio-Tek, Norcross, GA) and the mutants were verified by sequencing at both ends (University of Georgia Sequencing and Synthesis Facility). Constructs containing both 1279G→A and 1309G→A mutations were created in a 1279G→A mutant template using 1309G→A primer sets.
Expression in Xenopus Oocytes
Synthesis of cRNAs and Xenopus oocyte expression study were conducted as described previously (4). The apparent Km and Vmax values were estimated by nonlinear regression analysis using Prism 5.0 software (GraphPad, La Jolla, CA). Values of adenosine influx in RNA-injected oocytes, corrected by subtracting the values of diffusional uptake in H2O-injected oocytes, were used to derive these parameters.
Cell Culture, Retroviral Production, and Stable Cell Line Generation
A retroviral packaging cell line (ATCC, CRL 9078) was maintained in Dulbecco's modified minimum essential medium (DMEM) supplemented with 5% fetal bovine serum. These cells were transfected with wild-type or mutant hENT3 constructs for viral production. Briefly, cells were incubated with a complex containing 8 μg of each of the plasmids, 24 μl of FuGENE 6 reagent (Roche Applied Science), and 800 μl of Opti-MEM serum-free medium (Invitrogen). After 48 h of transfection, viruses in the supernatant were collected and filtered. NIH 3T3 fibroblasts were virally infected few times, and polyclonal cultures were used for immunocytochemical analysis and pulse-chase experiments.
Immunostaining Analysis
Immunostaining analysis was performed as described previously (7). Images were captured with Nikon Eclipse Ti fluorescence microscope fitted with a 14-bit CCD camera (Nikon Instruments, Inc.). For double immunolocalization experiments, images were deconvoluted using Ti-E software (Nikon) and merged to identify the extent of colocalization.
35S-Labeled Pulse-Chase Experiment
Cells in 10-cm dishes were incubated in cysteine- and methionine-free DMEM containing 5% dialyzed FBS at 37 °C for 30 min (starvation period). Then, 15 μl of Tran35S-label (1 mCi/ml) was added to each plate, and the plates were incubated for 30 min at 37° C. The radioactive media was removed by washing the cells with 2 ml of warm PBS. After removing PBS, cells were chased for different time points with 5 ml of warm DMEM containing 5% FBS, 2 mm methionine, and 2 mm cysteine (chase period). The media was then removed, and the cells were washed and scraped with 0.5 ml of ice-cold PBS. The cells were transferred to microfuge tubes and homogenized using a syringe and a 21-gauge needle in lysis buffer (100 mm NaCl, 20 mm Tris-HCl, pH 7.6, and 1% Triton X-100 containing 2 mm phenylmethylsulfonyl fluoride and 5 μg of each pepstatin, leupeptin, and aprotinin) for immunoprecipitation and Western blotting analysis.
Immunoprecipitation, Western Blotting, and Phosphorimaging Analysis
One μl of anti-hENT3 goat polyclonal antibody was added to various protein normalized cell lysates and were incubated for 8 h at room temperature. Subsequently, the complex was treated with protein A-Sepharose beads for 4 h, and the beads were washed three times with 1× TBST (137 mm sodium chloride, 20 mm Tris, 0.1% Tween 20, pH 7.6). For Western blotting analysis, the beads were incubated with 1× SDS gel loading buffer at 100 °C for 4 min, and the immunoprecipitated protein samples were loaded on a 12% SDS gel. The proteins were transferred to a polyvinylidene fluoride membrane (Pall Co., Pensacola, FL) with a blotting apparatus. The membrane was exposed to a phosphorimaging screen for 4 days at −80 °C. Image capture and quantitation of protein bands were done by phosphorimaging (STORM 865, GE Biosciences) using ImageQuant TL software (GE Biosciences).
Statistical Analyses
Student's t test was used to identify significant differences, and each experiment was repeated at least three times. p < 0.05, p < 0.01, and p < 0.001 compared with transport/kinetics parameters values derived from Δ36hENT3-expressing oocytes were represented by one, two, and three asterisks, respectively.
