Abstract
Functional inactivation of the tumor susceptibility gene tsg101 in NIH 3T3 fibroblasts results in cellular transformation and the ability to form metastatic tumors in nude mice. The N-terminal region of tsg101 protein is structurally similar to the catalytic domain of ubiquitin-conjugating enzymes, suggesting a potential role of tsg101 in ubiquitin-mediated protein degradation. The C-terminal domain of TSG101 can function as a repressor of transcription. To investigate the physiological function of tsg101, we generated a null mutation of the mouse gene by gene targeting. Homozygous tsg101−/− embryos fail to develop past day 6.5 of embryogenesis (E6.5), are reduced in size, and do not form mesoderm. Mutant embryos show a decrease in cellular proliferation in vivo and in vitro but no increase in apoptosis. Although levels of p53 transcripts were not affected in tsg101−/− embryos, p53 protein accumulated dramatically, implying altered posttranscriptional control of p53. In addition, transcription of the p53 effector, cyclin-dependent kinase inhibitor p21WAF-1/CIP-1, was increased 5- to 10-fold, whereas activation of MDM2 transcription secondary to p53 elevation was not observed. Introduction of a p53 null mutation into tsg101−/− embryos rescued the gastrulation defect and prolonged survival until E8.5. These results demonstrate that tsg101 is essential for the proliferative burst before the onset of gastrulation and establish a functional connection between tsg101 and the p53 pathway in vivo.
The tumor susceptibility gene 101 (tsg101) was discovered in mouse fibroblasts by using regulated antisense RNA initiated within a randomly located, chromosomally integrated, retrovirus-based gene search vector (1). Functional inactivation of tsg101 by antisense transcripts complementary to tsg101 mRNA leads to transformation of NIH 3T3 cells characterized by colony formation in soft agar and their ability to form metastatic tumors when injected into nude mice (1).
Sequence analysis of tsg101 cDNA indicates that the gene encodes a 43-kDa protein. Mouse and human TSG101 proteins are 94% identical (2) and contain putative DNA-binding motifs characteristic of transcription factors (1), suggesting that tsg101 may control gene expression. This presumption was supported by the findings that separate domains of tsg101 can act as transcriptional cofactors that are able to activate or repress nuclear hormone receptor-mediated transactivation (3–5). In addition, the N-terminal region of tsg101 resembles a group of apparently inactive homologues of ubiquitin-conjugating enzymes, suggesting a possible role for tsg101 in the regulation of ubiquitin-mediated protein degradation (6, 7). However, the mechanisms by which interference with tsg101 expression leads to neoplastic transformation remain unknown.
To investigate the physiological role of tsg101 in vivo, we generated tsg101-deficient mice by gene targeting in embryonic stem cells.
Materials and Methods
Gene Targeting.
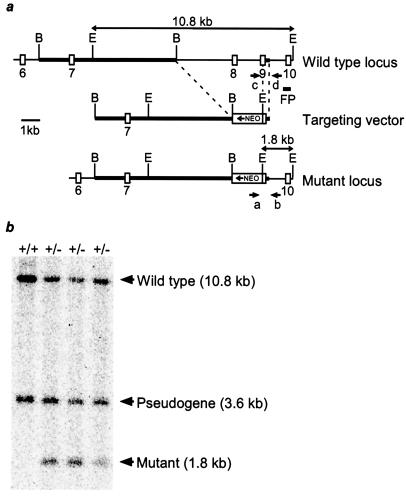
A genomic DNA clone containing exons 6–10 of tsg101 was isolated from a 129/J mouse genomic library. A targeting vector was designed to replace a 4.4-kb genomic fragment containing exon 8 and the 5′ part of exon 9 with the PGKneo resistance expression cassette in reverse orientation to tsg101 transcription. The vector was introduced into E14K embryonic stem (ES) cells by electroporation. Cells were subsequently cultured in the presence of 300 μg/ml of G418 (Sigma) for 10 days. Homologous recombinants were identified by PCR and verified by Southern blotting with a PCR-generated 3′ flanking probe containing exon 10 (primer, 5′-AAG TCC AAG AAA GAG AAA AAT and 5′-GGA TTG CTA GAT GCT GTC TGG). ES cell culture and generation of chimeras were performed as described (8). Two independently targeted ES cell clones transmitted the tsg101 mutation into the germ line.
