Abstract
In humans the mitochondrial inner membrane protein Oxa1L is involved in the biogenesis of membrane proteins and facilitates the insertion of both mitochondrial- and nuclear-encoded proteins from the mitochondrial matrix into the inner membrane. The C-terminal ∼100-amino acid tail of Oxa1L (Oxa1L-CTT) binds to mitochondrial ribosomes and plays a role in the co-translational insertion of mitochondria-synthesized proteins into the inner membrane. Contrary to suggestions made for yeast Oxa1p, our results indicate that the C-terminal tail of human Oxa1L does not form a coiled-coil helical structure in solution. The Oxa1L-CTT exists primarily as a monomer in solution but forms dimers and tetramers at high salt concentrations. The binding of Oxa1L-CTT to mitochondrial ribosomes is an enthalpy-driven process with a Kd of 0.3–0.8 μm and a stoichiometry of 2. Oxa1L-CTT cross-links to mammalian mitochondrial homologs of the bacterial ribosomal proteins L13, L20, and L28 and to mammalian mitochondrial specific ribosomal proteins MRPL48, MRPL49, and MRPL51. Oxa1L-CTT does not cross-link to proteins decorating the conventional exit tunnel of the bacterial large ribosomal subunit (L22, L23, L24, and L29).
Keywords: Mammal, Membrane, Mitochondria, Protein Synthesis, Ribosomes, Translation, Oxa1
Introduction
Mammalian mitochondria synthesize 13 proteins, all of which are inserted into the respiratory chain complexes in the inner membrane. These hydrophobic polypeptides are integrated into the membrane during or immediately after their synthesis on mitochondrial ribosomes. Members of the Oxa1 evolutionarily conserved family of proteins including mammalian mitochondrial Oxa1L, yeast mitochondrial Oxa1p, chloroplast Alb3, and bacterial YidC are involved in the insertion of hydrophobic proteins into membranes (1, 2). In yeast several proteins play a role in this process including Oxa1p and Mba1 (3–7). Both of these proteins bind to yeast mitochondrial ribosomes and facilitate the insertion of mitochondrial translation products into the inner membrane. Mammalian systems have a clear homolog of Oxa1p (Oxa1L). Mammals do not have a convincing member of the Mba1 family, although it has been suggested that the role of yeast Mba1 may be played by ribosomal protein MRPL45 in mammals (8).
A number of studies have been carried out on yeast and Neurospora crassa Oxa1p. These proteins contain an N-terminal region located in the intermembrane space, 5 transmembrane helices, and a C-terminal tail of about 90 amino acids located in the mitochondrial matrix (4, 9) (Fig. 1A). Genetic and biochemical studies indicate that the C-terminal tail binds to mitochondrial ribosomes and plays an important role in the insertion of newly synthesized mitochondrial polypeptides into the inner membrane (4). Cross-linking studies indicate that yeast Oxa1p is located close to Mrp20 (a homolog of bacterial L23) and MrpL40 (the homolog of L24), both of which are located near the exit tunnel on the large subunit (6, 10). It can also be cross-linked to nascent chains (11).
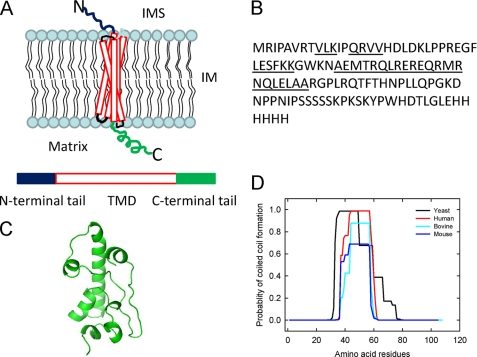
FIGURE 1.
A, hypothetical structural organization of Oxa1L in the inner membrane (IM) is shown. Oxa1L is composed of an N-terminal domain in the intermembrane space (IMS), a transmembrane domain (TMD) with five transmembrane helices, and a C-terminal domain located in the matrix. B, the primary sequence of the Oxa1L-CTT expressed and purified from E. coli is shown. Regions predicted to be α-helical by the secondary structure prediction programs on Biology Workbench using the PELE collection of programs are underlined. The methionine (M) at the beginning of the sequence and the LEHis6 at the C terminus of the sequence are from the vector. C, the Rosetta structure prediction protocol was used to generate a model of Oxa1L-CTT (26). The structure shown is the lowest free-energy structure and is displayed using PyMOL. D, prediction of coiled-coil formation is shown. The coiled-coil structure was predicted using the COILS program with two different windows. The figure shows the comparison of coiled-coil-forming tendency in a 14-residue window of the C-terminal tail of yeast, human, bovine, and mouse Oxa1. The amino acid listed as zero in the figure corresponds to residues 317, 334, 334, and 330 residues in full-length yeast, human, bovine, and mouse Oxa1, respectively.
Mitochondrial ribosomes from different organisms are structurally quite diverse (12, 13). Mammalian mitochondrial ribosomes are composed of only 25–30% RNA contributed by two rRNAs, 12 S in the small subunit and 16 S in the large subunit. The bulk of the mammalian mitochondrial ribosome consists of proteins (14). Mammalian mitochondrial ribosomes are 55 S particles and have ∼79 proteins, of which 42 have homologs in prokaryotic ribosomes, whereas 35 are specific for mitochondrial ribosomes. Twenty of these proteins have no clear homologs in yeast mitochondrial ribosomes (15), indicating a significant divergence between the protein composition of mitochondrial ribosomes from higher and lower eukaryotes. These differences may lead to significant alterations in the interaction of mitochondrial ribosomes from different sources with the machinery required for the insertion of mitochondrial translation products into the inner membrane.
Stiburek et al. (16) recently reported that human Oxa1L exists as a 600–700-kDa heterooligomeric complex in mitochondria from human embryonic kidney cells. Knockdown of human Oxa1L damages the biogenesis of the F1F0-ATP synthase and of Complex I (NADH:ubiquinone oxidoreductase) without altering the content of Complexes III or IV. These effects are distinct from those observed in yeast in which the assembly of Complex IV is strongly affected by mutations in Oxa1p (17). In the current report, we have examined the structure of the C-terminal tail of human Oxa1L (Oxa1L-CTT) and analyzed its interaction with the mitochondrial ribosome.
