Abstract
Tissue factor pathway inhibitor (TFPI) inhibits tissue factor-induced coagulation, but may, via its C terminus, also modulate cell surface, heparin, and lipopolysaccharide interactions as well as participate in growth inhibition. Here we show that C-terminal TFPI peptide sequences are antimicrobial against the Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa, Gram-positive Bacillus subtilis and Staphylococcus aureus, as well as the fungi Candida albicans and Candida parapsilosis. Fluorescence studies of peptide-treated bacteria, paired with analysis of peptide effects on liposomes, showed that the peptides exerted membrane-breaking effects similar to those seen for the “classic” human antimicrobial peptide LL-37. The killing of E. coli, but not P. aeruginosa, by the C-terminal peptide GGLIKTKRKRKKQRVKIAYEEIFVKNM (GGL27), was enhanced in human plasma and largely abolished in heat-inactivated plasma, a phenomenon linked to generation of antimicrobial C3a and activation of the classic pathway of complement activation. Furthermore, GGL27 displayed anti-endotoxic effects in vitro and in vivo in a mouse model of LPS shock. Importantly, TFPI was found to be expressed in the basal layers of normal epidermis, and was markedly up-regulated in acute skin wounds as well as wound edges of chronic leg ulcers. Furthermore, C-terminal fragments of TFPI were associated with bacteria present in human chronic leg ulcers. These findings suggest a new role for TFPI in cutaneous defense against infections.
Keywords: Antimicrobial Peptides, Blood Coagulation Factors, Complement, Innate Immunity, Skin
Introduction
To control our microbial flora, humans and virtually all life forms are armored with rapidly acting host defense systems based on various antimicrobial peptides (1–3). The majority of these peptides is characterized by an amphipathic structure and comprise linear peptides, many of which adopt an α-helical and amphipathic conformation upon bacterial binding, peptides forming cysteine-linked antiparallel β-sheets, as well as cysteine-constrained loop structures. In addition, antimicrobial peptides may be found among peptides not displaying such ordered structures as long as these are characterized by an over-representation of certain amino acids (1, 4–6). Although interactions with bacterial membranes are fundamental for antimicrobial peptide function, the exact modes of action are complex and can be divided into membrane disruptive and non-membrane disruptive (1, 3, 7, 8). During recent years it has also become increasingly evident that many cationic and amphipathic antimicrobial peptides, such as defensins and cathelicidins, are multifunctional, also mediating immunomodulatory roles and angiogenesis (9–11), thus motivating the recent and broader definition host defense peptides for these members of the innate immune system.
Tissue factor pathway inhibitor (or TFPI)2 is a Kunitz-type proteinase inhibitor that reversibly inhibits the tissue factor-factor VII (TF-VII) complex in a factor X (FX)-dependent manner, leading to inhibition of both FX and FIX activation. TFPI consists of a highly negatively charged N terminus, three tandemly linked Kunitz-type domains, and a highly positively charged C terminus. In plasma, TFPI exists in both full-length and various C-terminal truncated forms (12). The first and second Kunitz domains are involved in binding and inhibition of the TF-VII complex and factor Xa, respectively (13). The third Kunitz domain, in turn, may interact with heparin via its cationic C-terminal end (14). This C-terminal region has also been implicated in interactions with plasma lipoproteins, thrombospondin-1, clearance receptors (15), and lipopolysaccharide (16) and may also inhibit cell growth (17) and blood coagulation (18, 19). Since various C-terminally truncated forms exist in vivo, a potential role of proteolysis of the C terminus has been implicated, and data indicate that TFPI can be cleaved by various proteinases such as thrombin (20), plasmin (21), and matrix metalloptoteinase-8 (22), thereby releasing C-terminal fragments. Up-regulators of TFPI expression include endotoxin, IL-1, TNF-α, platelet-derived growth factor, heparin, and basic fibroblast growth factor, all molecules involved in infection, inflammation, and growth (15).