PCR Analysis of tsg101 and p53 Genotypes.
Genomic DNA from ES cells, embryos, and neonate tails was isolated and used for PCR or Southern blot analysis as described (8).
tsg101.
Primers “c” (5′-CCG TCT GAG GTT GAG TTG TAG) specific for targeted intronic sequence and “d” (5′-GAG AAG GGC TGA GGA GAA ACG) were used to detect the wild-type allele, whereas primers “a” (5′-CGG AAG GCA GTG GTA GAA CCT), specific for sequences in the neo resistance gene, and “b” (5′-TAA AGC GCA T GC TCC AGA CTG), derived from the tsg101 gene downstream of the targeting construct (Fig. 1), were used to detect the recombinant allele.
Figure 1.
Targeted disruption of the murine tsg101 locus. (a) Partial genomic organization of the mouse tsg101 locus (Top) and structure of the targeting vector (Middle). Coding exons are depicted as open boxes and homologous regions in the targeting vector are indicated by thickened lines. In the mutant allele (Bottom), a PGKneo cassette replaced 4.4 kb of the tsg101 locus, encompassing exon 8 and most of exon 9, in opposite orientation to tsg101 gene transcription. Positions of the PCR primers (a–d), 3′ flanking probe (FP) used in Southern blot analysis, and predicted sizes of restriction fragments for genotyping are shown. B, BamHI; E, EcoRI. (b) Southern blot analysis of ES cell clones generated by homologous recombination at the tsg101 locus. Genomic DNA from wild-type (+/+) and heterozygous ES cell clones (+/−) was digested with EcoRI and hybridized to the indicated 3′ flanking probe. The 1.8-kb fragment is diagnostic of the mutant allele.
p53.
Primers 5′-GTG TTT CAT TAG TTC CCC ACC TTG AC and 5′-ATG GGA GGC TGC CAG TCC TAA CCC specific for the target sequence in p53 were used to detect the wild-type allele, whereas primers 5′-GTG GGA GGG ACA AAA GTT CGA GGC C and 5′-TTT ACG GAG CCC TGG CGC TCG ATG T were used for identification of the p53 recombinant allele.
Histological Analysis.
Tsg101 heterozygous males and females were intercrossed. Deciduae were isolated in ice-cold PBS at day 5.5 of embryogenesis (E5.5), E6.5, and E7.5, fixed overnight in 4% paraformaldehyde at 4°C, dehydrated, and embedded in paraffin. Sections 7 μm thick were cut and stained with hematoxylin and eosin.
In Situ Hybridization.
Deciduae were isolated and processed as for histological analysis. The probes used were full-length cDNAs of tsg101, Brachyury (9), and HNF-4 (10). Probes were labeled and processed as described (11).
BrdUrd Labeling of Embryos.
BrdUrd labeling of the cells in the S phase of the cell cycle was performed as described (8). BrdUrd (100 mg/g of body weight) was injected i.p. into pregnant females at E6.5. The females were killed 45 min after injection; the deciduae were fixed in 4% paraformaldehyde at 4°C overnight and processed for immunohistochemistry. The sections were incubated with an anti-BrdUrd monoclonal antibody (Roche Molecular Biochemicals) at 1:10 dilution.
In Vitro Culture of Preimplantation Embryos.
E3.5 embryos were collected and individually cultured as described (8). Photographs of cultured embryos were taken every 24 h. After 6 days in culture, the morphology of the embryos was recorded and their genotypes were determined by PCR analysis of their DNAs.
Immunohistochemistry.
Immunohistochemistry was performed as described (8). Anti-p53 antiserum (NovoCastra, Newcastle, U.K., NCL-p53-CM5p) was used at a 1:200 dilution.
Reverse Transcription–PCR Analysis.