EXPERIMENTAL PROCEDURES
Materials
High purity chemicals were purchased from Sigma or Fisher. A rabbit polyclonal primary antibody to Oxa1L-CTT was made by Pacific Immunology Corp. Secondary antibody, HRP-labeled anti-rabbit antibody, was purchased from GE Healthcare. Protein free blocking agent and dimethyl suberimidate (DMS)2 were purchased from Pierce. Bovine mitochondria, mitochondrial 55 S ribosomes and ribosomal subunits (28 S and 39 S) were prepared as described previously (18). The cDNA clone of human Oxa1L was obtained from American Type Culture Collection (ATCC number 10961183, IMAGE 40017377). This cDNA corresponds to the major isoform of Oxa1L in humans (UniProtKB/Swiss-Prot Q15070; NCBI Q15070). This isoform is present in two variants in humans. One of these has a series of 4 Ser residues near the C terminus (residues 419–422), whereas the other has a series of 5 Ser residues in this location. The construct used in these studies has 436 amino acid residues in the coding region including the import signal and 5 Ser residues (amino acids 419–423) in the C-terminal tail.
Cloning, Expression, and Purification of Human Oxa1L-CTT
The cDNA (encompassing amino acids 334–436 of Oxa1L) was amplified out of the pCR-Blunt II TOPO vector using forward primer GGAATTGGCCATATGCGGATTCCAGCAGTACGC and reverse primer CCTTAACTCGAGGCCAAGTGTGTCGTGCCAGGG. The forward primer provides an N-terminal Met. The PCR product was purified and digested with NdeI and XhoI and cloned into the pET21 (c+) vector (Novagen), providing a His6 tag at the C terminus of the expressed protein. This construct was transformed into Escherichia coli DH5α, and the nucleotide sequence of the inserted DNA was confirmed by sequencing. The plasmid was subsequently transformed into E. coli BL21(DE3) pArgU218 (19) for expression.
The purification of Oxa1L-CTT was carried out basically as described previously for IF3mt (19–21) using Ni-NTA resins and followed by HPLC purification on a TSKgel SP-5PW column (supplemental Materials and Methods and Fig. S1). The isolated protein is greater than 98% pure.
Detection of Dimer and Tetramer Forms of Oxa1L-CTT by DMS Cross-linking
Oxa1L-CTT (14 μm) was incubated in the presence and absence of DMS (1.44 mm) in a 15-μl reaction mixture at the indicated concentrations of KCl in a buffer containing 20 mm HEPES-KOH, pH 7.6, and 2 mm MgCl2 for 1 h at 25 °C. The reactions were quenched by the addition of 1.5 μl of 1 m Tris-HCl, pH 7.6, giving a final Tris concentration of 91 mm. An equal volume of 2× SDS loading dye was added to the reaction mixtures. Samples were run on a 15% SDS-PAGE. Proteins were transferred to PVDF membranes (Millipore) and probed with Oxa1L-CTT polyclonal antibody using the ECL Western blotting method as described (22).
Determination of the Secondary Structure of Oxa1L-CTT Using Circular Dichroism (CD) Spectroscopy
CD spectra were obtained using an Applied Photophysics PiStar-180 spectropolarimeter (Surrey, UK) with a thermostated quartz cell of 1.0-mm path length located in the University North Carolina Macromolecular Interactions Facility. CD spectra were obtained by averaging 50,000 acquisitions with intervals of 0.5 nm from 260 to 190 nm. For CD measurements, protein (0.5 mg/ml) was exchanged into a buffer containing 10 mm sodium phosphate, pH 7.6. Control buffer spectra were subtracted from the spectra of the protein samples.
Determination of Oligomer Formation of the Oxa1L-CTT by Sedimentation Equilibrium Analysis Using Analytical Ultracentrifugation
Sedimentation equilibrium experiments were performed in a Beckman XL-1 analytical ultracentrifuge using 6 sector cells at 20 °C by recording the absorbance at 280 nm. Protein concentrations ranged from 0.3 to 1.0 mg/ml (24–78 μm), and the corresponding buffer blanks were run in parallel in sample cells at 24,000 rpm until equilibrium was reached (4 h). The offset values were determined by meniscus depletion method by spinning the samples at 48,000 rpm. The absorbance versus the radial distribution profiles were analyzed globally using least squares nonlinear curve fitting (program NONLIN). The absorption equilibrium constant units were converted to molar units using the molar extinction coefficient (12,700 mol−1 cm−1) based on the amino acid composition of Oxa1L-CTT.
Binding of Oxa1L-CTT to 39 S Subunits Using Surface Plasma Resonance
The interaction of Oxa1L-CTT with the large subunit of the mammalian mitochondrial ribosome (39 S) was determined using a Biacore 2000 biosensor instrument (located in the UNC Macromolecular Interactions Facility) with an L1 sensor chip (23). Before use, the L1 chip was cleaned with 20 mm CHAPS in running buffer (20 mm HEPES-KOH, pH 7.6, 5 mm MgCl2, 100 mm KCl, and 1 mm dithiothreitol) by injecting 10 μl at 10 μl/min. After surface cleaning, different amounts of ligands (39 S, 28 S, and bovine serum albumin (BSA)), diluted in running buffer, were injected into three different flow cells. Samples (70 μl of 0.01 μm 39 S, 100 μl of 0.009 μm 28 S, or 60 μl of 3.75 μm BSA) were injected at 5 μl/min until the surfaces were saturated with the ligands as indicated by no further increases in resonance units (RU). All of these materials (ligands) stick on the L1 surface without any loss of RU even after prolonged washing. For analyte binding to the ligands, different amounts of Oxa1L-CTT were injected into the cells. The RU values were recorded for the interaction of Oxa1L-CTT with 39 S subunits, 28 S subunits, and BSA as a function of Oxa1L-CTT concentration. The nonspecific binding surface BSA was used as the control surface. The RU change due to the binding Oxa1L to 39 S or 28 S subunits was obtained by subtracting the RU of the control surface (BSA).
Isothermal Titration Calorimetry
Isothermal titration calorimetry was performed using a ITC200 MicroCalorimeter (MicroCal, Inc., Northampton, MA) with 4 μm 55 S ribosomes and 80 μm Oxa1L-CTT. Ribosome and protein samples were extensively dialyzed in buffer (20 mm HEPES-KOH, pH 7.6, 50 mm KCl, 20 mm MgCl2, and 6 mm β-mercaptoethanol) immediately before the experiments were performed. All solutions were degassed before being loaded into the cell and the titration syringe. After an initial 90-s delay, a series of 2-μl samples of the Oxa1L-CTT solution was injected into the cell containing the ribosomes. The temperature was kept constant at 25 °C, and the mixing speed was 1000 rpm. The reference power was set to 5 μcal/s. Each peak in the power versus time plot was integrated and normalized per mol of injected protein, corrected for heats of ligand dilution by averaging the last three data points and subtracting the average, and plotted against the molar ratio of Oxa1L-CTT to ribosomes. The data were analyzed using MicroCal origin provided by the MicroCal system with a one site binding model.