RESULTS
All H and PHID Syndrome Mutations in hENT3 Reduce Nucleoside Transport Activity
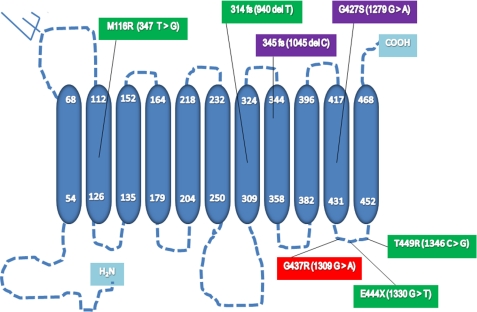
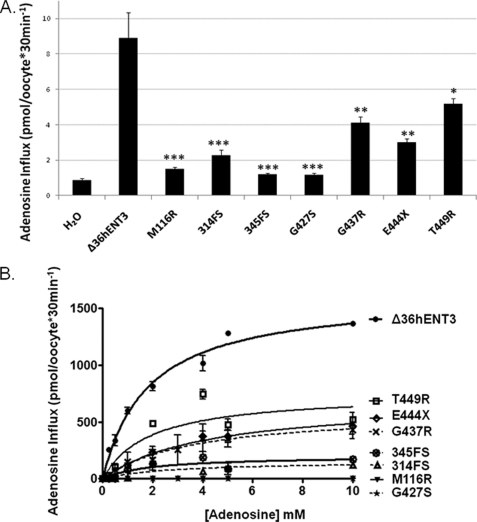
Positional analyses of H and PHID syndrome mutations in hENT3 show that all mutations except the M116R affect the C terminus of the protein (Fig. 1). To characterize the functional consequences of these H and PHID syndrome mutations, we generated mutant SLC29A3 constructs in a Xenopus oocyte expression vector through site-directed mutagenesis (see “Experimental Procedures” for details). We generated cRNAs for these mutants by in vitro transcription and injected them into Xenopus oocytes to examine the alterations in [3H]adenosine transports (Fig. 2, A and B). Because full-length hENT3 is retained intracellularly (6, 7), cRNAs for mutants were generated from an N-terminal-deleted pOXΔ36hENT3 construct (7), such that the mutant proteins also lacked the first 36 amino acids at the N terminus. As reported earlier (6, 7), oocytes expressing Δ36hENT3 exhibited an acidic pH-dependent [3H]adenosine transport. This transport was linear until ∼45 min and was maximal at pH 5.5. As depicted in Fig. 2A, all H and PHID syndrome mutants exhibited a severe reduction in [3H]adenosine transport at pH 5.5. Δ36hENT3-M116R, Δ36hENT3–345FS, and Δ36hENT3- G427S exhibited almost total losses in [3H]adenosine transport, whereas Δ36hENT3–314FS, Δ36hENT3-G437R, Δ36hENT3E444X, and Δ36hENT3-T449R retained some transport activity that were approximately one-half to one-fourth lower than the Δ36hENT3 transport.
FIGURE 1.
Positional localization of hENT3 syndromes mutations in hENT3. Secondary structure prediction of hENT3 shows the arrangement of the protein into 11 transmembrane segments (blue cylinders). The nucleotide (parentheses) changes and the corresponding amino acid changes of H and PHID syndromes mutations in SLC29A3 are shown in purple and green, respectively. The common mutation identified in H, PHID, Rosai-Dorfman disease, and SHML (familial histiocytosis, FHC) syndromes is shown in red. Two mutations that cause a frameshift and premature termination of hENT3 translation are marked fs after mutational positions.
FIGURE 2.
Mutant hENT3 proteins exhibit reduced nucleoside transport activity. Uptake of [3H]adenosine (0.026 μm) in oocytes after 24 h of injection of Δ36hENT3 or various H and PHID syndrome mutant cRNAs. A, pH-dependent uptake at pH = 5.5. B, concentration-dependent (0–10 mm) uptake at pH = 5.5. Data represent mean ± S.E. from three independent experiments. *, p < 0.05, **, p < 0.01, and ***, p < 0.001.
H and PHID Syndrome Mutations in hENT3 Alter the Kinetics of Adenosine Transport
To probe into the possible mechanisms of nucleoside transport alterations, we next determined the alterations in the maximal influx rates (Vmax) and apparent affinities (measured by Km) for [3H]adenosine transport by H and PHID syndrome mutants (Fig. 2B). Our analysis indicated that Δ36hENT3 transported adenosine at an apparent Km and Vmax of 1.8 ± 0.5 mm and 1621.0 ± 93.7 pmol/oocyte/30 min, respectively. Results in Table 1 show that the Vmax and/or the Km for adenosine transport were severely diminished for the frameshift mutants, Δ36hENT3–314FS and Δ36hENT3–345FS. The same was the case with all the mutants occurring in the sixth intracytoplasmic domain of hENT3, with an exception of Δ36hENT3-T449R which had no significant change in Km. An analysis of the activity loss caused by each mutation (calculated in Vmax/Km; Table 1) showed that the frameshift mutations in hENT3 (314FS and 345FS) were associated with 8–16-fold decreases in activities, whereas the three mutations in the last intracytoplasmic domain of hENT3 brought about 2–5-fold decreases.
TABLE 1.
Kinetics of adenosine transport by H and PHID mutants
Vmax, Km, and Vmax/Km values of adenosine influx determined by nonlinear regression analysis (Michalis-Menten). Data (Vmax, Km) represent mean ± S.E. from three independent experiments.
| Vmax | Km | Vmax/Km | |
|---|---|---|---|
| pmol/oocyte/30 min | mm | (pmol/oocyte/30 min)/mm | |
| Δ36hENT3 | 1621.0 ± 93.7 | 1.8 ± 0.5 | 900.5 |
| M116R | |||
| 314FS | 155.6 ± 73.4a | 2.9 ± 3.8 | 53.7 |
| 345FS | 202.4 ± 63.7a | 1.9 ± 2.0 | 106.3 |
| G427S | |||
| G437R | 598.7 ± 72.9a | 3.6 ± 1.0c | 166.3 |
| E444X | 683.7 ± 88.7a | 4.1 ± 1.7c | 166.8 |
| T449R | 750.3 ± 198.9b | 1.9 ± 1.3 | 394.9 |
a p < 0.001.
b p < 0.01.
c p < 0.05.