The above reported multifunctionality of TFPI and presence of an exposed cationic and heparin-binding C terminus made us raise the question as to whether the C-terminal region of TFPI could exert antimicrobial activity. We here show that C-terminal TFPI peptides, found to be expressed in skin wounds, and detected in fibrin of chronic leg ulcers, may indeed function as host defense peptides. Furthermore, killing of the Gram-negative Escherichia coli, but not Pseudomonas aeruginosa, was markedly boosted by peptide-mediated complement activation, including formation of the membrane attack complex (MAC) and antimicrobial C3a.
EXPERIMENTAL PROCEDURES
Peptides
The TFPI-derived peptides (see Fig. 1) and the control peptides GGL27(S) (GGLISTSSSSSSQRVKIAYEEIFVKNM) and DSE25 (DSEEDEEHTIITDTELPPLKLMHSF) were synthesized by Biopeptide Co., San Diego, CA, whereas LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES), was obtained from Innovagen AB, Lund, Sweden. The purity (>95%) of these peptides was confirmed by mass spectral analysis (MALDI-ToF Voyager).
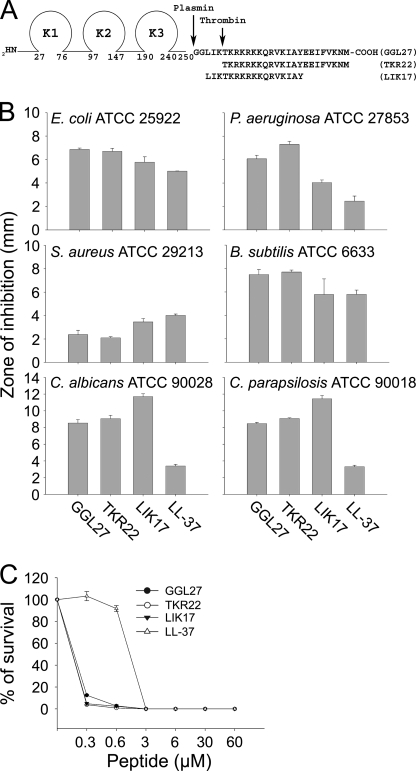
FIGURE 1.
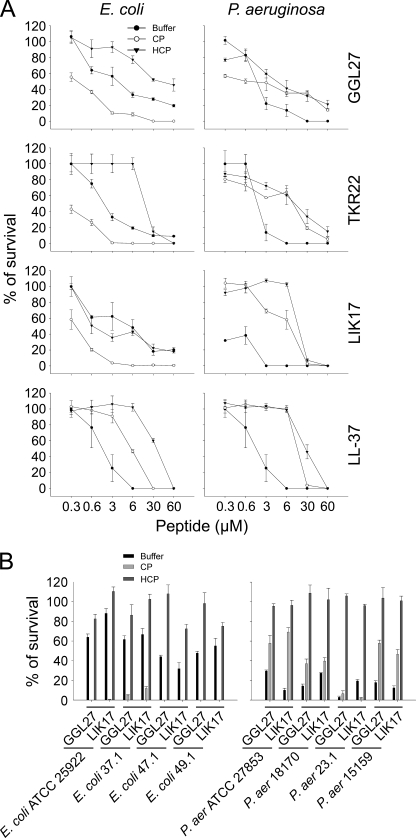
Antimicrobial activities of TFPI-derived peptides. A, schematic illustrating the structure of TFPI. Enzymatic cleavage sites are indicated. B, antimicrobial activity (using RDA) of selected peptides against the indicated microbes. For determination of antimicrobial activities, E. coli ATCC 25922, P. aeruginosa ATCC 27853, S. aureus ATCC 29213, or B. subtilis ATCC 6633 isolates (4 × 106 cfu) or C. albicans ATCC 90028 and C. parapsilosis ATCC 90018 (1 × 105 cfu) were inoculated in 0.1% TSB agarose gel. Each 4-mm-diameter well was loaded with 6 μl of peptide (at 100 μm). The zones of clearance correspond to the inhibitory effect of each peptide after incubation at 37 °C for 18–24 h (mean values are presented, n = 3). C, antibacterial effects of the TFPI-derived peptides and LL-37 against E. coli ATCC 25922 in viable count assays. 2 × 106 cfu/ml bacteria were incubated in 50 μl with peptides at the indicated concentrations in 10 mm Tris, pH 7.4, buffer, and the cfu were determined.