Identification of Ribosomal Proteins Cross-linked to Oxa1L-CTT by DMS Cross-linking Followed by Mass Spectrometry
A sample of 39 S ribosomal subunits (20 pmol) was mixed with 400 pmol (4 μm final concentration) of Oxa1L-CTT in cross-linking buffer (20 mm HEPES-KOH, pH 7.6, 50 mm KCl, 10 mm MgCl2) and incubated in 100 μl for 1 h at room temperature in the presence of a 50-fold molar excess of DMS (0.2 mm). The reaction was stopped by adding 5 μl of 100 mm Tris-HCl, pH 7.6. The cross-linked products were separated from uncross-linked Oxa1L-CTT by layering the reaction mixture on a 30% sucrose cushion in 20 mm Tris-HCl, pH 7.6, 50 mm KCl, 5 mm MgCl2, and 1 mm Tris(2-carboxyethyl)phosphine hydrochloride buffer followed by centrifugation (Ti70.1 rotor, 42,000 rpm for 12 h). The pellet was dissolved in 100 μl of Buffer A (20 mm HEPES-KOH, pH 7.6, 500 mm NH4Cl, 1 mm Tris(2-carboxyethyl)phosphine hydrochloride and 8 m urea) and incubated for 30 min at room temperature. After incubation, 20 μl of Ni-NTA resin in Buffer A was added to the solution followed by rocking for 1 h at 4 °C. The ribosomal proteins cross-linked to Oxa1L-CTT bound to the Ni-NTA, and unbound proteins were removed by 3 washes of 400 μl each with Buffer A and 2 washes of 400 μl each with Buffer B (20 mm HEPES-KOH, pH 7.6, 500 mm NH4Cl, and 1 mm Tris(2-carboxyethyl)phosphine hydrochloride). The bound proteins were eluted with 2 aliquots of 50 μl each of Buffer C (20 mm HEPES-KOH, pH 7.6, and 250 mm imidazole). A control sample for the identification of ribosomal proteins bound nonspecifically to the Ni-NTA resin was prepared in an identical manner except that Oxa1L-CTT was omitted from the incubation mixture. The cross-linked proteins from both the control sample and the Oxa1L-CTT sample were digested with trypsin overnight at 37 °C (24). The digested samples were lyophilized using a speed vacuum concentrator. The digested peptides (control sample and Oxa1L-CTT cross-linked sample) were dissolved in 20 μl of 5% acetonitrile and 0.1% formic acid solution and subjected to tandem mass spectrometry analysis as described previously (24, 25). Tandem MS spectra obtained by fragmenting a peptide by collision-induced dissociation were acquired using a capillary liquid chromatography-nanoelectrospray ionization-tandem mass spectrometry (LC/MS/MS) system that consisted of a Surveyor HPLC pump, a Surveyor Micro AS autosampler, and an LTQ linear ion trap mass spectrometer (ThermoFinnigan). The spectra (peaks) were analyzed using a site-licensed Mascot data base searching program. Individual ion scores of greater than 35 indicate identity or extensive sequence homology when MS/MS data are queried against a data base consisting of mitochondrial ribosomal protein sequences.
RESULTS
Secondary Structure of Oxa1L-CTT
To explore the structure of human Oxa1L-CTT, a construct encompassing amino acids 333–436 (Fig. 1B) was cloned into a vector carrying a C-terminal His6 tag and expressed in E. coli. The protein was highly expressed (0.8 mg/g of cells or 6.5 mg/liter of culture) and purified to near homogeneity by Ni-NTA affinity and strong cation exchange chromatography (supplemental Fig. S1).
Secondary structure predictions for Oxa1L-CTT (Fig. 1B) indicated that there would be a long α-helical region encompassing residues 40–61 (underlined sequence in Fig. 1B). In addition, several short helical segments were predicted. Further insight into the possible structure of the CTT was obtained using the Rosetta structure prediction protocol (26) to develop a potential three-dimensional structure for Oxa1L-CTT (Fig. 1C). The predicted structure includes the major helical segment encompassing residues 40–61 of the tail as well as several shorter helical segments. Significant regions were not predicted to form traditional secondary structural elements.
Previous work (4) suggested that the CTT of yeast Oxa1p forms a coiled-coil structure. The tendency of the C-terminal tails to form coiled-coil structures was predicted for Oxa1 from several organisms including yeast, human, bovine, and mouse using the COILS program (27). The coiled-coil-forming tendency of the four proteins in a 14-residue window is plotted as a function of the position of the amino acid in the primary sequence (Fig. 1D). Yeast Oxa1p is predicted to have nearly a 100% probability of forming a coiled-coil. In mammals, the probability of coiled-coil helix-forming propensity varied from 0.7 to 1 even though human, bovine, and mouse Oxa1 possess more than 90% identity. Very similar results were obtained using a 21-residue window. However, with the MultiCoil (28), PairCoil (29), or PairCoil2 (30) programs and a 21-residue window, the probability of coiled-coil formation ranges from 0.4 for the yeast protein to 0.1 for human Oxa1L-CTT. Thus, these programs suggest that the propensity for coiled-coil formation of mammalian Oxa1L-CTT is quite low.
To obtain experimental evidence for or against the tendency of human Oxa1L-CTT to form a coiled-coil, a CD analysis was carried out. The molar ellipticity ratio [θ222/θ208] has been used as a criterion in many proteins to determine the presence of coiled-coil helices. The ratio [θ222/θ208] is about 0.83 for non-interacting helices, whereas it is 1.03 for two-stranded coiled-coil helices (31, 32). As indicated in Fig. 2A, the molar ellipticity ratio [θ222/θ208] for the Oxa1L-CTT examined at several different protein concentrations is 0.52, clearly indicating that this region of Oxa1L does not form a coiled-coil. Similar results were obtained at a variety of salt concentrations (Fig. 2B). This idea is compatible with the modeled structure of the Oxa1L-CTT (Fig. 1C), which predicts a long helical segment that does not interact in solution. Generally, coiled-coil-forming proteins have a tendency to induce helical content with increasing protein concentrations and increasing ionic strength (33). Thus, our results suggest that Oxa1L-CTT does not behave like a typical coiled-coil protein.