Mutations M116R, 314FS, and 345FS Cause Mistrafficking of hENT3 to Endoplasmic Reticulum or Golgi
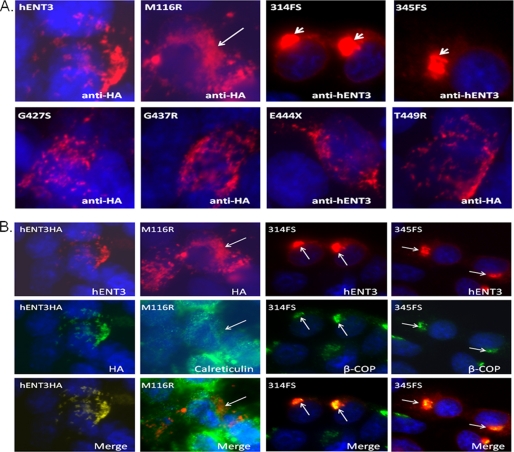
Because a reduction in Vmax of substrate transport could be due to defects in cellular trafficking (8), we determined whether some of the hENT3 mutant proteins exhibited a lack of proper targeting to mitochondria or to lysosomes. We recreated the H and PHID syndrome mutations on full-length hENT3 with an epitope (hemagglutinin (HA)) tag at their C or N termini. The HA-tagged mutants were then expressed in the NIH 3T3 fibroblasts using retroviral gene transfer (see “Experimental Procedures” for details), and their immunolocalization patterns were examined (Fig. 3A). As reported earlier (6, 7), wild-type hENT3 exhibited vesicular and slightly elongated staining patterns (Fig. 3A) and showed partial colocalization with HSP70 mitochondrial marker and LAMP2 lysosomal marker (data not shown). Furthermore, the immunostaining pattern of four of the mutants (hENT3-G427S, hENT3-G437R, hENT3-E444X, and hENT3-T449R) did not differ considerably from wild-type hENT3 staining (Fig. 3A) (6, 7). However, hENT3-M116R mutant exhibited a somewhat diffuse/reticular staining pattern, whereas the two frameshift mutants (hENT3–314FS and hENT3–345FS) exhibited distinct “button-like” staining patterns at the perinuclear area (Fig. 3A). To investigate the subcellular localization of the latter three mutants, we further examined their colocalization with markers of several subcellular organelles. Fig. 3B shows that the hENT3-M116R mutant was retained partially in the endoplasmic reticulum (ER) as evidenced by some colocalization with calreticulin (an ER marker). Conversely, hENT3–314FS and hENT3–345FS mutant hENT3 proteins were mistargeted extensively to the Golgi, which was indicated by their complete colocalization with β-COP (β-coatomer protein; a Golgi marker) (Fig. 3B). These results suggest that mistargeting to the ER or Golgi resulted in impaired nucleoside transport activities of M116R, 314FS, and 345FS mutants.
FIGURE 3.
Mistargeting of mutant hENT3 proteins. A, localization of mutant hENT3 proteins in NIH 3T3 fibroblasts. Cells were infected with retroviruses harboring hENT3 or mutant hENT3 proteins, and polyclonal cultures were examined. Cells were fixed, permeabilized, and immunostained with 1:1000 anti-hENT3 rabbit/goat polyclonal or 1:10,000 anti-HA rabbit polyclonal antibodies. The arrowheads indicate a button-like staining pattern at the paranuclear area with 314FS and 345FS mutants, and an arrow indicates a diffuse/reticular staining pattern with M116R mutant. The original magnifications are ×60. B, subcellular localization of hENT3 mutants. NIH 3T3 fibroblasts expressing M116R, 314FS, or 345FS mutants were coimmunostained with a goat anti-hENT3 polyclonal (mouse anti-HA monoclonal for cells expressing M116R) and a rabbit anti-calreticulin or an anti-β-COP polyclonal antibody. hENT3HA expressing cells coimmunostained with a rabbit anti-hENT3 polyclonal and a mouse anti-HA monoclonal antibody is shown as a control (left). Nuclei stained with DAPI (blue). The original magnification is ×60.
Because earlier studies indicate that cellular mistargeting/mislocalization defects of certain disease-causing mutations (e.g. cystic fibrosis) in transmembrane proteins can be corrected by chemical chaperones or their transport substrates (9, 10, 11), we next investigated whether hENT3 subcellular targeting defects due to mutations can be corrected by treatment with a purine (adenosine; 10 μm) or a pyrimidine (cytidine; 10 μm) nucleoside (6, 7). However, our results indicated pretreatment (16 h) of NIH 3T3 fibroblasts or Xenopus oocytes expressing M116R, 314FS, or 345FS mutants with one or both of these substrates neither rescued normal mitochondrial/lysosomal localization nor increased [3H]adenosine transport (data not shown).