Microorganisms
Bacterial isolates E. coli ATCC 25922, P. aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, Bacillus subtilis ATCC 6633, Candida albicans ATCC 90028, and Candida parapsilosis ATCC 90018 were obtained from the Department of Bacteriology, Lund University Hospital. Other clinical isolates of E. coli and P. aeruginosa were from patients with skin infections.
Biological Materials
Fibrin slough was collected from chronic venous leg ulcers (chronic wound slough/surface) with a sterile spatula and immediately fixed for electron microscopy. Tissue sections from three patients with chronic venous ulcers (duration > 6 months) were analyzed. 4-mm biopsies were taken from the edge of the wound and a control area on the thigh. For acute wounds, a 4-mm biopsy was taken from the thigh of one individual, and subsequent biopsies from the wound edges were taken at days 5 and 8. Biopsies from normal skin (n = 3, thigh) were also taken for analysis. The research project was approved by the Ethics committee, Lund University Hospital. Written consent was obtained from the patients.
Radial Diffusion Assay
Essentially as described earlier (23, 24), bacteria were grown to mid-logarithmic phase in 10 ml of full-strength (3% w/v) Trypticase soy broth (TSB) (Becton-Dickinson). The microorganisms were then washed once with 10 mm Tris, pH 7.4. Subsequently, 4 × 106 cfu were added to 15 ml of the underlay agarose gel, consisting of 0.03% (w/v) TSB, 1% (w/v) low electroendosmosis type agarose (Sigma-Aldrich) and 0.02% (v/v) Tween 20 (Sigma-Aldrich). The underlay was poured into a 144-mm diameter Petri dish. After agarose solidification, wells of 4-mm diameter were punched and 6 μl of peptide solution of required concentration was added to each well. Plates were incubated at 37 °C for 3 h to allow peptide diffusion. The underlay gel was then covered with 15 ml of molten overlay (6% TSB and 1% low electroendosmosis type agarose in distilled H2O). Antimicrobial activity of a peptide was visualized as a clearance zone around each well after 18–24 h of incubation at 37 °C.
Viable Count Analysis
E. coli strains were grown to mid-logarithmic phase in Todd-Hewitt agar. P. aeruginosa strains were grown in Todd-Hewitt agar overnight. Bacteria were washed and diluted in 10 mm Tris, pH 7.4, containing 5 mm glucose. 2 × 106 cfu/ml bacteria were incubated in 50 μl, at 37 °C for 2 h with the C-terminal TFPI-derived peptides GGL27, TKR22, or LIK17 at the indicated concentrations. Additional experiments were performed in 10 mm Tris, pH 7.4, containing 0.15 m NaCl, either alone or with 20% normal or heat inactivated citrate-plasma. Serial dilutions of the incubation mixture were plated on Todd-Hewitt agar, followed by incubation at 37 °C overnight and cfu determination.
Flow Cytometry Analysis
50 μl of bacteria (1–2 × 109 cfu) were incubated with 450 μl of human plasma either alone or supplemented with GGL27 (at 3 μm). Samples were incubated for 30 min or 1 h at 37 °C, divided into two equal parts, centrifuged, washed with PBS, and resuspended in 100 μl of PBS with rabbit polyclonal antibodies against either LGE27 (25), a C-terminal epitope of human C3a, or rabbit polyclonal antibodies against C1q (both at 1:100). The mixtures were subsequently incubated for 1 h at room temperature. Bacteria were pelleted and washed twice with PBS, incubated in 100 μl of PBS with goat anti-rabbit IgG FITC-labeled antibodies (1:500, Sigma) for 30 min at room temperature, and washed twice with PBS. In another experiment bacteria were incubated for 1 h in citrate plasma together with 3 μm 5-carboxytetramethylrhodamine- labeled GGL27 peptide, then pelleted and washed twice with PBS. Flow cytometry analysis (BD Biosciences, Franklin Lakes, NJ) was performed using a FACSCalibur flow cytometry system equipped with a 15-milliwatt argon laser set at 488 nm. The bacterial population was selected by gating with appropriate settings of forward scatter and sideward scatter.
