Abstract
G-protein-coupled receptors mediate the senses of taste, smell, and vision in mammals. Humans recognize thousands of compounds as bitter, and this response is mediated by the hTAS2R family, which is one of the G-protein-coupled receptors composed of only 25 receptors. However, structural information on these receptors is limited. To address the molecular basis of bitter tastant discrimination by the hTAS2Rs, we performed ligand docking simulation and functional analysis using a series of point mutants of hTAS2R16 to identify its binding sites. The docking simulation predicted two candidate binding structures for a salicin-hTAS2R16 complex, and at least seven amino acid residues in transmembrane 3 (TM3), TM5, and TM6 were shown to be involved in ligand recognition. We also identified the probable salicin-hTAS2R16 binding mode using a mutated receptor experiment. This study characterizes the molecular interaction between hTAS2R16 and β-d-glucopyranoside and will also facilitate rational design of bitter blockers.
Keywords: Calcium Imaging, G-protein-coupled Receptors (GPCR), Membrane Proteins, Receptor Structure-Function, Signal Transduction, Bitter, Taste Receptor, Binding Site, Molecular Dynamics, Molecular Modeling
Introduction
There are five basic and distinct taste sensations: sweetness, bitterness, saltiness, sourness, and umami. Recent progress in the molecular biology of taste has revealed that sweet, umami, and bitter tastes are mediated by G-protein-coupled receptors (1). The TAS1R family of taste receptors mediates two taste sensations; TAS1R2/TAS1R3 responds to sweetness (2, 3) and TAS1R1/TAS1R3 responds to umami taste (2, 4), whereas the TAS2R family responds to bitterness (5–14).
Bitterness is generally considered a sign of toxic substances; on the other hand, appropriate use of certain bitter food ingredients can provide a unique and favorable character to foods, as in the case of tea, coffee, and beer. It is, thus, important for the food industry to use bitter taste ingredients properly. Information on the ligand binding structure of the bitter taste receptors with tastants is useful for controlling bitterness. Using the crystal structure of bovine rhodopsin as a template, several in silico computational modeling for hTAS2Rs have been demonstrated (15, 16). In these reports, the three-dimensional structure of hTAS2R38 was predicted, and docking simulations with 6-propyl-2-thiouracil and N-phenylthiourea were performed to clarify the differences in their sensitivity to the hTAS2R38 receptor variants (13). However, these previous analyses lacked functional studies to compensate for the ambiguity of the computational simulations and validate the accuracy of the predictions.
On the other hand, to clarify the ligand binding sites in TAS1Rs, several studies have been conducted using chimeric and mutated receptor experiments. For example, the large extracellular amino-terminal domain of TAS1R2 is required for recognition of aspartame and neotame as sweet tastants (17, 18), and the hTAS1R3 amino-terminal domain is required for response to neoculin, a sweet protein with taste-modifying activity (19–22). Molecular modeling is also effective for identifying binding sites, and a combination of functional analysis along with molecular modeling has identified the neohesperidin dihydrochalcone binding site in TAS1R3 and glutamate binding site in the TAS1R1 Venus flytrap domain (23, 24).
In the present study we performed both a precise ligand docking simulation and functional analysis of a series of point mutants to determine the molecular basis of bitter ligand discrimination by hTAS2R16 and have successfully identified the putative salicin binding site in hTAS2R16. This study provides new insights into bitter taste reception by hTAS2Rs.
EXPERIMENTAL PROCEDURES
Compounds
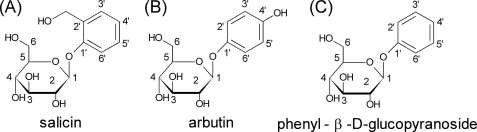
Salicin, arbutin, and phenyl-β-d-glucopyranoside were selected as test ligands for hTAS2R16 (Fig. 1) (7). Salicin was purchased from Nacalai Tesque, Inc. (Kyoto, Japan), arbutin was from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan), and phenyl β-d-glucopyranoside was from Sigma.
FIGURE 1.
The molecular structures of salicin (A), arbutin (B), and phenyl-β-d-glucopyranoside (C). Numbers denote the positions of carbon atoms.
Construction of a Structural Model of hTAS2R16
Seven TM3 helical regions were deduced based on White and Wimley parameters (25), and a homology alignment between the amino acid sequences of bovine rhodopsin and hTAS2R16 was constructed. The structure of the TM helical regions and the extra- and intracellular loops were constructed using the Homology module installed in Insight II (Accelrys Inc., San Diego, CA) and an alignment from a previously reported three-dimensional structural model of a rhodopsin photointermediate, metarhodopsin (26). Insertion and deletion sites were selected in the loop regions and constructed using the loop structure data base provided in Insight II.