Glycine at Position 427 in hENT3 Is Critical for Nucleoside Transport Activity: Dual Mutations G427S and G437R Exhibit Null Activity
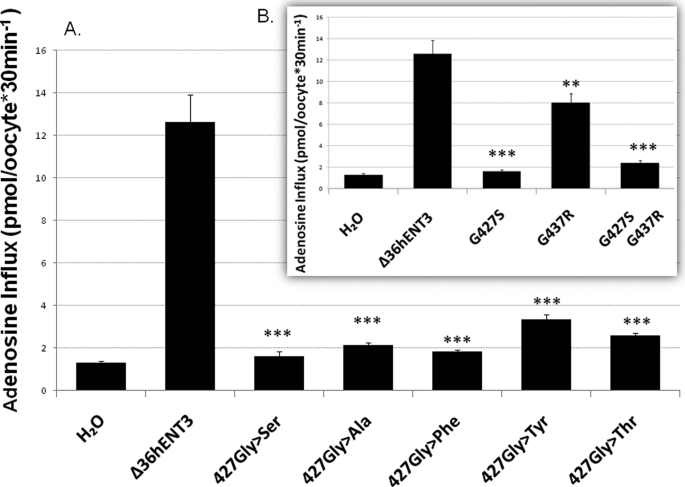
Interestingly, hENT3-G427S exhibited a total loss of transport activity, despite expression in oocytes, as judged by the presence of a band corresponding to hENT3-G427S in Western blots of oocyte extracts (data not shown) and no change in localization (Fig. 3A). Because this mutation involves substitution of glycine by serine in the middle of the 10th TMD of hENT3, we hypothesized that the hydroxyl group of Ser blocks the transport activity of hENT3 by altering the packing of the 10th TMD with other helices, thereby altering the transport of the nucleoside within the translocation pore. To test this, we first studied the effects of changing the Gly to an amino acid with no hydroxyl group (Ala), with a bulky phenyl group (Phe), phenyl and hydroxyl groups (Tyr), or with methyl and hydroxyl groups (Thr). As seen in Fig. 4A, none of these substitutions in Δ36hENT3 regained adenosine transport activity, suggesting the glycine at position 427 in hENT3 is a critical residue for nucleoside transport function.
FIGURE 4.
Scanning mutagenesis and transport activity of Δ36hENT3 mutants with various substitutions at position 427. A, uptake of [3H]adenosine (0.026 μm) in oocytes expressing the indicated mutants. B, uptake of [3H]adenosine in Δ36hENT31279G→A, Δ36hENT31309G→A, and in the dual mutant-injected oocytes. Data represent mean ± S.E. from three independent experiments. **, p < 0.01, and ***, p < 0.001.
Because one of the H syndrome patients was a compound heterozygote for both G427S and G437R (1), we also studied the effect of these dual mutations on the transport activity of hENT3. As expected, the double mutant showed a total loss of activity (Fig. 4B, inset).
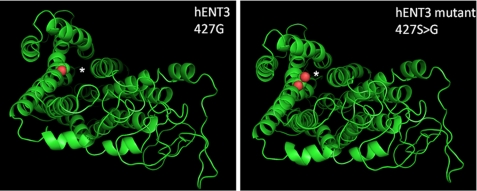
Furthermore, structural models of hENT3, built based on the crystal structure of glycerol-3-phosphate transporter (Protein Data Bank code 1PW4; Fig. 5), indicated that the TM10 domain can potentially form one of the pore-lining helices for nucleoside transport and that mutation of Gly427 located in the TM10 domain by serine or any other amino acid can potentially alter the size of the pore by disrupting the orientation of the pore-lining helices.
FIGURE 5.
Structural model of hENT3 and predicted location of G427S mutation. Left, structural model of hENT3 showing the location of Gly427 (in spheres representation; red). The model was built using the MUSTER (25) threading program. Because glycerol 3-phosphate transporter (Protein Data Bank code 1pw4 (26)) showed the highest Z-score (Z-score = 6) with hENT3, it was used as the structural template for threading. Right, model of hENT3 showing the location of G427S mutant. G427S mutation in hENT3 (red) was modeled using the MODELLER program (27) and displayed using the PyMOL program. The membrane spanning helical segments and loop regions are shown by schematic representation (green), and the predicted translocation pore (based on the crystal structure of glycerol 3-phosphate transporter) is shown by an asterisk.