Poly(A) RNA was isolated from individual E6.5 embryos by using the MicroFastTrack Kit (Invitrogen). cDNAs were generated by using a cDNA Synthesis Kit (Invitrogen). Quantitative reverse transcription–PCR was performed on cDNAs from single embryos by using specific primers for p53, p21, mdm2, and β-actin as described (12). Amplified PCR products were electrophoresed, followed by Southern blotting and hybridization, to a radiolabeled oligonucleotide internal to the amplified product. Signals were quantified by using a PhosphorImager and IMAGEQUANT software (Molecular Dynamics) and normalized to β-actin. One-quarter of the cDNA preparation from each individual embryo was used for genotyping using primers 5′-GGC GGA TGA AGG AGG AAA TG and 5′-GTG GGG CTG TGG GAA TGA TAA and a probe (5′-AAA CTG GAA GAG ATG GTC ACC CGC T) specific for the deleted tsg101 exons 8 and 9.
Results
Generation and Characterization of tsg101−/− Embryos.
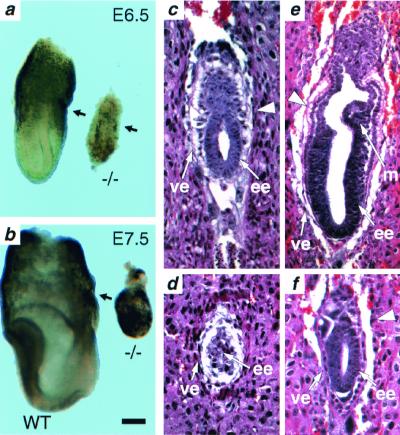
The murine tsg101 gene was disrupted by homologous recombination in ES cells (Fig. 1). The mutation was introduced into both C57BL/6J and CD1 genetic backgrounds with no discernable difference in resulting phenotypes. Mice heterozygous for the tsg101 mutation were phenotypically normal up to 14 mo of age. Heterozygous mice were intercrossed but homozygous tsg101−/− neonates were not observed in more than 400 offspring, indicating that homozygous mutation of tsg101 results in embryonic lethality. To determine the lethality phase of the tsg101 mutation, embryos from heterozygous intercrosses were analyzed at different days of gestation. At E6.5, approximately 25% of all embryos were phenotypically abnormal. These were genotypically homozygous mutants (Fig. 2a). The tsg101−/− embryos were smaller than their wild-type littermates and had a poorly defined boundary between the embryonic and extraembryonic regions. At E7.5, mutant embryos had not significantly increased in size or progressed in their development (Fig. 2b). By E8.5, most mutant embryos were either in resorption or degenerating within the yolk sac (data not shown). These data indicate that tsg101 is essential for postimplantation development at the time of initiation of gastrulation.
Figure 2.
Severe developmental delay in tsg101 mutant embryos. Morphology and histological analysis. (a) E6.5 tsg101−/− embryos (−/−) are smaller and less organized than their wild-type littermates (WT). The arrows point to the separation between the embryonic and extraembryonic regions. (b) E7.5 tsg101−/− embryos have failed to progress and are starting to be resorbed. (c) E5.5 wild-type embryo at the early egg cylinder stage. (d) E5.5 tsg101−/− embryo showing a poorly defined extraembryonic region and disorganized visceral endoderm. (e) E6.5 wild-type egg cylinder stage embryo. Both the embryonic and extraembryonic regions are well-organized and nascent mesoderm tissue can be distinguished. (f) E6.5 tsg101 mutant embryo. The embryonic and extraembryonic regions are severely underdeveloped and no mesoderm is observed. The large arrowhead in c, e, and f points to the separation between the embryonic and extraembryonic regions. ee, embryonic ectoderm; ve, visceral endoderm; m, mesoderm. (Bar = 60 μm in a, c–f; 120 μm in b.)