Energy minimization was carried out using a molecular mechanics/molecular dynamics program, Discover 3, within Insight II, until the root mean square deviation became less than 0.1 kcal/mol-Å2. Subsequent molecular dynamics calculations for the structural optimization were performed using Discover 3 at 300 K for 0.2 ns by sampling the structures every 2 ps. The resulting 100 structures were energy-minimized using Discover 3. Among the minimized structures, the lowest energy structure was selected. During the calculations, the main-chain atoms in the TM regions were tethered at their original positions.
Construction of Complex Structural Models of the TM Region
The ligand binding sites in the TM region were deduced using the binding site module of Insight II. Salicin was docked in the binding site to simulate conformational changes in the receptor structure. These structures were energy-minimized using Discover 3 followed by structural optimization using molecular dynamics calculations.
Construction of Expression Plasmids for hTAS2R16 and the Chimeric G Protein
To construct an expression plasmid for hTAS2R16, DNA fragments encoding hTAS2R16 (NCBI refseq number NM_016945) were subjected to PCR amplification from human genomic DNA (BD Clontech, Mountain View, CA). The coding region of hTAS2R16 (1–876) was tagged at the amino terminus with the first 45 amino acids of rat somatostatin receptor type 3 (ssr3) (27) and subcloned into the EcoRI-NotI site of the pEAK10 expression vector (Edge Biosystems, Gaithersburg, MD). Expression plasmids for the chimeric G-protein subunit Gα16gust44, subcloned into pcDNA3.1 (Invitrogen), were kindly gifted by Dr. Ueda and Dr. Shimada (Nagoya City University, Nagoya, Japan) (28).
Site-directed Mutagenesis
Mutations were introduced into ssr3-hTAS2R16 cDNA using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Fragments containing the desired mutation product were digested with EcoRI and NotI and ligated into the EcoRI-NotI site of the pEAK10 expression vector. All mutations were checked by DNA sequencing using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA).
Cell Culture and Transfection
Human embryonic kidney 293T (HEK293T) cells were cultured at 37 °C in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen). Cells were seeded onto 35-mm dishes and transiently transfected with the plasmid expressing the ssr3-hTAS2R16 or ssr3-hTAS2R16 mutants along with Gα16gust44 at a 4:1 ratio using Lipofectamine 2000 (Invitrogen).
Cell-based Assay
Transfected cells were transferred to a 96-well black, clear-bottomed CellBIND surface plate (Corning Inc., Bedford, MA) 6 h after transfection. The cells were incubated for an additional 18–20 h, then rinsed with an assay buffer (130 mm NaCl, 10 mm glucose, 5 mm KCl, 2 mm CaCl2, 1.2 mm MgCl2, and 10 mm HEPES, pH 7.4), loaded with 5 μm Fluo-4 AM calcium indicator dye (Dojindo Laboratories, Kumamoto, Japan) diluted with the assay buffer, and incubated for an additional 30 min at 27 °C. The cells were then rinsed with the assay buffer and incubated in 100 μl of assay buffer for 10 min at 27 °C before the plate was loaded onto a FlexStation 3 (Molecular Devices, Inc., Sunnyvale, CA) for fluorescence detection. Fluorescence (excitation at 485 nm, emission at 525 nm, and cutoff at 515 nm) was monitored at 2-s intervals at 27 °C; 100 μl of assay buffer supplemented with 2× test compound solution was added at 20 s, and scanning was continued for an additional 100 s. Fluorescence responses were measured from 20 to 30 s after the addition of the ligand and corrected for background fluorescence measured before ligand application. Stimuli were tested at concentrations that did not elicit calcium responses from Gα16gust44-transfected cells (data not shown). Final concentrations of the test ligands (salicin, arbutin, and phenyl β-d-glucopyranoside) were each set at 0.01, 0.03, 0.1, 0.3, 1, 3, 10, and 30 mm. Data were collected from at least three independent experiments.
For calculation of EC50 values, plots of amplitude versus concentration were prepared in Clampfit Version 9.2.0.09 (Molecular Devices). Nonlinear regression of the plots produced the function f(x) = Imin + (Imax − Imin)/(1 + (x/EC50)h), where x is the ligand concentration and h is the Hill coefficient, which was used to calculate the EC50 values for ligand-receptor interactions.