FIGURE 2.
Effect of protein concentration, salt, and TFE on the secondary structure of Oxa1L-CTT determined by CD. A, shown is the effect of Oxa1L-CTT concentration (0.1, 0.2, and 0.34 mg/ml) on the CD spectra. B, shown is the effect of salt concentration on the CD spectra of Oxa1L-CTT. The protein concentration was 0.1 mg/ml. C, shown is the effect of TFE concentration on the CD spectra of Oxa1L-CTT. Protein concentration was 0.2 mg/ml.
The % α-helix and β-sheet content were calculated from the CD data using the CDPro program (34). The CD data were fit using three algorithms (CDSSTR, CONTIL, and SELCON) in CDPro (34). The outputs obtained from all three algorithms gave very similar values (supplemental Table 1) and indicate that the C-terminal tail is 16–20% α-helical in aqueous solutions at all of the protein and salt concentrations tested. This value is in reasonable agreement with α-helices predicted by computational methods, which suggested a helical content of 25–30%.
Further investigation into the potential for Oxa1L-CTT to form a coiled-coil structure was obtained using trifluoroethanol (TFE). This co-solvent is known to disrupt coiled-coil interactions in proteins leading to non-interacting single helices. It can also induce more stable helices in single α-helical segments (35, 36). The helical content of Oxa1L-CTT increased with increasing concentration of TFE (Fig. 2C and supplemental Table 1). The ratio of θ222/θ208 increased from 0.5 to 0.75 with increasing concentrations of TFE. If a coiled-coil were present, then the tertiary structure should be disrupted at high TFE concentration. These results support the conclusion that Oxa1L-CTT does not contain a coiled-coil structural motif.
As indicated in supplemental Table S1, TFE induces additional secondary helical structure in Oxa1L-CTT perhaps by stabilizing the helices predicted to form in this protein. This result is supported by tryptophan anisotropy changes upon the addition of TFE to Oxa1L-CTT as described in supplemental Materials and Methods and Fig. S2. Finally, Oxa1L-CTT displays non-cooperative thermal and denaturant unfolding profiles, which also suggest the presence of a straight helix rather than a coiled-coil helical structure (supplemental Results and Fig. S3).
Oligomerization State of Oxa1L-CTT
Studies in N. crassa have indicated that Oxa1p forms oligomers and may exist as a tetramer (37). Cryo-EM studies have suggested that yeast Oxa1p binds to E. coli ribosomes in a dimeric form (38). The oligomerization state of mammalian Oxa1L is unknown. In the mitochondrial inner membrane it exists as a complex with a molecular mass of ∼700 kDa (16) most likely consisting of an Oxa1L oligomer complexed with other proteins. It is not known whether this protein binds to the ribosome as a monomer or oligomer. To assess the potential ability of Oxa1L-CTT to promote the oligomerization of Oxa1L, we have analyzed its oligomeric state and the strength of the oligomerization by sedimentation equilibrium using analytical ultracentrifugation.
The oligomeric state of Oxa1L-CTT was initially tested at low salt concentrations (50 mm KCl and 10 mm MgCl2) with data collected at two rotor speeds and at three different protein concentrations (Fig. 3A). Good agreement was obtained when the data were fit globally to the equation for a single species. The molecular mass varied from 12.5 and 13.0 kDa at 18,000 and 24,000 rpm, respectively. Thus, at low salt concentrations, the C-terminal tail of Oxa1L is predominantly monomeric.
FIGURE 3.
Oligomerization of Oxa1L-CTT. A, shown is the effect of KCl on oligomerization of Oxa1L-CTT as examined by analytical ultracentrifugation. The panels show the representative equilibrium sedimentation profiles at three different KCl concentrations at 10 mm MgCl2. The data are plotted as the absorption of Oxa1L-CTT (0.6 mg/ml) at 280 nm versus the distance from the center of the axis of rotation (radius). The lower section of each panel shows the raw data in closed circles. The lines represent the best fits. The upper panels show the residual for the corresponding given fit. The global fit of three protein concentrations (0.3, 0.6, and 0.9 mg/ml) is not shown. The data presented here were obtained at 24,000 rpm. B, detection of dimer and tetramer formation of Oxa1L-CTT by DMS cross-linking followed by Western blotting as described in “Experimental Procedures” is shown.
As the KCl concentration was raised to 200 and 300 mm, there was a clear increase in the molecular mass (Fig. 3A and supplemental Table S2). The data from these runs were fit to a single species with reasonable variance values (testing goodness of fit) but there was a clear increase in the molecular mass of Oxa1L-CTT with increasing KCl concentrations (supplemental Table S2). At higher salt concentrations, significant improvements in the fits were obtained using a monomer-dimer-tetramer model (considering the theoretical mass of Oxa1L-CTT as 12,700 based on the amino acid composition). The strength of oligomerization of Oxa1L-CTT is in the μm range and clearly depends on the salt concentration (supplemental Table S2). Because Oxa1L-CTT is highly basic (pI = 10.7), it is reasonable that its oligomerization would be stabilized by monovalent ions.
To obtain further insights into the oligomerization of Oxa1L-CTT, chemical cross-linking has been carried out using DMS, a homobifunctional cross-linker that reacts with amine groups. For optimization, cross-linking was done with different concentrations of DMS at a fixed concentration of Oxa1L-CTT (data not shown). The salt-dependent formation of Oxa1L-CTT oligomers (dimer and tetramer bands) was visualized by Western blotting (Fig. 3B). This analysis clearly shows the formation of significant amounts of the dimeric species and a lower amount of the tetramer. The dimeric and tetrameric products increased with increasing KCl concentrations as observed for oligomer formation in analytical ultracentrifugation (Fig. 3A).
We further characterized the solution behavior of the Oxa1L-CTT and the effects of salt on its hydrodynamic radius using size-exclusion chromatography followed by multiangle light scattering measurements (supplemental Materials and Methods and Fig. S4). The light scattering results are consistent with the analytical ultracentrifugation and DMS cross-linking results and indicate that Oxa1L-CTT can form oligomers.