Fluorescence Microscopy
Fluorescein isothiocyanate (FITC, Sigma-Aldrich) was used for monitoring of bacterial membrane permeabilization. E. coli ATCC 25922 bacteria were grown to mid-logarithmic phase in TSB medium. Bacteria were washed and resuspended in buffer (10 mm Tris, pH 7.4, 0.15 m NaCl, 5 mm glucose) to yield a suspension of 1 × 107 cfu/ml. 100 μl of the bacterial suspension was incubated with 30 μm of the respective peptides at 30 °C for 30 min. Microorganisms were then immobilized on poly (l-lysine)-coated glass slides by incubation for 45 min at 30 °C, followed by addition onto the slides of 200 μl of FITC (6 μg/ml) in buffer and a final incubation for 30 min at 30 °C. The slides were washed, and bacteria were fixed by incubation, first on ice for 15 min, then in room temperature for 45 min in 4% paraformaldehyde. The glass slides were subsequently mounted on slides using Prolong Gold Antifade reagent mounting medium (Invitrogen). Bacteria were visualized using an Eclipse TE300 (Nikon) inverted fluorescence microscope equipped with a Hamamatsu C4742-95 cooled charge-coupled device camera (Bridgewater, CT) and a Plan Apochromat ×100 objective (Olympus, Orangeburg, NY). Differential interference contrast (Nomarski) imaging was used for visualization of the microbes themselves.
Histochemistry
For immunostaining, wound biopsies were fixed in 10% formalin, rehydrated, and embedded in paraffin. Sections of 5-μm thickness were placed on polylysine-coated glass slides, deparaffinized in xylene, and rehydrated in graded alcohols. The slides were then treated with Dako antigen retrieval solution (Dako) for 40 min at 97 °C and incubated for 24 h at room temperature in a 1:50 dilution of polyclonal antibodies against TFPI (Sigma-Aldrich, in TBS with 1% BSA, 5% goat serum, 0.05% Tween 20). After three 20-min washes in TBS with 0.05% Tween 20, the sections were incubated with alkaline phosphatase-conjugated secondary goat anti-rabbit IgG (Dako) diluted 1:1000 in the same buffer as the primary antibody and incubated for another 24 h, followed by three 20-min washes. Sections were developed with Vulcan Fast Red chromogen (Biocare Medical, Concord, CA), and the slides were counterstained with Harris Hematoxylin (EM Science, Gibbstown, NJ). For histological evaluation of lungs derived from the in vivo LPS models in mice, tissues were embedded as above, sectioned, and stained with hematoxylin and eosin by routine procedures (Histocenter, Gothenburg, Sweden).