The structural organization of tsg101−/− embryos was characterized in detail by histological analyses of serially sectioned E5.5 and E6.5 embryos obtained from heterozygous intercrosses. Embryos were classified morphologically as wild type or mutant. At the E5.5 egg cylinder stage, wild-type embryos showed a well-organized ectoderm region (Fig. 2c), whereas mutant embryos were smaller and poorly organized (Fig. 2d). At E6.5, phenotypic differences were even more pronounced. Wild-type embryos exhibited a well-organized ectoderm, visceral endoderm, a primitive streak region, and a developing mesoderm (Fig. 2e). In contrast, mutant embryos had no detectable primitive streak, poorly defined visceral and parietal endoderm, and abnormal organization of the extraembryonic region (Fig. 2f). Thus, embryos were unable to progress significantly beyond E5.5 in the absence of tsg101, a developmental block whose timing coincides with the dramatic increase in embryo size that occurs at E5.5–6.5 during normal mouse development (13).
Tsg101 Expression and the Effects of Its Absence in Mutant Embryos.
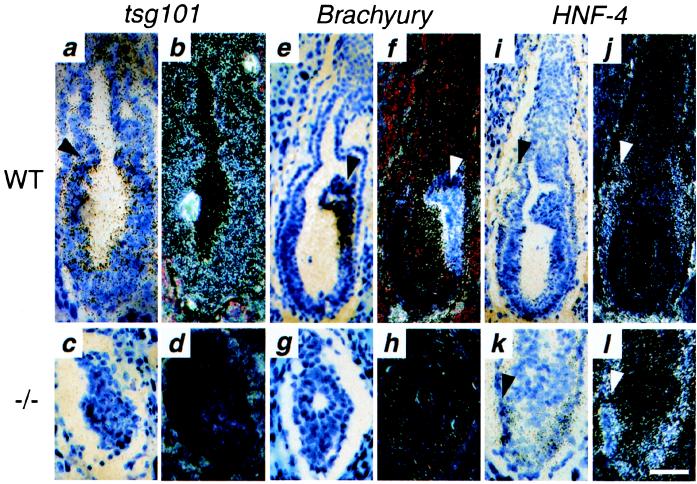
Northern blot analysis showed that tsg101 transcripts are expressed in wild-type ES cells, throughout normal mouse development, and in all adult tissues analyzed (data not shown; see also refs. 2 and 14). The spatial pattern of tsg101 expression at E6.5 was analyzed by in situ hybridization in tissue sections using a full-length cDNA antisense probe. Tsg101 expression was ubiquitous in wild-type embryos, detected throughout the epiblast, developing mesoderm, visceral endoderm, extraembryonic ectoderm, and endoderm (Fig. 3 a and b). No tsg101 transcripts were detected in mutant embryos (Fig. 3 c and d), demonstrating that the tsg101 mutation is a null mutation.
Figure 3.
Spatial expression of tsg101, Brachyury, and HNF-4 in E6.5 tsg101−/− embryos as determined by in situ hybridization: bright-field (a, c, e, g, i, k) and dark-field views (b, d, f, h, j, and l). (a and b) Ubiquitous tsg101 expression in sections of a wild-type E6.5 embryo hybridized to an antisense tsg101 probe. The arrow points to the separation between embryonic and extraembryonic regions. (c and d) No tsg101 expression is detected in a tsg101−/− E6.5 embryo. (e and f) Brachyury expression in a wild-type E6.5 embryo. Strong expression is detected in the nascent streak (arrow). (g and h) No Brachyury expression is detected in the tsg101−/− embryo. (i and j) HNF-4 expression in the visceral endoderm (arrow) of a wild-type E6.5 embryo. (k and l) Normal HNF-4 expression in a tsg101−/− E6.5 embryo. (Bar = 50 μm in a–d, g, h, k, l; 80 μm in e, f, i, j.)