Oxa1L-CTT Binding to the Large Ribosomal Subunit (39 S) of the Mitochondrial Ribosome
The interaction between Oxa1L-CTT and mitochondrial ribosomal subunits was examined using surface plasmon resonance. Normally one would like to couple the small protein ligand (Oxa1L-CTT) to the surface and flow the larger binding partner (the ribosomal subunits) through. However, Oxa1L-CTT was deactivated after amine coupling, and the interaction of 39 S subunits with Oxa1L-CTT could not be measured using the traditional CM5 chip. As an alternative, the L1 chip was used. These chips are generally used to capture liposomes or lipid bilayers. However, we observed that it was possible to capture mitochondrial 28 S or 39 S subunits on L1 chips. In addition, a control chip was prepared using BSA. Ribosomes and BSA bound tightly to the L1 chip, and there was very little change in RU over a prolonged period of washing in the running buffer.
Different concentrations of Oxa1L- CTT were then injected onto the surface, and the RU change was recorded. The sensogram profiles (data not shown) indicated that Oxa1L-CTT interacts with the large ribosomal subunit (39 S), but no interaction was detected with the 28 S subunit (Fig. 4A). The RU response is sigmoidal as a function of protein concentration, and the data could not be fit to a single site binding model. The data were fit to a sigmoidal curve using Sigmaplot 10, which indicates that Oxa1L-CTT binds to 39 S subunits cooperatively with an approximate affinity of 0.23 μm. This result is compatible with the observation that Oxa1L-CTT has a tendency to dimerize and leads to the suggestion that two molecules of Oxa1L-CTT are bound to each 39 S subunit. Data obtained by isothermal titration calorimetry support this conclusion as discussed below. This idea is in accord with the cryo-EM studies of yeast Oxa1p bound to E. coli 70 S ribosomes in which a dimer appears to be bound to the ribosome (38).
FIGURE 4.
Interaction of the mammalian mitochondrial large ribosomal subunit (39 S) with Oxa1L-CTT analyzed by surface plasma resonance. A, shown is the RU change of Oxa1L-CTT binding to 39 S (circles) and 28 S (triangles) as a function of Oxa1L-CTT concentrations. Ribosomes (39 S and 28 S) and BSA were immobilized on a L1 sensor chip, and Oxa1L-CTT was flowed through the cell as described under “Experimental Procedures.” The RU values were recorded for each injection after 15 s of buffer exchange. The value of the RU from the cell carrying BSA has been subtracted from each value. The solid circles represent experimental data, and the line represents a sigmoidal fit. B, the salt dependence of the RU change when Oxa1L-CTT (20 μl at 0.18 μm) was used in buffer containing different KCl concentrations and injected at a flow rate of 10 μl/min, and RU values were noted from the base line after 15 s of buffer exchange.
The binding of Oxa1L-CTT to the large ribosomal subunit is highly salt-dependent (Fig. 4B). This result indicates that the interaction between 39 S and Oxa1L-CTT is electrostatic as might be expected. Yeast Oxa1p binds to yeast mitochondrial ribosomes at low salt concentrations, but the binding is undetectable above 200 mm KCl (4).
The approximate strength of the Oxa1L-CTT binding to 39 S subunits determined from surface plasmon resonance was verified using a dot blot method (39, 40). In this method, Oxa1L-CTT·39 S complexes are centrifuged through Microcon membranes with a molecular mass cut-off of 100 kDa. The Oxa1L-CTT·ribosome complexes do not pass through the filter, and the amount of Oxa1L-CTT retained with ribosomes is determined using anti-Oxa1L-CTT antibodies on a dot blot (data not shown). Using this method, the Kd for the interaction of Oxa1L-CTT with ribosomes falls within the range of 0.3–0.5 μm.
Isothermal titration calorimetry was used to monitor the binding of Oxa1L-CTT to mitochondrial 55 S ribosomes and to obtain an estimate of the thermodynamics of the binding interaction. The disadvantage of this method is that significant amounts of material are required due to the sample concentrations and volumes needed. Mitochondrial ribosomes are difficult to obtain in these amounts. To reduce this problem an ITC200 was used. This instrument has a cell volume 200 μl and a syringe volume of 40 μl, thus, requiring smaller amounts of material. In this experiment, 55 S ribosomes were used instead of 39 S subunits as they are obtained in higher yields during isolation procedures. A fixed amount of Oxa1L-CTT in the syringe was sequentially injected into the cell containing purified ribosomes, and the raw heat change data were recorded as a function of time (Fig. 5A). The normalized heat change (per mol of injected protein) as a function of molar ratio of Oxa1L-CTT to ribosomes is shown in Fig. 5B. Analysis of these data (using a binding model with one binding site on the 55 S ribosome) indicates that the interaction between Oxa1L-CTT and ribosomes is exothermic characterized by a large and negative enthalpy change (-28.6 ± 1.8 kcal/mol) and negative entropy (−68.4 cal/mol-deg) of binding. These values indicate that the formation of the Oxa1L-CTT·ribosome complex is an enthalpically favorable and entropically unfavorable process. The stoichiometry of binding was estimated to be 2.2 ± 0.08 Oxa1L-CTT to 55 S ribosomes. The Ka value for the binding of Oxa1L-CTT to 55 S is 1.3 ± 0.35 × 106 m−1, corresponding to an apparent Kd ∼0.78 ± 0.22 μm. The apparent Kd for the binding of Oxa1L-CTT to 55 S is the same range as that determined for 39 S subunits as described above.
FIGURE 5.
Estimation of the thermodynamic parameters governing the interaction of Oxa1L-CTT with mitochondrial 55 S ribosomes using isothermal titration calorimetry. A, raw data for the binding of Oxa1L-CTT to 55 S ribosomes provided as the power output (μcal/s) as a function of time are shown. The protein concentration was 80 μm (syringe), and the 55 S ribosome concentration was 4 μm (cell). The first injection is only for the purpose of the experimental setup and is ignored for data analysis. B, shown is the amount of heat evolved at each injection normalized to the number of moles of Oxa1L-CTT injected (kcal/mol) versus the molar ratio of Oxa1L-CTT to 55 S ribosome. The solid line represents the nonlinear least squares fit for the data.