RESULTS
Molecular Modeling of hTAS2R16 and Ligand-Receptor Complexes
We constructed a molecular model of hTAS2R16 based on the previously reported three-dimensional structural model of a rhodopsin photointermediate, metarhodopsin (26). According to the homology alignment between bovine rhodopsin and hTAS2R16, the TM domains and extra- and intracellular loops of hTAS2R16 were constructed, and the ligand binding sites in the TM region were deduced. Salicin docked in the binding site was then obtained after structural optimization of the initial complexes. Salicin was bound in the putative ligand binding pocket formed by TM3, TM5, and TM6 and interacted with eight amino acid residues (Table 1).
TABLE 1.
Predicted amino acid residues involved in salicin recognition
| Location | Residues | |||
|---|---|---|---|---|
| TM3 | Glu-86 | Asn-89 | Phe-93 | Trp-94 |
| TM5 | Gln-177 | His-181 | ||
| TM6 | Phe-240 | Ile-243 | ||
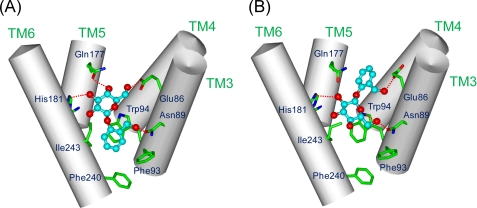
Through the above approach, two binding modes of salicin in the ligand binding region were suggested as candidates for the salicin-receptor complex structure (Figs. 2, A and B). In both complex models, salicin was located in the same putative binding pocket with different binding modes. In one mode (model A), the aromatic ring of salicin was directed to the cytoplasmic site and the sugar moiety to the extracellular site of the putative ligand binding pocket (Fig. 2A), whereas in the other mode (model B), it was located upside down in the binding site (Fig. 2B).
FIGURE 2.
Two putative salicin-hTAS2R16 binding structures and a detailed map of the salicin-hTAS2R16 interaction. Numbered amino acid positions were predicted to be involved in salicin recognition. A, shown is a side view of one proposed salicin-hTAS2R16 binding model (model A). Salicin is bound within the binding pocket with the aromatic ring of salicin oriented toward the cytoplasm. B, shown is a side view of another proposed salicin-hTAS2R16 model (model B). Salicin is bound within the binding pocket with the aromatic ring of salicin oriented toward extracellular space.
To clarify the salicin-hTAS2R16 complex structure, Glu-86, Trp-94, and Gln-177 were selected as key amino acid residues in the constructed models. In model A, Glu-86 and Gln-177 formed hydrogen bonds with the sugar moiety, and Trp-94 interacted with the aromatic ring of salicin (Fig. 2A). On the other hand, in model B, Glu-86 hydrogen-bonded with the hydroxymethyl group on the aromatic ring, and Trp-94 interacted with the sugar moiety of salicin, whereas Gln-177 did not have any interaction with salicin (Fig. 2B).
Clarification of the Salicin-hTAS2R16 Binding Mode Using Point Mutations of Key Amino Acid Residues
To clarify the salicin-hTAS2R16 binding mode, two key amino acid residues, Trp-94 and Gln-177, were targeted for point mutagenesis, and their responses to salicin were examined using a cell-based assay for HEK293T cells cotransfected with the ssr3-hTAS2R16 mutants along with Gα16gust44.
In model A, Trp-94 predicted that salicin had aromatic-aromatic interactions only. In model B, Trp-94 seemed to be involved in not only CH-π interactions but also in hydrogen bonding with the sugar moiety of salicin. To examine its interaction with salicin, Trp-94 was mutated to a Phe (W94F) or Tyr (W94Y) residue. If salicin binding is mediated as in model A, the response of these mutants to salicin would not change, because both Phe and Tyr residues have aromatic rings to make hydrophobic interactions. However, if salicin binds as in model B, the response of W94F to salicin would be much more drastically reduced than W94Y mutants because Phe residue has no hydroxyl group to make a hydrogen-bonding interaction with salicin.