Electron Microscopy
For transmission electron microscopy and visualization of peptide effects on bacteria, P. aeruginosa ATCC 27853 (1–2 × 106 cfu/sample) was incubated for 2 h at 37 °C with the peptide GGL27 at 30 μm. LL-37 (30 μm) was included as a control. P. aeruginosa sample suspensions were adsorbed onto carbon-coated copper grids for 2 min, washed briefly on two drops of water, and negatively stained on two drops of 0.75% uranyl formate. The grids were rendered hydrophilic by glow discharge at low pressure in air. In vivo experiment; fibrin slough from patients with chronic venous ulcers was fixed (1.5% paraformaldehyde, 0.5% glutaraldehyde, in 0.1 m phosphate buffer, pH 7.4) for 1 h at room temperature, followed by washing with 0.1 m phosphate buffer, pH 7.4. The fixed and washed samples were subsequently dehydrated in ethanol and further processed for Lowicryl embedding (26). Sections were cut with an LKB ultratome and mounted on gold grids. For immunostaining, the grids were floated on top of drops of immune reagents displayed on a sheet of parafilm. Free aldehyde groups were blocked with 50 mm glycine, and the grids were then incubated with 5% (v/v) goat serum in incubation buffer (0.2% acetylated bovine serum albumin (BSA-c) in PBS, pH 7.6) for 15 min. This blocking procedure was followed by overnight incubation at 4 °C with C-terminal TFPI goat polyclonal antibodies (1 μg/ml, Abcam, UK) alone, or in combination with rabbit polyclonal antibodies against the C-terminal part of C3a (LGE27 antibodies, 1 μg/ml, Innovagen AB) (25). Controls without primary antibodies were included. The grids were washed in a large volume (200 ml) of incubation buffer, floating on drops containing the gold conjugate reagents. For detection of TFPI-peptides, 1 μg/ml EM rabbit anti-goat IgG 10 nm Au (BBI) in incubation buffer was added, and the incubation was performed for 2 h at 4 °C. For simultaneous detection of TFPI and C3a, 1 μg/ml EM rabbit anti-goat IgG 20 nm Au (BBI) and 1 μg/ml EM goat anti-rabbit IgG 10 nm Au (BBI) were used. After further washes by an excess volume of incubation buffer, the sections were postfixed in 2% glutaraldehyde. Finally, sections were washed with distilled water and post-stained with 2% uranyl acetate and lead citrate. All samples were examined with a JEOL JEM 1230 electron microscope operated at 80 kV accelerating voltage. Images were recorded with a Gatan Multiscan 791 charge-coupled device camera.
SDS-PAGE and Immunoblotting
Human citrate plasma (450 μl) was incubated with 50 μl of bacteria (1–2 × 109 cfu) either alone or supplemented with the peptide GGL27 at 3 μm. The mixture was incubated for 30 min or 1 h at 37 °C and centrifuged, and supernatants and bacteria were collected. The bacterial pellet was washed with PBS, and bound proteins were eluted with 0.1 m glycine-HCl, pH 2.0. The pH of the eluted material was raised to 7.5 with 1 m Tris. Eluted proteins were precipitated by addition of 1 volume of TCA to 4 volumes of sample, followed by incubation for 30 min on ice and centrifugation at 15,000 × g (4 °C for 20 min). Precipitated material and supernatants were dissolved in SDS sample buffer and analyzed under reducing conditions by SDS-PAGE on 16.5% Tris-Tricine gels (Clear PAGETM, C.B.S. Scientific). Proteins and peptides were transferred to nitrocellulose membranes (Hybond-C). Membranes were blocked by 3% (w/v) skimmed milk, washed, and incubated for 1 h with rabbit polyclonal antibodies against the C-terminal part of C3a (LGE27 antibodies, 1:1000), rabbit polyclonal antibodies to C1q (1:1000, Dako), goat polyclonal antibodies recognizing the C-terminal TFPI sequence VKIAYEEIFVKNM (Abcam), or rabbit polyclonal antibodies to C5b-9 (1:1000, Abcam, England). The membranes were washed three times for 10 min and incubated (1 h) with HRP-conjugated secondary antibodies (1:2000, Dako), and washed (3 × 10 min). Proteins were visualized by an enhanced chemiluminescent substrate (LumiGLO®) developing system (Upstate Cell Signaling Solutions).