Identification of 39 S Ribosomal Proteins Interacting with Oxa1L-CTT
The exit tunnel of the bacterial ribosome is formed by large segments of rRNA and is decorated with proteins L22, L23, L24, and L29. In contrast, the exit tunnel of the mammalian mitochondrial ribosomes is enriched by mitochondrial-specific ribosomal proteins in addition to three ribosomal proteins with bacterial homologs, MRPL22, MRPL23, and MRPL24 (41, 42). A clear homolog of L29 is missing in mammalian mitochondrial ribosomes, although it was been suggested that MRPL47 may be an ortholog of L29 (8). To identify proteins at or near the Oxa1L-CTT binding site on the 39 S subunit, a multistep process was used (Fig. 6). In this procedure, Oxa1L-CTT was bound to 39 S subunits and cross-linked to nearby proteins using DMS. Excess Oxa1L-CTT was removed using centrifugation, and the cross-linked complexes were denatured by the addition of urea. The His-tagged Oxa1L-CTT and its cross-linked protein partners were selectively retained on a Ni-NTA column. The proteins present in the Ni-NTA eluates were cleaved by trypsin and identified by LC/MS/MS. In the cross-linked samples, peptides from six different proteins were identified as being at or close to the Oxa1L-CTT binding site (Table 1). Three of these proteins have bacterial homologs (MRPL13, MRPL20, and MRPL28), whereas the other three are mitochondria-specific ribosomal proteins (MRPL48, MRPL49, and MRPL51). These results are surprising as conventional wisdom indicates that Oxa1L-CTT would bind to ribosomes near the exit tunnel and would cross-link to ribosomal proteins corresponding to L22, L23, and L24, which are located near to the exit tunnel of bacterial ribosomes. The cross-linking experiment was repeated four independent times with different preparations of material to ensure the accurate identification of the proteins present. No proteins were identified in the control sample, which did not contain Oxa1L-CTT indicating that the proteins identified do not simply bind to the Ni-NTA resin non-specifically.
FIGURE 6.
Strategy used to identify ribosomal proteins near the Oxa1L binding site. Oxa1L-CTT was incubated with 39 S subunits and cross-linked to nearby proteins using DMS as described under “Experimental Procedures.” Cross-linked complexes were purified by centrifugation through a sucrose cushion. The ribosomes were then denatured, and ribosomal proteins cross-linked to Oxa1L-CTT were recovered on Ni-NTA. Cross-linked proteins were digested with trypsin, and the proteins present were identified by LC/MS/MS.
TABLE 1.
Peptides and ion scores of ribosomal proteins cross-linked to Oxa1L-CTT
Ion scores of greater than 35 are considered significant.
| Protein | Sequence | Scorea | m/z | Mr (experimental) |
|---|---|---|---|---|
| MRPL13 | APQQWATFAR | 73 | 588.5 | 1174.9 |
| NLTEELPQPR | 51 | 598.9 | 1195.7 | |
| VWSPPEDYRL | 61 | 631.7 | 1260.8 | |
| VYSSHTGYPGGFK | 82 | 700.7 | 1399.5 | |
| MRPL20 | VLADLAIYEPK | 54 | 617.1 | 1232.3 |
| MRPL28 | ELYSEILDTR | 43 | 620.2 | 1238.4 |
| LLEEKDPVPLFK | 69 | 714.8 | 1427.6 | |
| MRPL48 | ARPELEELLAK | 59 | 635.3 | 1268.7 |
| MLLDSVLTTHER | 88 | 708.18 | 1414.3 | |
| TMEVLQLQDQGNK | 119 | 752.5 | 1503.1 | |
| MRPL49 | FLESVDEYR | 48 | 1157.7 | 1156.7 |
| VEGDIWALQK | 63 | 580.5 | 1158.9 | |
| DVEDFLSPLLGK | 78 | 668.0 | 1334.0 | |
| TPITQVNEVTGTLR | 105 | 766.5 | 1531.0 | |
| MRPL51 | MFIDDLHNLNKR | 51 | 758.0 | 1514.0 |
a This is a representative score. Actual values varied in the analysis of different preparations.
DISCUSSION
The Oxa1 family of proteins including yeast Oxa1p, chloroplast Abl3, and bacterial YidC is widely distributed in nature. Much remains to be learned about the mechanism of action of Oxa1 in the insertion of proteins into membranes. Most of the studies on the role of this protein in mitochondria have been carried out in yeast. In this organism the C-terminal tail has been shown to play an important role in binding to the large subunit of the ribosome and in promoting the insertion of mitochondrial translation products into the inner membrane. Experiments using CD suggested that the C-terminal tail of yeast Oxa1p forms a coiled-coil structure (4). The data presented here strongly indicate that human Oxa1L-CTT does not form this type of structure.
The oligomeric behavior of Oxa1 has been controversial. N. crassa Oxa1 is thought to exist as a homotetramer (43). In contrast, human Oxa1L is present in a hetero-oligomeric complex with a mass of 600–700 kDa of unknown composition (16). Recent studies indicate that yeast Oxa1p and E. coli YidC exist as monomers, dimers, and higher oligomers at low detergent concentrations, whereas increasing detergent concentrations promote the formation of mostly monomers (38). The role of the C-terminal tail of Oxa1 in the oligomerization state is unclear. The C-terminal tail of human Oxa1L is largely monomeric in solution but does tend to oligomerize at increasing salt concentrations. However, like yeast Oxa1p, the data presented here indicate that two molecules of human Oxa1L-CTT bind to a single ribosome.
It is clear from studies of yeast Oxa1p that the C-terminal tail is necessary for ribosome binding and that this binding is independent of the presence of a nascent chain on the ribosome (4). Yeast Oxa1p is believed to bind near the exit tunnel of the mitochondrial ribosome where it can help direct the nascent chain insertion into the inner membrane (11).
Our data indicate that human Oxa1L-CTT binds to mammalian 55 S ribosomes quite differently from that observed in the interaction of yeast Oxa1p with either yeast mitochondrial 74 S ribosomes or bacterial 70 S ribosomes. One striking difference between prokaryotic and mammalian mitochondrial ribosomes is found in the region of the large subunit corresponding to the exit tunnel. In bacterial ribosomes, this region contains large amounts of rRNA from domains I and III of the 23 S rRNA. Yeast Oxa1p contacts helices H24 and H59 when it is bound to E. coli ribosomes (38). Yeast mitochondrial ribosomes have a segment equivalent to H24 and a region encompassing H59, although secondary structure predictions of this region of the large subunit rRNA do not correspond to the structure observed in E. coli 23 S rRNA. In contrast, the large subunit rRNA of mammalian mitochondria (16 S rRNA) has almost completely lost the regions of the rRNA corresponding to domains I and III. Both helices H24 and H59 are missing in mammalian mitochondrial ribosomes, suggesting that mammalian Oxa1L will interact with ribosomes in quite a different way.