As was shown in Fig. 2f, no histological signs of mesoderm formation could be detected in E6.5 tsg101−/− embryos. The absence of mesoderm was confirmed at the molecular level by in situ hybridization in tissue sections for expression of Brachyury (T), one of the earliest marker of mesoderm formation expressed at the onset of gastrulation at E6.5 (9). Intense Brachyury expression was detected in the nascent primitive streak of wild-type embryos (Fig. 3 e and f) but not in mutant embryos (Fig. 3 g and h). In contrast, expression of Hnf-4, a transcription factor whose expression is initially restricted to the extraembryonic visceral endoderm (10), was comparable in wild type (Fig. 3 i and j) and tsg101−/− (Fig. 3 k and l) embryos. Terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) assays of tissue sections revealed only a few TUNEL-positive nuclei close to the amniotic cavity in both wild-type and mutant embryos, indicating that the reduced size of tsg101−/− embryos probably is not caused by an excess of apoptosis (data not shown).
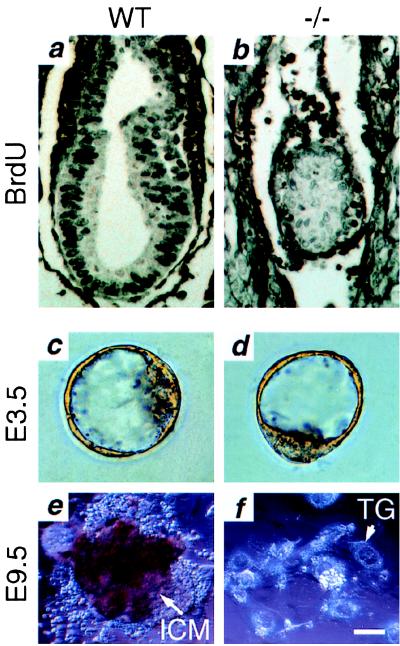
The effect of the tsg101 mutation on cellular proliferation in vivo was examined by BrdUrd incorporation. The ratio of the number of proliferating cells (indicated by BrdUrd-positive nuclei) to the total cell number was taken as the mitotic index. As exemplified in Fig. 4a, 70–80% of the nuclei from phenotypically wild-type E6.5 embryos were BrdUrd-positive, producing a mitotic index of 0.7–0.8. However, all mutant embryos analyzed showed <30% nuclear staining (Fig. 4b) and a mitotic index of 0.2–0.3. These data indicate that the proliferative capacity of tsg101−/− embryos is severely impaired at E6.5 in vivo.
Figure 4.
Reduced cellular proliferation in tsg101−/− embryos in vivo and in vitro. (a and b) BrdUrd incorporation. (a) Strongly BrdUrd-positive nuclei can be seen throughout an E6.5 wild-type embryo. (b) Fewer BrdUrd-positive cells with a much weaker signal are seen in an E6.5 tsg101−/− embryo. (c—f) ICM outgrowth. (c) Wild-type E3.5 blastocyst. (d) Tsg101−/− E3.5 blastocyst. (e) Wild-type outgrowth after 6 days of culture. The ICM is surrounded by trophoblast giant cells (TG). (f) Outgrowth of tsg101−/− blastocysts after 6 days of culture. Only TG cells remain. (Bar = 80 μm in a and b; 20 μm in c and d; 40 μm in e and f.)
To further investigate the growth capability of mutant tsg101 embryos, E3.5 blastocysts from heterozygous matings were individually cultured in vitro (Fig. 4 c–f). At E3.5, tsg101−/− blastocysts were indistinguishable from the wild type (Fig. 4 c and d), indicating that the tsg101 mutation does not affect preimplantation development. However, after 6 days in culture, the inner cell mass (ICM) failed to grow in mutant blastocysts, in contrast to extensive ICM proliferation in wild-type blastocysts (Fig. 4 e and f). In addition, numerous attempts to generate tsg101−/− ES cells were unsuccessful (data not shown), further suggesting that a generalized failure of cell cycle progression occurs in the absence of tsg101.
Altered Regulation of p53 and Its Effectors in tsg101−/− Embryos.