LPS Effects on Macrophages in Vitro
3.5 × 105 cells were seeded in 96-well tissue culture plates (Nunc, 167008) in phenol red-free DMEM (Invitrogen) supplemented with 10% FBS containing 1% Anti-Anti (Invitrogen). Following 20 h of incubation to permit adherence, cells were washed and stimulated with 10 ng/ml E. coli (0111:B4) or P. aeruginosa LPS (Sigma), with and without the peptides GGL27, GGL27(S), and DSE25 of various doses. The levels of NO in culture supernatants were determined after 24 h from stimulation using the Griess reaction (27). Briefly, nitrite, a stable product of NO degradation, was measured by mixing 50 μl of culture supernatants with the same volume of Griess reagent (Sigma, G4410) and reading absorbance at 550 nm after 15 min. Phenol-red free DMEM with FBS and antibiotics were used as a blank. A standard curve was prepared using 0–80 μm sodium nitrite solutions in ddH2O.
Animal Infection Model
Animals were housed under standard conditions of light and temperature and had free access to standard laboratory chow and water. P. aeruginosa 15159 bacteria were grown to logarithmic phase (A620 ∼ 0.5), harvested, washed in PBS, diluted in the same buffer to 2 × 108 cfu/ml, and kept on ice until injection. 100 μl of the bacterial suspension was injected intraperitoneally into female Balb/c mice. Sixty minutes after the bacterial injection, 0.5 mg of GGL27 or buffer alone was injected subcutaneously into the mice. In a corresponding E. coli infection model, bacteria were grown to early logarithmic phase (A620 ∼ 0.4), harvested, washed in PBS, diluted in the same buffer to 1 × 108 cfu/ml, and kept on ice until injection. 100 μl of the bacterial suspension was injected intraperitoneally into female Balb/c mice. Thirty minutes after the bacterial injection, 0.2 mg of GGL27 peptide or buffer alone was injected intraperitoneally. Data from two independent experiments were pooled.
LPS Model in Vivo
Male C57BL/6 mice (8–10 weeks, 22 ± 5 g) were injected intraperitoneally with 18 mg of E. coli 0111:B4 LPS (Sigma) per kg of body weight. Thirty minutes after LPS injection, 0.5 mg of GGL27, GGL27(S), DSE25, or buffer alone was injected intraperitoneally into the mice. Survival and status were followed over 7 days. For blood collection and histochemistry, mice were sacrificed 20 h after LPS challenge, and lungs were removed and fixed. These experiments were approved by the Laboratory Animal Ethics Committee of Malmö/Lund.
Cytokine Assay
The cytokines IL-6, IL-10, MCP-1, INF-γ, and TNF-α were measured in plasma from mice subjected to LPS (with or without peptide treatment) using the cytometric bead array and mouse inflammation kit (BD Biosciences) according to the manufacturer's instructions.
Hemolysis Assay
EDTA-blood was centrifuged at 800 × g for 10 min, thereafter plasma and buffy coat were removed. The erythrocytes were washed three times and resuspended in PBS, pH 7.4, to get a 5% suspension. The cells were then incubated with end-over-end rotation for 60 min at 37 °C in the presence of peptides (60 μm). 2% Triton X-100 (Sigma-Aldrich) served as positive control. The samples were then centrifuged at 800 × g for 10 min, and the supernatant was transferred to a 96-well microtiter plate. The absorbance of hemoglobin release was measured at 540 nm and expressed as % of Triton X-100-induced hemolysis.
LDH Assay
HaCaT keratinocytes were grown to confluency in 96-well plates (3000 cells/well) in serum-free keratinocyte medium supplemented with bovine pituitary extract and recombinant EGF (Invitrogen). The medium was then removed, and 100 μl of the peptides was investigated (at 60 μm, diluted in serum-free keratinocyte medium/bovine pituitary extract-recombinant EGF or in keratinocyte-serum-free keratinocyte medium supplemented with 20% human serum) were added. The lactate dehydrogenase (LDH)-based TOX-7 kit (Sigma-Aldrich) was used for quantification of LDH release from the cells. Results represent mean values from triplicate measurements and are given as fractional LDH release compared with the positive control consisting of 1% Triton X-100 (yielding 100% LDH release).