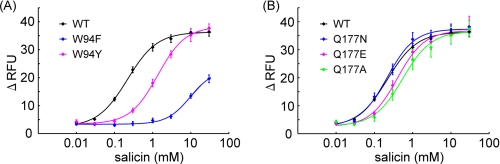
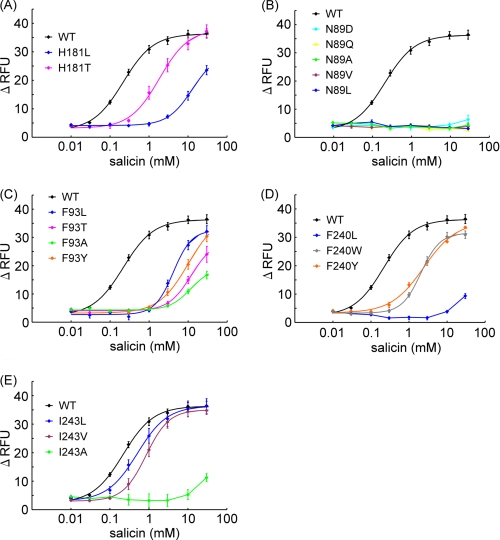
When these mutants were applied to the assay, the W94Y mutant resulted in a relatively small difference in the EC50 value to salicin (1.36 mm) compared with wild-type (WT) ssr3-hTAS2R16-salicin (0.22 mm), whereas W94F had a 40-fold higher EC50 value (9.68 mm) than the WT (Fig. 3A, Table 2). This result suggests that Trp-94 interacts with salicin not only through hydrophobic interactions such as aromatic-aromatic or CH-π interactions but also through hydrogen bonds.
FIGURE 3.
Point mutation analyses of the responses to salicin of key amino acid residues. The dose-response curves show the results of the analyses for Trp-94 mutants (A) and Gln-177 mutants (B). The dose-response relationship to salicin was obtained using a cell-based assay of HEK293T cells cotransfected with ssr3-hTAS2R16 or ssr3-hTAS2R16 mutants along with Gα16gust44. Each point represents the mean ± S.E. from at least three independent measurements. RFU, relative fluorescence units.
TABLE 2.
Effect of point mutations on hTAS2R16-salicin EC50 values obtained from cell based assay
| Location | Mutants | EC50 (mm) | EC50 mutant/EC50 WT |
|---|---|---|---|
| WT | 0.22 | 1.0 | |
| TM3 | E86D | 2.13 | 9.8 |
| E86Q | 1.25 | 5.7 | |
| N89Q | No response | ||
| N89D | No response | ||
| N89V | No response | ||
| N89L | No response | ||
| N89A | No response | ||
| F93T | 12.0 | 55.1 | |
| F93Y | 10.0 | 45.9 | |
| F93L | 4.15 | 19.1 | |
| F93A | 11.5 | 52.8 | |
| W94F | 9.68 | 44.5 | |
| W94Y | 1.36 | 6.2 | |
| TM5 | Q177N | 0.39 | 1.8 |
| Q177E | 0.22 | 1.0 | |
| Q177A | 0.55 | 2.5 | |
| H181T | 1.84 | 8.5 | |
| H181L | 12.9 | 59.3 | |
| TM6 | F240Y | 2.44 | 11.2 |
| F240L | >30 | – | |
| F240W | 2.05 | 9.4 | |
| I243L | 0.50 | 2.3 | |
| I243V | 0.84 | 3.9 | |
| I243A | >30 | – |
Next, we observed the role of Gln-177, which was predicted to form a hydrogen bond with the sugar moiety in model A but not to form any hydrogen bonds with salicin in model B (Fig. 2). None of the Gln-177 mutants tested (Q177N, Q177E, and Q177A) significantly affected the activity of salicin (Fig. 3B, Table 2). Thus, Gln-177 is not predicted to be an important residue for interaction with salicin. Based on the functional analysis of Trp-94 and Gln-177 mutants above, the binding mode for salicin in hTAS2R16 of model B is preferred over that of model A (Fig. 2B).
Functional Analysis of Glu-86 Mutants with Various β-Glucopyranosides
Next, we performed a functional analysis of the Glu-86 mutants E86D and E86Q using arbutin and phenyl-β-d-glucopyranoside as the ligands to verify whether or not hTAS2R16-salicin binding mode accords with model B.
In model B, Glu-86 was expected to interact with hydroxymethyl group on the aromatic ring of salicin. On the other hand, arbutin and phenyl-β-d-glucopyranoside could not interact with Glu-86, because the para-hydroxy group on the phenyl group of arbutin would be too far from it and phenyl-β-d-glucopyranoside has no hydroxy group on its aromatic ring (Fig. 1). For these reasons, only salicin can form a hydrogen bond with Glu-86 to facilitate the binding.
In model A, Glu-86 was predicted to interact with the sugar moiety, not the hydroxy group on the aromatic ring, so there are no specific interactions between Glu-86 and these phenyl-β-d-glucopyranosides to facilitate the binding. E86D mutant was considered as a feasible mutant to clarify the putative interaction, because Asp-86 of E86D mutant was predicted to maintain hydrogen-bonding distance from the hydroxymethyl group on the aromatic ring of salicin. E86Q was designed to examine the effect of length of the side chain of Glu-86.