Human ENT3 Is Predominantly Degraded by the Lysosomal Pathway: Mutations G437R and E444X Cause Accelerated Turnover of hENT3 Protein
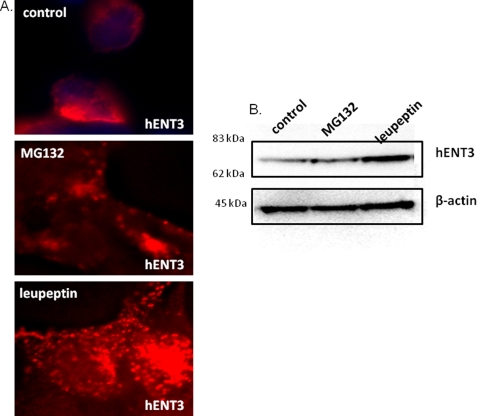
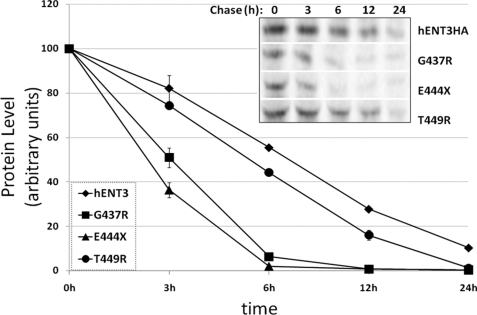
The results so far suggest all mutants except those occurring in the last cytoplasmic loop of hENT3 bring a complete loss of hENT3 nucleoside transport. Because loss of protein stability could be a possible cause for reduction in transport activity, and moreover, an earlier study demonstrated loss of protein stability for the T449R mutant in patients (2), we hypothesized that the three mutants closely positioned in the last cytoplasmic loop of hENT3 (G437R, E444X, and T449R) would exhibit altered protein stabilities. To address this, we first examined the pathways of degradation of wild-type hENT3 in mammalian cells. To do so, we treated NIH 3T3 fibroblasts expressing hENT3HA with a proteasomal inhibitor (20 μm MG132) or a lysosomal inhibitor (10 μm leupeptin), and the levels of accumulation of hENT3 were judged by immunocytochemical (Fig. 6A) and Western blotting analyses (Fig. 6B). Treatment of leupeptin increased the staining intensity of hENT3 and displayed a more vesicular staining pattern for hENT3 compared with the vehicle-treated cells (Fig. 6A). These results were consistent with accumulation of hENT3 protein in the lysosomes. However, the MG132 treatment exhibited no discernable alterations in hENT3 staining pattern and was comparable to vehicle-treated cells (Fig. 6A). Consistently, Western blotting analysis indicated increased accumulation of hENT3 protein only in leupeptin-treated cells and not in MG132-treated cells (Fig. 6B). Together, these data indicate that the wild-type hENT3 was predominantly subjected to lysosomal degradation. Consequently, we investigated the turnover of three mutant proteins (hENT3-G437R, hENT3-E444X, and hENT3-T449R) by performing 35S-labeled pulse-chase experiments (see “Experimental Procedures” for details). After radiolabeling the proteins (pulse), we examined the disappearance of mutant hENT3 proteins for different time intervals (chase) and compared the results with that seen in wild-type hENT3. The results depicted in Fig. 7 clearly show an accelerated turnover of two of the three hENT3 mutants examined (G437R and E444X), suggesting decreased stability as an additional cause of aberrant nucleoside transport in H and PHID syndrome patients. T449R mutant turnover was only slightly increased (Fig. 7) compared with the wild-type hENT3.
FIGURE 6.
hENT3 is predominantly degraded by the lysosomal pathway. NIH 3T3 fibroblasts expressing hENT3HA were treated with dimethyl sulfoxide (control), 20 μm MG132 (proteasomal inhibitor), or 10 μm leupeptin (lysosomal inhibitor) for 16 h, and hENT3 was accumulation examined. A, immunostaining analysis of hENT3 (red) using 1:1000 rabbit anti-HA polyclonal antibody indicated increased staining intensity of hENT3 in leupeptin-treated cells (bottom panel) but not in MG132-treated cells (middle panel). The original magnification is ×40. B, Western blotting analysis indicated a higher accumulation of hENT3 (∼65 kDa) in leupeptin-treated cells (right lane) than seen in MG132-treated cells (middle lane). β-Actin (45 kDa) was used as a loading control.
FIGURE 7.
Accelerated turnover of intracytoplasmic mutant hENT3 proteins. NIH 3T3 fibroblasts expressing the wild-type or mutant hENT3 proteins were subjected to [35S]methionine labeled pulse-chase experiments, and turnover rates were examined (see “Experimental Procedures”). The intensity of hENT3 and mutant hENT3 protein bands was quantified by densitometry and normalized to the control. Results are average ± S.D. of two independent experiments. A representative blot is shown (inset).
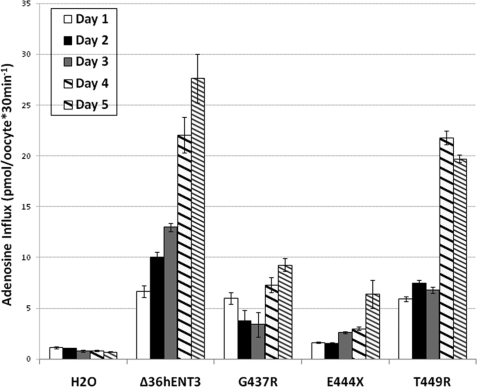
Finally, to test whether differences in the protein stability of hENT3 mutants can be reflected in the transport activity of Xenopus oocytes expressing these mutants, we performed a timed analysis of [3H]adenosine uptake in G437R, E444X, and T449R mutant cRNA-injected Xenopus oocytes. Results shown in Fig. 8 confirmed parallel reductions in [3H]adenosine influx rates in oocytes expressing these mutants compared with those expressing Δ36hENT3.
FIGURE 8.