Mutations of the mdm2 protooncogene, whose protein product has ubiquitin ligase activity that negatively regulates the tumor suppressor p53 (15), lead to embryonic lethality before gastrulation with a phenotype resembling that of tsg101−/− mutants (16, 17). Mdm2 both represses p53 transcriptional activity and mediates degradation of p53. p53 is capable of blocking cell cycle progression at the G1 phase through transcriptional up-regulation of the cyclin-dependent kinase (cdk) inhibitor p21WAF-1/CIP-1 (14, 18, 19). Additionally, p53 induces mdm2 transcription in an autoregulatory feedback loop. Mutational disruption of mdm2 results in activation and accumulation of p53 (15), generating a proliferative block that leads to early embryonic death around E6.5. However, double mutation of both p53 and mdm2 completely rescues this lethality, indicating that accumulation of p53 is the sole cause of the developmental block occurring in mdm2 mutant embryos (16, 17).
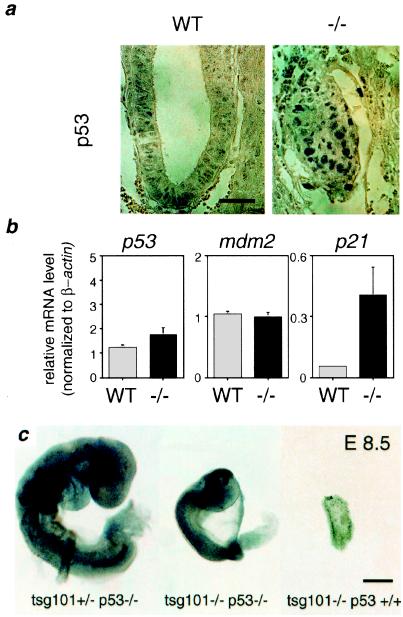
The similar phenotypes observed for both tsg101 and mdm2 mutant embryos led us to investigate, by using quantitative reverse transcription–PCR and immunohistochemistry for individual E6.5 embryos, whether the expression of p53, mdm2, or p21 was altered in tsg101−/− embryos. Immunohistochemical staining showed that, whereas weak anti-p53 staining was observed in a few cells in the epiblast of wild-type embryos, all tsg101−/− embryos analyzed showed strong nuclear anti-p53 staining throughout the embryonic and extraembryonic regions (Fig. 5a), demonstrating an accumulation of p53 protein. Interestingly, the levels of p53 and mdm2 mRNA were unaffected by the absence of tsg101 (Fig. 5b), indicating that the observed p53 accumulation occurs posttranscriptionally, and also indicating the absence of normal p53 activation of mdm2 transcription in these embryos. However, with the accumulation of p53 protein, transcription of its downstream effector p21 was dramatically increased (Fig. 5b), suggesting that cell cycle arrest in tsg101−/− embryos may result from p53-mediated activation of p21 expression.
Figure 5.
Deregulated p53 signaling is responsible for the gastrulation defect in tsg101−/− embryos. (a) Immunohistochemical analysis of p53 protein levels. A few weakly p53-positive cells can be detected in a wild-type E6.5 embryo (WT), whereas strongly p53-positive nuclei occur throughout the tsg101−/− embryo (−/−). (Bar = 50 μm.) (b) Levels of p53, mdm2, and p21 mRNA expression. Quantitative reverse transcription–PCR on mRNA from individual E6.5 wild type (WT) and mutant (−/−) embryos (see Materials and Methods). Expression levels are calculated relative to β-actin. p21 expression is increased 5- to 10-fold in tsg101−/− embryos. (c) Partial rescue of tsg101−/− embryos by null mutation of p53. Whole mount preparations of embryos from tsg101+/−/p53+/− intercrosses at E8.5. Although less advanced than their tsg101+/−/p53−/− littermates (Left), tsg101−/−/p53−/− embryos at E8.5 are developing an anterior–posterior pattern with a head, trunk, and tail region (Middle). A remnant of a tsg101−/−/p53+/+ single mutant embryo at E8.5 is shown on the Right. (Bar = 100 μm.)