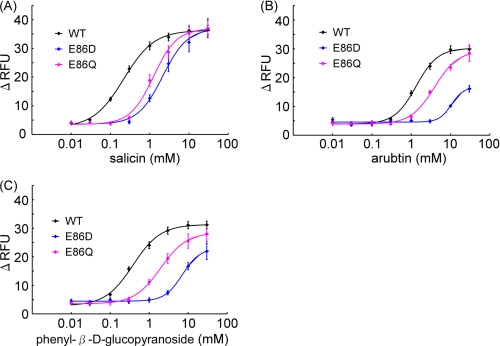
The mutants did exhibit reduced responses to salicin and had similar EC50 values (E86D, 2.13 mm; E86Q, 1.25 mm) compared with the WT (0.22 mm) (Fig. 4A, Table 2). On the other hand, E86D had a large reduction in its response to arbutin and phenyl-β-d-glucopyranoside (arbutin, 10.6 mm; phenyl-β-d-glucopyranoside, 7.09 mm), whereas E86Q had a response similar to that of the WT (E86Q, arbutin, 3.89 mm; phenyl-β-d-glucopyranoside, 1.96 mm; WT, arbutin, 1.34 mm, phenyl-β-d-glucopyranoside, 0.38 mm) (Figs. 4, B and C, and supplemental Tables 1 and 2).
FIGURE 4.
Dose-response relationships for various phenyl-β-d-glucopyranosides obtained using a cell-based assay of HEK293T cells cotransfected with ssr3-hTAS2R16 or ssr3-hTAS2R16 Glu-86 mutants along with Gα16gust44. Each point represents the mean ± S.E. from at least three independent measurements. Dose-response curves are shown for salicin (A), arbutin (B), and phenyl-β-d-glucopyranoside (C). RFU, relative fluorescence units.
Our data obtained from the Glu-86 mutant analysis suggested that the length of the side chain of the Glu-86 amino acid residue is important for ligand recognition. In addition, only salicin forms a hydrogen bond with Glu-86 to facilitate the binding. These results are also consistent with our putative model B.
Verification of the hTAS2R16-β-Glucopyranoside Binding Mode
Based on the functional analysis of Glu-86, Trp-94, and Gln-177 mutants described above, model B was supported as the salicin-hTAS2R16 binding model (Fig. 2B). To further verify this model, we performed additional functional analyses using other amino acid residues expected to be involved in ligand recognition.
In model B, Asn-89 and His-181 formed hydrogen bonds with the sugar moiety and were considered important for ligand recognition (Fig. 2B). To examine the putative hydrogen bond between His-181 and the sugar moiety of salicin, we constructed two mutants, H181L and H181T. The H181T mutant responded to salicin with a small increase in EC50 value (1.84 mm) compared with that of WT ssr3-hTAS2R16 (0.22 mm), whereas H181L had a 50-fold higher EC50 value (12.9 mm) than the WT (Fig. 5A, Table 2). These results suggest that His-181 forms a hydrogen bond with salicin. Unfortunately, all of the mutations tested for Asn-89 completely lost the ability to respond to salicin (Fig. 5B), and therefore, it is unclear whether Asn-89 is involved in ligand recognition.
FIGURE 5.
Dose-response relationships for various hTAS2R16 mutants to salicin obtained using a cell-based assay of HEK293T cells cotransfected with ssr3-hTAS2R16 or ssr3-hTAS2R16 mutants along with Gα16gust44. Each point represents the mean ± S.E. from at least three independent measurements. Dose-response curves are shown for His-181 mutants (A), Asn-89 mutants (B), Phe-93 mutants (C), Phe-240 mutants (D), and Ile-243 mutants (E). RFU, relative fluorescence units.
Three other hydrophobic amino acid residues, Phe-93, Phe-240, and Ile-243, were predicted to form the salicin binding site through hydrophobic interactions (Fig. 2B). All of the tested Phe-93 mutants had a greatly reduced response to salicin (F93T, 12.0 mm; F93Y, 10.0 mm; F93L, 4.15 mm; F93A, 11.5 mm) compared with the WT (Fig. 5C, Table 2). We examined three Phe-240 mutants, F240L, F240W, and F240Y. The F240L mutant had a substantially reduced response to salicin (F240L, >30 mm) in comparison with the F240Y and F240W mutants (F240Y, 2.44 mm; F240W, 2.05 mm) (Fig. 5D, Table 2). Finally, the I243A mutant, with a change in the size of the amino acid residue, had a greatly reduced response to salicin (I243A, >30 mm) in comparison with the I243L and I243V mutants (I243L, 0.50 mm; I243V, 0.84 mm) (Fig. 5E, Table 2). These results indicate that at least these three amino acid residues play important roles in forming the salicin binding pocket.