Decreased [3H]adenosine influx rates by Xenopus oocytes expressing intracytoplasmic mutant hENT3 proteins. Uptake of [3H]adenosine (0.026 μm) in oocytes expressing Δ36hENT3 or intracytoplasmic mutant Δ36hENT3 proteins was measured after allowing expression of proteins for various time intervals (days 1–5). Uptake in oocytes injected with H2O was used as a control. The bars represent mean ± S.E. (n = 3).
DISCUSSION
Earlier studies in Xenopus oocytes demonstrated hENT3 as an acidic pH-dependent nucleoside transporter (6, 7). This characterization was performed after deletion of 36 N-terminal amino residues (Δ36hENT3) or after replacement of the dileucine motif at positions 31 and 32 with alanine residues to allow redirection of hENT3 to the oocyte cell surface (6, 7). At the beginning of the study, we have tried extensively to redirect hENT3 to the mammalian (Madin-Darby canine kidney) cell surface by the aforementioned approaches to retrieve functional hENT3 transport activity at the cell surface. Despite successful targeting of both Δ36hENT3 and hENT3LL-AA to the Madin-Darby canine kidney cell surface, so far, all of our attempts to retrieve hENT3 transport activity at the mammalian cell surface have failed. Because of this technical issue, we characterized mutations in hENT3 in Xenopus oocytes after removing the first 36 amino acids in mutant hENT3 proteins. Kinetic characterization of mutations in hENT3 in oocytes identified all hENT3 mutations exhibiting severe reductions in the maximal influx rates (decrease in Vmax) and apparent affinities (increase in Km) for adenosine transport with an overall impairment in intrinsic transport activities (decrease in Vmax/Km). Four of seven hENT3 mutations (M116R, 314FS, 345FS, and G427S) exhibited almost a complete reduction in adenosine transport activity, whereas three mutations occurring at the C terminus end of hENT3 (G437R, E444X, and T449R) displayed a partial reduction in nucleoside transport activity. These data provide direct evidence of aberrations in nucleoside transport occurring as a consequence of disease-causing mutations in hENT3.
Baldwin et al. (6) reported a partial localization of hENT3 in acidic late endosomal/lysosomal compartments. Subsequently, we reported that in addition to lysosomes, endogenous hENT3 is localized substantially to mitochondria in a number of human cell types (7). Because loss or reduction of nucleoside transport functions due to reduction in Vmax can occur due to the mistargeting of mutant proteins away from mitochondria or lysosomes, we further examined mislocalization as a possible outcome for mutations in hENT3. Because our previous studies demonstrate that tagging yellow florescence protein (YFP) to the N terminus of hENT3 largely diverts the fusion protein to lysosomes (7), we recreated the disease mutations on full-length hENT3 proteins that contained a small peptide HA tag at their C or N termini. Our results demonstrate that wild-type hENT3 tagged to HA faithfully reproduced endogenous hENT3 localization in several human cell types (data not shown). However, two frameshift mutants in the TM8 and TM10 domains (314FS and 345FS) and the mutation replacing a hydrophobic residue by a basic amino acid in the TM2 domain of hENT3 (M116R) displayed a partial or complete mistargeting to the ER or Golgi. Although the precise reasons for the retention of M116R mutant in the ER is unclear, it is likely that the large irrelevant peptide sequence replacing the transmembrane domains 8–11 of hENT3 consequent to frame shifts at positions 314 and 345 resulted in misfolding and mistargeting of these two mutants. Nevertheless, mislocalization seemed to be mainly responsible for the very low activity or inactivity of these three mutants in Xenopus oocytes. If such perturbations also occur in mammalian cells, the abundance of hENT3 (hence Vmax) targeted to mitochondria or lysosomes also will be reduced. Loss of mitochondrial or lysosomal nucleoside transport coupled with the possible gain of function in other subcellular organelles (e.g. Golgi) could result in severe alterations in subcellular compartmentalization of nucleosides. These outcomes likely are to result in abnormal cellular signaling and/or aberrant cellular metabolism exhibited as severe disease manifestations. The wide-ranging spectrum of deformities seen in all hENT3 spectrum disorders as well as their clinical heterogeneities and severities (1–3) support this possibility very well.
Mutants 314FS, 345FS, and M116R are clearly mislocalized to the ER/Golgi and, as expected, had only very low or very minimal Vmax levels (∼10-fold lesser). Unlike these three mutants, the complete inactivity of the G427S mutant was quite surprising despite the lack of any obvious change in the subcellular localization. Because loss of nucleoside transport activity could be due to the lack of expression and/or low intrinsic specific activity, we first examined whether the lack of expression in oocytes caused a problem in detecting transport activity for G427S mutant. However, our results verified the expression of this mutant in oocytes by identifying a band corresponding to del36hENT3–1279 in oocyte extracts. This suggests that expression of this mutant is not the reason for inactivity. One likely possibility is that the G427S mutation allows proper expression and mitochondrial/lysosomal targeting but renders the transporter dysfunctional by other mechanisms such as improper packing of TM domains lining the translocation pore or altering the local transporter-substrate interactions. Although we do not have definitive evidence for the latter, the following findings support this assertion. First, none of the experimented amino acid substitutions at this mutational position (427), except for the original glycine, could retain nucleoside transport activity (Fig. 4A). The fact that minor substitutions such as alanine or other amino acids with or without a hydroxyl group made no difference in activity level indicated that it was not the hydroxyl group in particular, but any side chain substitution cannot be tolerated at the Gly427 position without compromising transport function. Second, another hENT3 disease mutant, G437R, which exhibited reduced nucleoside transport compared with wild-type hENT3, became completely inactive when the G427S mutation was additionally introduced to the G437R mutation (see Fig. 4B). Third, several examples in literature (9), including another member of the ENT family (ENT1) (12), have shown that mutations replacing glycine residues in the transmembrane domains can affect functionality without perturbing cellular expression or membrane targeting. Fourth, a molecular modeling approach identified Gly427 as a putative translocation pore-lining residue in hENT3 and that any substitutions could potentially alter the size of the translocation pore. Consistently, several previous studies on membrane proteins have highlighted the importance of conserved glycines in helix packing (13) and ligand transport across the membrane (14). These data, together with the fact that Gly427 is highly conserved across species (1), support that Gly427 per se is an absolute requirement for facilitating hENT3-mediated nucleoside transport activity.