In addition to large amounts of rRNA, several proteins are located at or near the exit tunnel of bacterial ribosomes including L22, L23, L24, and L29 (Fig. 7A). Homologs of all of these proteins are present in yeast mitochondrial ribosomes. In addition, the region near the exit tunnel of yeast mitochondrial ribosomes contains several mitochondrial specific ribosomal proteins including Mrpl3, Mrpl13, and Mrpl27 (44). Yeast Oxa1p can be cross-linked to mitochondrial ribosomal proteins that are homologs of bacterial L23 (Mrp20) and L24 (MrpL40) at the exit tunnel (6, 10).
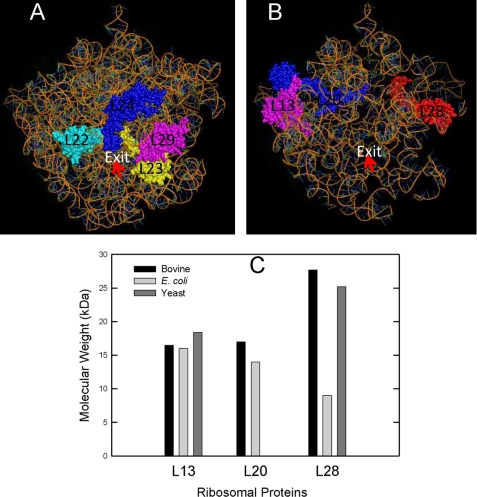
FIGURE 7.
A, structural representation the putative binding site of Oxa1L mapped onto the structure of the Thermus thermophilus 50 S subunit (PDB coordinate 2WRL) using PyMOL. A, shown is a representation of the exit tunnel on bacterial ribosomes showing the traditional proteins (L22, L23, L24, and L29) near to exit tunnel of the 50 S ribosomal subunit. B, shown is a representation of the mammalian mitochondrial ribosomal proteins homologous to bacterial L13, L20, and L28 (space-filled) modeled onto the bacterial 50 S subunit. The regions of the rRNA missing in the mammalian mitochondrial ribosome have been manually removed from the coordinates for the 50 S subunit. In E. coli, L28 is almost completely covered by rRNA, but these segments of rRNA are missing in the 39 S subunit, leaving L28 more exposed to solvent. C, shown is a comparison of the sizes of bacterial and yeast L13, L20, and L28 with mitochondrial homologs. L20 is absent in yeast. The molecular weights of the MRPs are estimated after the removal of import signals predicated by MitoProt II.
The exit tunnel of mammalian mitochondrial ribosomes has two solvent-accessible openings (42). One site corresponds to the traditional exit site located about 88 Å from the peptidyltransferase site. The other, termed the polypeptide accessible site, is located closer (about 68 Å from the peptidyltransferase). It is possible that some nascent chains may emerge from the large subunit through the polypeptide accessible site. Both of these regions are dominated by ribosomal proteins rather than rRNA (42). Ribosomal proteins corresponding to bacterial L22, L23, and L24 are present in mammalian mitochondrial ribosomes, but significant fractions of these proteins are obscured by mitochondrial-specific ribosomal proteins. There is no clear homolog of L29, although it has been suggested that MRPL47 may be a distant ortholog of L29 (8, 41). Two of the mitochondrial specific ribosomal proteins found near the exit tunnel in yeast are observed in mammalian mitochondrial ribosomes; yeast Mrpl27 is homologous to mammalian mitochondrial MRPL41, and yeast Mrpl3 corresponds to MRPL44. No homolog of yeast Mrpl13, found near the exit tunnel, has been observed in mammalian mitochondrial ribosomes. Despite some similarity in the protein composition between yeast and mammalian mitochondrial ribosomes, very different ribosomal partner proteins were observed for human Oxa1L-CTT than seen with yeast Oxa1p. Neither MRPL23 nor MRPL24 was observed to cross-link to human Oxa1L-CTT. In some respects this observation is surprising as they cross-link to yeast Oxa1p. However, large portions of MRPL23 and MRPL24 are covered by mitochondrial-specific ribosomal proteins in mammals (42) potentially making them inaccessible for cross-linking to Oxa1L.
Oxa1L-CTT is cross-linked to homologs of bacterial ribosomal protein L13, L20, and L28 (Fig. 7B). These homologs are all located on the solvent side of the large subunit spanning both sides of the exit tunnel and lie somewhat above the tunnel (Fig. 7B). MRPL28 is considerably larger than its bacterial homolog (Fig. 7C) and may cover a significant fraction of the back of the subunit. Furthermore, the lack of significant portions of Domains I and III of the large subunit rRNA in mitochondrial 39 S subunits exposes a larger portion of this protein on the solvent side of the ribosome (Fig. 7B). It is noteworthy that MRPL20, which cross-links to human Oxa1L-CTT, is missing in yeast mitochondrial ribosomes.
In addition to the bacterial homologs, three mitochondrial-specific proteins (MRPL48, MRPL49, and MRPL51) were identified close to Oxa1L-CTT. Neither MRPL48 nor MRPL51 has a homolog in yeast. The locations of MRPL48, MRPL49, and MRPL51 are not known, but the yeast homolog of MRPL49 (Img2) has been reported to interact with the homologs of L23 and L19 placing it on the solvent side of the large subunit and relatively close to the exit tunnel in mitochondrial ribosomes (45). Of six proteins cross-linked to human Oxa1L-CTT, three are missing in yeast (MRPL20, MRPL48, and MRPL51). Therefore, the decoration of the exit tunnel and the interaction of Oxa1 is clearly different between yeast and mammalian mitochondrial ribosomes.
Although we cannot rule out the possibility that other ribosomal proteins will cross-link to the full-length Oxa1L when it is organized in the membrane, the data presented here indicate that Oxa1L-CTT binds to the solvent side of the large subunit where it is located close to the exit tunnel and to a number of proteins that have no bacterial homologs. These proteins may play a role in the interaction of the mitochondrial ribosome with the nascent chain and may facilitate the integration of mitochondrial translation products into the respiratory chain complexes into the inner membrane.
Supplementary Material
Acknowledgments
We thank Dr. Ramesh Jha for creating the three-dimensional structure of Oxa1L-CTT based on de novo structure prediction protocols. We thank Dr. Carol Parker for technical assistance with the mass spectrometry results.
This work was supported, in whole or in part, by National Institutes of Health Grants GM 32734 (to L. L. S.) and GM071034 (to E. C. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Materials and Methods and Tables S1–S3 and Figs. S1–S4.