DISCUSSION
Humans recognize thousands of compounds as bitter, and response to bitterness is mediated by the hTAS2R family, which are G-protein-coupled receptors composed of only 25 receptors. Heterologous expression experiments have identified many of their cognate bitter ligands (29), and receptor-based assays have been developed as novel in vitro approaches for taste assessment (30). In principle, this method could be used to screen taste modulators such as bitter blockers that act on taste receptors. Using such assays, we found that Glu-Glu significantly reduced the response of several hTAS2Rs to these ligands, suggesting that acidic pH may be one of the critical factors responsible for the previously reported bitter-masking effect of Glu-Glu (31, 32). Information on the ligand binding structure of taste receptors with tastants is essential for rational design of taste modulators such as bitter blockers. However, most G-protein-coupled receptors have not been crystallized, and to date only five independent x-ray crystal structures have been determined (33–38). In silico computational methods employing receptor-based modeling are useful tools for predicting ligand-receptor complex structures. However, because virtual docking simulations using homology-based structural models still have low predictive power, the accuracy of this approach is improved by combining it with functional studies using mutants of the receptor. In this study we experimentally identified the putative β-glucopyranoside binding site in hTAS2R16 predicted from computational molecular modeling using functional analysis.
Because salicin is an agonist of hTAS2R16, the structural model should be constructed from its activated form. Although crystal structures of G-protein-coupled receptors such as adrenergic receptors and adenosine receptor have been reported, all the structures are of an inactive form (33–38). Thus, we selected the previously reported, active form three-dimensional structural model of a rhodopsin photointermediate, metarhodopsin (26), as a template.
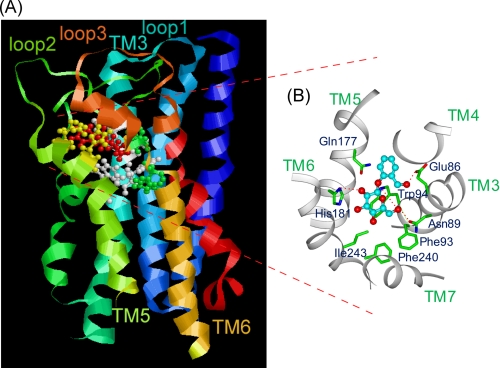
The results of the docking model suggested two binding modes as candidates for the complex structure of salicin-hTAS2R16 (Fig. 2), and certain amino acid residues were deduced to be directly involved in salicin recognition (Table 1). Based on the two models, Glu-86, Trp-94, and Gln-177 were selected as key amino acid residues for clarifying the salicin-hTAS2R16 complex structure (Fig. 2). In our functional analysis of these three mutants, interactions between Glu-86 and the hydroxy methyl group on the aromatic ring and between Trp-94 and the sugar moiety of salicin were experimentally confirmed, with little interaction observed between Gln-177 and salicin (Figs. 3A and 4). These results suggest that the salicin-hTAS2R16 complex structure is that of model B rather than model A (Fig. 6). Our functional analysis was able to experimentally confirm the salicin-hTAS2R16 binding mode, which could not be definitively determined based on the molecular modeling study alone.
FIGURE 6.
Predicted salicin binding structure of hTAS2R16. A, shown is a side view of the predicted salicin-binding model of hTAS2R16. B, shown is a close-up of the top view of the predicted salicin-binding model of hTAS2R16. The indicated amino acid positions are predicted to influence receptor activation by salicin. Hydrogen bonds are indicated by dashed lines.
We further identified at least five additional amino acid residues involved in salicin recognition through functional analysis using a series of hTAS2R16 point mutants. In model B, Asn-89 forms a hydrogen bond with the sugar hydroxy group of salicin (Fig. 2B). Although all the Asn-89 mutants tested abolished the Ca2+ response to salicin and other β-d-glucopyranosides (Fig. 5B and supplemental Figs. 1 and 2A), from the modeling, Asn-89 may be a critical amino acid residue for ligand recognition though the hydrogen bonds with the sugar hydroxyl group of salicin (Fig. 6). However, it still remains unclear whether Asn-89 affects the ligand binding or ligand-evoked receptor activation.