Membrane proteins can be targeted to both proteasomal and lysosomal degradation pathways and mutations in membrane proteins can increase the turnover of these proteins by these pathways. Although the degradation pathways of equilibrative and concentrative nucleoside transporters are largely unknown, an earlier study has shown the third member of the concentrative nucleoside transporter family (hCNT3) is not sensitive to inhibition by a proteasomal inhibitor (MG132) (15). Consistently, our studies identified the degradation of hENT3 to be predominantly via the lysosomal pathway and not the proteasomal pathway. Cliffe et al. (2) recently showed that the reduction of activity in Δ36hENT3-T449R, positionally localized in the last cytoplasmic domain in hENT3, was associated with the presence of low levels of mutated hENT3 mRNA and proteins in patients. As mutations that are closely located in the cytoplasm domain are likely to have common outcomes, we tested whether the three closely positioned mutations in the last cytoplasmic domain of hENT3 would trigger rapid hENT3 protein turnover. We found that hENT3-T449R turnover was increased only slightly (Fig. 7) compared with the wild-type hENT3. These data suggest that the reduced hENT3 mRNA levels, as observed in patients (2), and not decreased protein stability, acted as a major cause for the decrease in transport of the T449R mutant. However, mutants G437R and the truncation mutant E444X both clearly exhibited an accelerated turnover compared with wild-type hENT3. These results show decreased hENT3 stability as an additional cause of dysfunction in H and PHID syndrome patients carrying mutations G437R and E444X. In addition, these data also suggest that the last cytoplasmic loop in hENT3 could act as an important regulatory domain governing the degradation and turnover of hENT3 protein.
In summary, this is the first study where the functional activity of mutations in hENT3 associated with a spectrum of human genetic disorders is characterized. We have identified defects in subcellular localization with M116R, 314FS, and 345FS mutants, impairments in protein stability with G437R and E444X mutants, and reductions in nucleoside transport activity with all H and PHID syndrome mutants. By identifying these defects, we have obtained evidence for the loss of hENT3 function in all H and PHID syndrome patients. The noticeable similarities between hENT3 spectrum disorders and their resemblances to mitochondrial DNA (mtDNA) mutations (e.g. short stature, cardiovascular problems) (16, 17), mtDNA rearrangements (e.g. diabetes, sensorineural hearing loss) (18, 19), mitochondrial respiratory chain defects (e.g. mental retardation) (20), and lysosomal defects (e.g. reticuloendothelial involvement) (1), suggest that the abnormalities arise from a common dysfunction that is potentially linked to hENT3 transport activities in the mitochondrial and lysosomal compartments. As hENT3 is normally functional in the membranes of mitochondria and lysosomes (6, 7), hENT3 syndrome mutations are likely to alter nucleoside pools in those organelles. These alterations could potentially perturb mitochondrial and lysosomal homeostatic functions dependent on hENT3 substrates (e.g. nucleosides, nucleobases) (6, 7). For instance, when the level of guanosine sensed by mitochondria decreases, apoptosis may not be initiated, and cell proliferation could be affected (21). Another likely consequence is the reduction in mitochondrial NTP production, which could lead to a decrease in the synthesis of mtDNA. The level of mitochondrial ATP goes down as well, decreasing the synthesis of other triphosphonucleosides such as GTP, which is important for protein synthesis (22). The lower level of ATP also means less energy to power the chaperones of the protein import machinery in mitochondria (23). Because the mitochondrial ATP level is also in control of the (excitatory) neurotransmission and/or intracellular signaling, many of the normal physiological roles mediated by ATP could become altered (24). Future studies are needed to further identify the effects of aberrant hENT3 nucleoside transport on the pathogenesis of these syndromes.
Acknowledgment
We thank Dr. Chung-Ming Tse for providing an anti-hENT3 rabbit polyclonal antibody.
The work was supported, in whole or in part, by NCI, National Institutes of Health Award 5 P50-CA128613-SPORE in Head-and-Neck Cancer (to R. G.). This work was also supported by the Department of Pharmaceutical and Biomedical Sciences (University of Georgia).