To investigate the role of p53 in the lethal block in cell proliferation occurring in tsg101−/− mutant embryos, we introduced the tsg101 mutation into a p53 null background (20). Sixty embryos from tsg101+/−/p53+/− intercrosses in a mixed 129J/C57BL6J/CD1 genetic backgrounds were retrieved at E8.5 and genotyped by PCR. Although no viable tsg101−/− embryos were identified in a p53+/+ or p53+/− background, three tsg101−/− embryos in a p53−/− background were still alive. Although smaller than their wild-type littermates, double mutant embryos developed past the gastrulation stage with a distinct anterior–posterior pattern (Fig. 5c). Death of the double mutants occurred between E8.5 and E10.5, indicating a failure of the p53−/− mutation to entirely rescue embryos from the effects of a tsg101 deficiency on development. However, tsg101−/−/p53−/− embryos lived longer and developed further than did tsg101−/− embryos in either a wild-type or p53 heterozygous background, supporting the hypothesis that developmental arrest in early (i.e., E6.5) tsg101−/− embryos is a consequence of p53 accumulation.
Discussion
In this study, we have generated a null mutation in the mouse tsg101 gene by gene targeting and shown that tsg101 is required for embryonic cellular proliferation before the onset of gastrulation and for modulation of the p53 pathway in vivo. Tsg101 mutant embryos showed signs of growth retardation as early as E5.5 and did not form mesoderm at E6.5, as revealed by morphological analysis and the absence of Brachyury expression. Gastrulation in the mouse embryo occurs at around E6.5 of gestation and requires rapid epiblast proliferation, resulting in a 100-fold increase in cell number between E5.5 and E7.5 (13). Tsg101−/− embryos showed reduced BrdUrd incorporation at E6.5, indicating a slowed cell cycle that ultimately culminated in cell cycle arrest and embryonic death. Furthermore, the ICM of tsg101−/− blastocysts failed to proliferate in vitro, additionally demonstrating a requirement for tsg101 in embryonic cellular proliferation.
We have shown that accumulation of the tumor suppressor protein p53 is at least in part responsible for the proliferative block observed in tsg101−/− embryos. During normal early mouse development, p53 activation is controlled by its negative regulator Mdm2 in a feedback control loop and p53 is not activated before E11 (21). However, in the absence of tsg101, p53 protein accumulates at E6.5 by a posttranscriptional mechanism. This is accompanied by transcriptional up-regulation of the p53 target gene p21, which is able to block the activity of cyclin-dependent kinases, resulting in slowed progression through the cell cycle (or cell cycle arrest) (18, 19, 22). The elevated transcription of p21 observed in tsg101−/− mutants together with the partial rescue of tsg101−/− mutants by deletion of the p53 gene suggests that the accumulation of p53 and activation of the p53 pathway are responsible for the proliferative block at E6.5 in tsg101−/− embryos. However, the subsequent death of tsg101−/− embryos at E8.5 indicates that tsg101 function is also required for other pathways important to later development.
Although our results establish the effect of null mutation of tsg101 on the p53 pathway, they leave open the possibility that mutation of tsg101 activates p53 indirectly in early embryos, as occurs following targeted disruption of Brca1 or Brca2 (8, 12, 23, 24). Brca1 and Brca2 are tumor suppressor genes thought to be involved in the maintenance of genome integrity. Brca1−/− and Brca2−/− embryos also exhibit a developmental block at gastrulation that is partially rescued by inactivation of p53.
Alternatively, tsg101 may be directly involved in the regulation of the p53 level in vivo. Cellular p53 levels normally are tightly regulated through ubiquitin-mediated proteolysis controlled by the ubiquitin ligase Mdm2 (15). Mdm2 levels are in turn controlled by p53 through feedback regulation and further by modulation of Mdm2 stability itself (25–27). Because of its resemblance to inactive ubiquitin conjugase homologs, it has been speculated that tsg101 has a role in the regulation of ubiquitin-mediated proteolysis (6, 7), and recently tsg101 has been found to participate with Mdm2 in a feedback control loop that parallels the p53/mdm2 autoregulatory loop (28). Overexpression of tsg101 inhibits Mdm2 ubiquitination and degradation and consequently decreases the cellular level of p53, whereas elevation of p53 and/or its transcriptionally activated target Mdm2 accelerate decay of tsg101 (28). These results are consistent with, and complementary to, our in vivo findings showing that tsg101 deficiency results in p53 accumulation, growth arrest, and early embryonic lethality. Additionally, our data show that, in the absence of tsg101, p53 accumulation fails to accomplish its normal stimulation of mdm2 transcription. Thus, the accumulation of p53 observed in tsg101−/− embryos may result from at least two separate mechanisms that act at transcriptional and posttranscriptional levels to decrease MDM2-mediated degradation of p53.