H181T, a hydrophilic amino acid mutant, exhibited a salicin response with a small change in EC50 value, whereas H181L, having a loose hydrogen-bonding interaction with salicin, had a greatly decreased response to salicin (Fig. 5A, Table 2). These results suggest that His-181 interacts with salicin through a hydrogen bond, supporting the molecular model (Fig. 6). Other hydrophobic residues, Phe-93, Phe-240, and Ile-243, are located at the salicin binding site in the model (Fig. 2B). Functional analysis of these mutants designed to disrupt the putative binding site produced a greatly reduced response to salicin (Figs. 5, C–E, Table 2). Thus, these residues likely contribute to formation of the binding site (Fig. 6).
The functional analysis using arbutin and phenyl-β-d-glucopyranoside supported direct interactions between Trp-94 and His-181 and these β-d-glucopyranosides and indirect participation in ligand recognition of Phe-93, Phe-240, and Ile-243 (supplemental Figs. 1 and 2 and Tables 1 and 2). Thus, these residues likely form the same binding pocket for these β-d-glucopyranosides. These results suggest that hTAS2R16 has a strict ligand binding site for the sugar moiety.
In summary, in silico modeling of hTAS2R16 suggested a putative mode of interaction with phenyl-β-glucopyranosides and that the binding site of hTAS2R16 is formed by TM3, TM5, and TM6 and that at least Glu-86, Trp-94, and His-181 interact with salicin directly, whereas Phe-93, Phe-240, and Ile-243 are involved in forming the binding pocket (Fig. 6). Although full validation of the present molecular model may require solving the crystal structure of hTAS2R16, our experimental evidence supported this prediction.
Very recently, the generation of chimeric and mutant receptors followed by functional analysis identified amino acids residues critical for agonist selectivity and activation of hTAS2R46, hTAS2R43, and hTAS2R31 (39). From this experiment, systematic mutational analysis might provide a powerful tool for exploring the binding mode of hTAS2Rs. Molecular characterization of the interactions between hTAS2Rs and bitter ligands may be useful in clarifying how a large number of bitter compounds can be detected by such a limited number of bitter sensors, facilitating the design of bitter blockers.
Supplementary Material
This study was performed with a grant from the Research and Development Program for New Bio-industry Initiatives of Bio-oriented Technology Research Advancement Institution. This work was also supported by Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan 19880008 and Grants 20780089 (to Y. I.), 18688005, 20688015, and 21658046 (to T. M.), 19300248 (to T. A.), and 20380183 (to K. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Tables 1 and 2.
- TM
- transmembrane
- ssr3
- somatostatin receptor type 3.
REFERENCES
- 1.Chandrashekar J., Hoon M. A., Ryba N. J., Zuker C. S. (2006) Nature 444, 288–294 [DOI] [PubMed] [Google Scholar]
- 2.Li X., Staszewski L., Xu H., Durick K., Zoller M., Adler E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson G., Hoon M. A., Chandrashekar J., Zhang Y., Ryba N. J., Zuker C. S. (2001) Cell 106, 381–390 [DOI] [PubMed] [Google Scholar]
- 4.Nelson G., Chandrashekar J., Hoon M. A., Feng L., Zhao G., Ryba N. J., Zuker C. S. (2002) Nature 416, 199–202 [DOI] [PubMed] [Google Scholar]
- 5.Maehashi K., Matano M., Wang H., Vo L. A., Yamamoto Y., Huang L. (2008) Biochem. Biophys. Res. Commun. 365, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens M., Brockhoff A., Kuhn C., Bufe B., Winnig M., Meyerhof W. (2004) Biochem. Biophys. Res. Commun. 319, 479–485 [DOI] [PubMed] [Google Scholar]
- 7.Bufe B., Hofmann T., Krautwurst D., Raguse J. D., Meyerhof W. (2002) Nat. Genet. 32, 397–401 [DOI] [PubMed] [Google Scholar]
- 8.Kuhn C., Bufe B., Winnig M., Hofmann T., Frank O., Behrens M., Lewtschenko T., Slack J. P., Ward C. D., Meyerhof W. (2004) J. Neurosci. 24, 10260–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pronin A. N., Xu H., Tang H., Zhang L., Li Q., Li X. (2007) Curr. Biol. 17, 1403–1408 [DOI] [PubMed] [Google Scholar]