- PHID
- pigmented hypertrichotic dermatosis with insulin-dependent diabetes
- hENT
- human equilibrative nucleoside transporter
- BCA
- bicinchoninic acid
- FS
- frameshift.
REFERENCES
- 1.Molho-Pessach V., Lerer I., Abeliovich D., Agha Z., Abu Libdeh A., Broshtilova V., Elpeleg O., Zlotogorski A. (2008) Am. J. Hum. Genet. 83, 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cliffe S. T., Kramer J. M., Hussain K., Robben J. H., de Jong E. K., de Brouwer A. P., Nibbeling E., Kamsteeg E. J., Wong M., Prendiville J., James C., Padidela R., Becknell C., van Bokhoven H., Deen P. M., Hennekam R. C., Lindeman R., Schenck A., Roscioli T., Buckley M. F. (2009) Hum. Mol. Genet. 18, 2257–2265 [DOI] [PubMed] [Google Scholar]
- 3.Morgan N. V., Morris M. R., Cangul H., Gleeson D., Straatman-Iwanowska A., Davies N., Keenan S., Pasha S., Rahman F., Gentle D., Vreeswijk M. P., Devilee P., Knowles M. A., Ceylaner S., Trembath R. C., Dalence C., Kismet E., Köseoğlu V., Rossbach H. C., Gissen P., Tannahill D., Maher E. R. (2010) PLoS Genet. 6, e1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. (2008) Xenobiotica 38, 995–1021 [DOI] [PubMed] [Google Scholar]
- 5.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. (2004) Pflugers Arch. 447, 735–743 [DOI] [PubMed] [Google Scholar]
- 6.Baldwin S. A., Yao S. Y., Hyde R. J., Ng A. M., Foppolo S., Barnes K., Ritzel M. W., Cass C. E., Young J. D. (2005) J. Biol. Chem. 280, 15880–15887 [DOI] [PubMed] [Google Scholar]
- 7.Govindarajan R., Leung G. P., Zhou M., Tse C. M., Wang J., Unadkat J. D. (2009) Am. J. Physiol. Gastrointest. Liver Physiol. 296, G910–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders C. R., Ismail-Beigi F., McEnery M. W. (2001) Biochemistry 40, 9453–9459 [DOI] [PubMed] [Google Scholar]
- 9.Brown C. R., Hong-Brown L. Q., Biwersi J., Verkman A. S., Welch W. J. (1996) Cell. Stress Chaperones 1, 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh O. V., Pollard H. B., Zeitlin P. L. (2008) Mol. Cell. Proteomics 7, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loo T. W., Bartlett M. C., Clarke D. M. (2005) Mol. Pharm. 2, 407–413 [DOI] [PubMed] [Google Scholar]
- 12.SenGupta D. J., Lum P. Y., Lai Y., Shubochkina E., Bakken A. H., Schneider G., Unadkat J. D. (2002) Biochemistry 41, 1512–1519 [DOI] [PubMed] [Google Scholar]
- 13.Javadpour M. M., Eilers M., Groesbeek M., Smith S. O. (1999) Biophys. J 77, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ermolova N. V., Smirnova I. N., Kasho V. N., Kaback H. R. (2005) Biochemistry 44, 7669–7677 [DOI] [PubMed] [Google Scholar]
- 15.Errasti-Murugarren E., Molina-Arcas M., Casado F. J., Pastor-Anglada M. (2009) FASEB J. 23, 172–182 [DOI] [PubMed] [Google Scholar]
- 16.DiMauro S., Schon E. A. (2001) Am. J. Med. Genet. 106, 18–26 [DOI] [PubMed] [Google Scholar]
- 17.Goto Y., Nonaka I., Horai S. (1990) Nature 348, 651–653 [DOI] [PubMed] [Google Scholar]
- 18.Ballinger S. W., Shoffner J. M., Gebhart S., Koontz D. A., Wallace D. C. (1994) Nat. Genet. 7, 458–459 [DOI] [PubMed] [Google Scholar]
- 19.Ballinger S. W., Shoffner J. M., Hedaya E. V., Trounce I., Polak M. A., Koontz D. A., Wallace D. C. (1992) Nat. Genet. 1, 11–15 [DOI] [PubMed] [Google Scholar]
- 20.Bindoff L. A., Desnuelle C., Birch-Machin M. A., Pellissier J. F., Serratrice G., Dravet C., Bureau M., Howell N., Turnbull D. M. (1991) J. Neurol. Sci. 102, 17–24 [DOI] [PubMed] [Google Scholar]
- 21.Flanagan S. A., Gandhi V., Meckling K. A. (2007) Leuk. Lymphoma 48, 1816–1827 [DOI] [PubMed] [Google Scholar]
- 22.Thomson M. (1998) Biochim. Biophysc. Acta. 1403, 211–218 [DOI] [PubMed] [Google Scholar]
- 23.Neupert W., Herrmann J. M. (2007) Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 24.Kann O., Kovács R. (2007) Am. J. Physiol. Cell Physiol 292, C641–657 [DOI] [PubMed] [Google Scholar]
- 25.Wu S., Zhang Y. (2008) Proteins 72, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 27.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]