Acknowledgments
We thank Michael Bezuhly, Vincent Tsui, Betty Hum, and Julia Potter for technical assistance; Atsushi Hirao, Razqallah Hakem, and Wen-Chen Yeh for comments and discussion; and Mary Saunders for scientific editing. S.N.C. and L.L. recieved support from the Helmut Horten Foundation and Chiron Corporation. J.R. was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
Abbreviations
- tsg101
tumor susceptibility gene 101
- ES
embryonic stem
- En
embryonic day n
- ICM
inner cell mass
- cdk
cyclin-dependent kinase
References
- 1.Li L, Cohen S N. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Li X, Francke U, Cohen S N. Cell. 1997;88:143–154. doi: 10.1016/s0092-8674(00)81866-8. [DOI] [PubMed] [Google Scholar]
- 3.Hittelman A B, Burakov D, Iniguez-Lluhi J A, Freedman L P, Garabedian M J. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Z, Pan J, Hope W X, Cohen S N, Balk S P. Cancer. 1999;86:689–696. doi: 10.1002/(sici)1097-0142(19990815)86:4<689::aid-cncr19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe M, Yanagi Y, Masuhiro Y, Yano T, Yoshikawa H, Yanagisawa J, Kato S. Biochem Biophys Res Commun. 1998;245:900–905. doi: 10.1006/bbrc.1998.8547. [DOI] [PubMed] [Google Scholar]
- 6.Koonin E V, Abagyan R A. Nat Genet. 1997;16:332–333. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- 7.Ponting C P, Cai Y D, Bork P. J Mol Med. 1997;75:467–469. [PubMed] [Google Scholar]
- 8.Hakem R, de la Pompa J L, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, et al. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 9.Kispert A, Hermann B G. EMBO J. 1993;12:4898–4899. doi: 10.1002/j.1460-2075.1993.tb06179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan S A, Manova K, Chen W S, Hoodless P, Weinstein D C, Bachvarova R F, Darnell J E J. Proc Natl Acad Sci USA. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hui C C, Joyner A L. Nat Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki A, de la Pompa J L, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, et al. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 13.Snow M. J Embryol Exp Morphol. 1977;42:293–303. [Google Scholar]
- 14.Wagner K U, Dierisseau P, Rucker E B, III, Robinson G W, Hennighausen L. Oncogene. 1998;17:2761–2770. doi: 10.1038/sj.onc.1202529. [DOI] [PubMed] [Google Scholar]
- 15.Prives C. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 16.Montes de Oca Luna R, Wagner D S, Lozano G. Nature (London) 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 17.Jones S N, Roe A E, Donehower L A, Bradley A. Nature (London) 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 18.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 19.Gu Y, Turck C W, Morgan D O. Nature (London) 1993;366:707–710. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- 20.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 21.Komarova E A, Chernov M V, Franks R, Wang K, Armin G, Zelnick C R, Chin D M, Bacus S S, Stark G R, Gudkov A V. EMBO J. 1997;16:1391–1400. doi: 10.1093/emboj/16.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 23.Hakem R, de la Pompa J L, Elia A, Potter J, Mak T W. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig T, Chapman D L, Papaioannou V E, Efstratiadis A. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 25.Buschmann T, Fuchs S Y, Lee C G, Pan Z Q, Ronai Z. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 26.Sherr C J, Weber J D. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Liao L, Ruland J, Mak T W, Cohen S N. Proc Natl Acad Sci USA. 2001;98:1619–1624. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]