- 10.Brockhoff A., Behrens M., Massarotti A., Appendino G., Meyerhof W. (2007) J. Agric. Food Chem. 55, 6236–6243 [DOI] [PubMed] [Google Scholar]
- 11.Pronin A. N., Tang H., Connor J., Keung W. (2004) Chem. Senses 29, 583–593 [DOI] [PubMed] [Google Scholar]
- 12.Sainz E., Cavenagh M. M., Gutierrez J., Battey J. F., Northup J. K., Sullivan S. L. (2007) Biochem. J. 403, 537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bufe B., Breslin P. A., Kuhn C., Reed D. R., Tharp C. D., Slack J. P., Kim U. K., Drayna D., Meyerhof W. (2005) Curr. Biol. 15, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behrens M., Brockhoff A., Batram C., Kuhn C., Appendino G., Meyerhof W. (2009) J. Agric. Food Chem. 57, 9860–9866 [DOI] [PubMed] [Google Scholar]
- 15.Miguet L., Zhang Z., Grigorov M. G. (2006) J. Recept. Signal Transduct. Res. 26, 611–630 [DOI] [PubMed] [Google Scholar]
- 16.Floriano W. B., Hall S., Vaidehi N., Kim U., Drayna D., Goddard W. A., 3rd (2006) J. Mol. Model 12, 931–941 [DOI] [PubMed] [Google Scholar]
- 17.Jiang P., Ji Q., Liu Z., Snyder L. A., Benard L. M., Margolskee R. F., Max M. (2004) J. Biol. Chem. 279, 45068–45075 [DOI] [PubMed] [Google Scholar]
- 18.Xu H., Staszewski L., Tang H., Adler E., Zoller M., Li X. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14258–14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakajima K., Asakura T., Oike H., Morita Y., Shimizu-Ibuka A., Misaka T., Sorimachi H., Arai S., Abe K. (2006) Neuroreport 17, 1241–1244 [DOI] [PubMed] [Google Scholar]
- 20.Koizumi A., Nakajima K., Asakura T., Morita Y., Ito K., Shmizu-Ibuka A., Misaka T., Abe K. (2007) Biochem. Biophys. Res. Commun. 358, 585–589 [DOI] [PubMed] [Google Scholar]
- 21.Shirasuka Y., Nakajima K., Asakura T., Yamashita H., Yamamoto A., Hata S., Nagata S., Abo M., Sorimachi H., Abe K. (2004) Biosci. Biotechnol. Biochem. 68, 1403–1407 [DOI] [PubMed] [Google Scholar]
- 22.Okubo S., Asakura T., Okubo K., Abe K., Misaka T., Akita T., Abe K. (2008) J. Plant Physiol. 165, 1964–1969 [DOI] [PubMed] [Google Scholar]
- 23.Winnig M., Bufe B., Kratochwil N. A., Slack J. P., Meyerhof W. (2007) BMC Struct. Biol. 7, 66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F., Klebansky B., Fine R. M., Xu H., Pronin A., Liu H., Tachdjian C., Li X. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20930–20934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White S. H., Wimley W. C. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 319–365 [DOI] [PubMed] [Google Scholar]
- 26.Ishiguro M., Oyama Y., Hirano T. (2004) Chembiochem 5, 298–310 [DOI] [PubMed] [Google Scholar]
- 27.Ammon C., Schäfer J., Kreuzer O. J., Meyerhof W. (2002) Arch. Physiol. Biochem. 110, 137–145 [DOI] [PubMed] [Google Scholar]
- 28.Ueda T., Ugawa S., Yamamura H., Imaizumi Y., Shimada S. (2003) J. Neurosci. 23, 7376–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyerhof W., Batram C., Kuhn C., Brockhoff A., Chudoba E., Bufe B., Appendino G., Behrens M. (2010) Chem. Senses 35, 157–170 [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Avila L., Ming D., Margolskee R. F. (2000) Chem. Senses 25, 361–368 [DOI] [PubMed] [Google Scholar]
- 31.Noguchi M., Yamashita M., Arai S., Fujimaki M. (1975) J. Food Sci. 40, 367–369 [Google Scholar]
- 32.Sakurai T., Misaka T., Nagai T., Ishimaru Y., Matsuo S., Asakura T., Abe K. (2009) J. Agric Food Chem. 57, 2508–2514 [DOI] [PubMed] [Google Scholar]
- 33.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 34.Murakami M., Kouyama T. (2008) Nature 453, 363–367 [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen S. G., Choi H. J., Rosenbaum D. M., Kobilka T. S., Thian F. S., Edwards P. C., Burghammer M., Ratnala V. R., Sanishvili R., Fischetti R. F., Schertler G. F., Weis W. I., Kobilka B. K. (2007) Nature 450, 383–387 [DOI] [PubMed] [Google Scholar]
- 36.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimamura T., Hiraki K., Takahashi N., Hori T., Ago H., Masuda K., Takio K., Ishiguro M., Miyano M. (2008) J. Biol. Chem. 283, 17753–17756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brockhoff A., Behrens M., Niv M. Y., Meyerhof W. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11110–11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.