- DMS
- dimethyl suberimidate
- RU
- resonance units
- TFE
- trifluoroethanol
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Luirink J., Samuelsson T., de Gier J. W. (2001) FEBS Lett. 501, 1–5 [DOI] [PubMed] [Google Scholar]
- 2.Ott M., Herrmann J. M. (2010) Biochim. Biophys. Acta 1803, 767–775 [DOI] [PubMed] [Google Scholar]
- 3.Bonnefoy N., Fiumera H. L., Dujardin G., Fox T. D. (2009) Biochim. Biophys. Acta 1793, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J. M. (2003) EMBO J. 22, 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ott M., Prestele M., Bauerschmitt H., Funes S., Bonnefoy N., Herrmann J. M. (2006) EMBO J. 25, 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia L., Dienhart M., Schramp M., McCauley M., Hell K., Stuart R. A. (2003) EMBO J. 22, 6438–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hell K., Neupert W., Stuart R. A. (2001) EMBO J. 20, 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smits P., Smeitink J. A., van den Heuvel L. P., Huynen M. A., Ettema T. J. (2007) Nucleic Acids Res. 35, 4686–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sääf A., Monné M., de Gier J. W., von Heijne G. (1998) J. Biol. Chem. 273, 30415–30418 [DOI] [PubMed] [Google Scholar]
- 10.Jia L., Kaur J., Stuart R. A. (2009) Eukaryot. Cell 8, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hell K., Herrmann J. M., Pratje E., Neupert W., Stuart R. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2250–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pel H. J., Grivell L. A. (1994) Mol. Biol. Rep. 19, 183–194 [DOI] [PubMed] [Google Scholar]
- 13.Kitakawa M., Isono K. (1991) Biochimie 73, 813–825 [DOI] [PubMed] [Google Scholar]
- 14.Pietromonaco S. F., Denslow N. D., O'Brien T. W. (1991) Biochimie 73, 827–835 [DOI] [PubMed] [Google Scholar]
- 15.Graack H. R., Wittmann-Liebold B. (1998) Biochem. J. 329, 433–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stiburek L., Fornuskova D., Wenchich L., Pejznochova M., Hansikova H., Zeman J. (2007) J. Mol. Biol. 374, 506–516 [DOI] [PubMed] [Google Scholar]
- 17.He S., Fox T. D. (1997) Mol. Biol. Cell 8, 1449–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews D. E., Hessler R. A., Denslow N. D., Edwards J. S., O'Brien T. W. (1982) J. Biol. Chem. 257, 8788–8794 [PubMed] [Google Scholar]
- 19.Koc E. C., Spremulli L. L. (2002) J. Biol. Chem. 277, 35541–35549 [DOI] [PubMed] [Google Scholar]
- 20.Bhargava K., Spremulli L. L. (2005) Nucleic Acids Res. 33, 7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasso D. G., Christian B. E., Spencer A., Spremulli L. L. (2007) Methods Enzymol. 430, 59–78 [DOI] [PubMed] [Google Scholar]
- 22.Emdadul, Haque M., Grasso D., Miller C., Spremulli L. L., Saada A. (2008) Mitochondrion 8, 254–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima H., Kiyokawa N., Katagiri Y. U., Taguchi T., Suzuki T., Sekino T., Mimori K., Ebata T., Saito M., Nakao H., Takeda T., Fujimoto J. (2001) J. Biol. Chem. 276, 42915–42922 [DOI] [PubMed] [Google Scholar]
- 24.Miller J. L., Koc H., Koc E. C. (2008) Protein Sci. 17, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J. L., Cimen H., Koc H., Koc E. C. (2009) J. Proteome Res. 8, 4789–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradley P., Misura K. M., Baker D. (2005) Science 309, 1868–1871 [DOI] [PubMed] [Google Scholar]
- 27.Lupas A., van Dyke M., Stock J. (1991) Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 28.Wolf E., Kim P. S., Berger B. (1997) Protein Sci. 6, 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger B., Wilson D. B., Wolf E., Tonchev T., Milla M., Kim P. S. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonnell A. V., Jiang T., Keating A. E., Berger B. (2006) Bioinformatics 22, 356–358 [DOI] [PubMed] [Google Scholar]
- 31.Choy N., Raussens V., Narayanaswami V. (2003) J. Mol. Biol. 334, 527–539 [DOI] [PubMed] [Google Scholar]
- 32.Zhou N. E., Kay C. M., Hodges R. S. (1992) J. Biol. Chem. 267, 2664–2670 [PubMed] [Google Scholar]
- 33.Cánaves J. M., Montal M. (1998) J. Biol. Chem. 273, 34214–34221 [DOI] [PubMed] [Google Scholar]
- 34.Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 35.Lau S. Y., Taneja A. K., Hodges R. S. (1984) J. Biol. Chem. 259, 13253–13261 [PubMed] [Google Scholar]
- 36.Frère V., Sourgen F., Monnot M., Troalen F., Fermandjian S. (1995) J. Biol. Chem. 270, 17502–17507 [DOI] [PubMed] [Google Scholar]
- 37.Raught B., Gingras A. C., Sonenberg N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7037–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohler R., Boehringer D., Greber B., Bingel-Erlenmeyer R., Collinson I., Schaffitzel C., Ban N. (2009) Mol. Cell 34, 344–353 [DOI] [PubMed] [Google Scholar]
- 39.Haque M. E., Grasso D., Spremulli L. L. (2008) Nucleic Acids Res. 36, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haque M. E., Spremulli L. L. (2008) J. Mol. Biol. 384, 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koc E. C., Burkhart W., Blackburn K., Moyer M. B., Schlatzer D. M., Moseley A., Spremulli L. L. (2001) J. Biol. Chem. 276, 43958–43969 [DOI] [PubMed] [Google Scholar]
- 42.Sharma M. R., Koc E. C., Datta P. P., Booth T. M., Spremulli L. L., Agrawal R. K. (2003) Cell 115, 97–108 [DOI] [PubMed] [Google Scholar]
- 43.Nargang F. E., Preuss M., Neupert W., Herrmann J. M. (2002) J. Biol. Chem. 277, 12846–12853 [DOI] [PubMed] [Google Scholar]
- 44.Gruschke S., Gröne K., Heublein M., Hölz S., Israel L., Imhof A., Herrmann J. M., Ott M. (2010) J. Biol. Chem. 285, 19022–19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. (2007) Mol. Cell. Proteomics 6, 439–450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.