Liposome Preparation and Leakage Assay
The liposomes investigated were anionic (1,2-dioleoyl-sn-glycero-3-phoshoetanolamine/1,2-dioleoyl-sn-glycero-3-phosphoglycerol, monosodium salt; 75/25 mol/mol). 1,2-Dioleoyl-sn-glycero-3-phosphoglycerol, monosodium salt and 1,2-dioleoyl-sn-glycero-3-phoshoetanolamine were both from Avanti Polar Lipids (Alabaster, AL) and of >99% purity. Due to the long, symmetric, and unsaturated acyl chains of these phospholipids, several methodological advantages are reached. In particular, membrane cohesion is good, which facilitates very stable, unilamellar, and largely defect-free liposomes (observed from cryo-TEM), allowing detailed studies on liposome leakage. The lipid mixtures were dissolved in chloroform, after which solvent was removed by evaporation under vacuum overnight. Subsequently, 10 mm Tris buffer, pH 7.4, was added together with 0.1 m carboxyfluorescein (CF, Sigma). After hydration, the lipid mixture was subjected to eight freeze-thaw cycles consisting of freezing in liquid nitrogen and heating to 60 °C. Unilamellar liposomes of ∼140 nm diameter were generated by multiple extrusions through polycarbonate filters (pore size, 100 nm) mounted in a LipoFast miniextruder (Avestin, Ottawa, Canada) at 22 °C. Untrapped CF was removed by two subsequent gel filtrations (Sephadex G-50, Amersham Biosciences) at 22 °C, with Tris buffer as eluant. CF release from the liposomes was determined by monitoring the emitted fluorescence at 520 nm from a liposome dispersion (10 μm lipid in 10 mm Tris, pH 7.4). An absolute leakage scale was obtained by disrupting the liposomes at the end of each experiment through addition of 0.8 mm Triton X-100 (Sigma-Aldrich). A SPEX-fluorolog 1650 0.22-m double spectrometer (SPEX Industries) was used for the liposome leakage assay. Measurements were performed in triplicate at 37 °C.
CD Spectroscopy
CD spectra of the peptides were measured on a Jasco J-810 Spectropolarimeter (Jasco, UK). The measurements were performed at 37 °C in a 10-mm quartz cuvette under stirring, and the peptide concentration was 10 μm. The effect on peptide secondary structure of liposomes at a lipid concentration of 100 μm was monitored in the range of 200–250 nm. The fraction of the peptide in α-helical conformation, Xα, was calculated from Equation 1,
where A is the recorded CD signal at 225 nm, and Aα and Ac are the CD signal at 225 nm for a reference peptide in 100% α-helix and 100% random coil conformation, respectively. 100% α-helix and 100% random coil references were obtained from 0.133 mm (monomer concentration) poly-l-lysine in 0.1 m NaOH and 0.1 m HCl, respectively (28, 29). For determination of effects of lipopolysaccharide on peptide structure, the peptide secondary structure was monitored at a peptide concentration of 10 μm, both in Tris buffer and in the presence of E. coli lipopolysaccharide (0.02 wt%) (E. coli 0111:B4, highly purified, less than 1% protein/RNA, Sigma). To account for instrumental differences between measurements the background value (detected at 250 nm, where no peptide signal is present) was subtracted. Signals from the bulk solution were also corrected for.
Phylogenetic Analyses of TFPI
The TFPI amino acid sequence was retrieved from the NCBI site. Each sequence was analyzed with Psi-Blast (NCBI) to find the ortholog and paralog sequences. Sequences that showed structural homology >70% were selected. These sequences were aligned using ClustalW using Blosum 69 protein weight matrix settings. Internal adjustments were made by taking the structural alignment into account utilizing the ClustalW interface. The level of consistency of each position within the alignment was estimated by using the alignment-evaluating software Tcoffee.
Statistical Analysis
Bar diagrams (radial diffusion assay (RDA) and visible count assay) are presented as mean and standard deviation, from at least three independent experiments.
Supplemental Data
RDA data on effects of GGL27 and control peptides, permeabilization studies, and kinetic data are available as supplemental data.
RESULTS
To elucidate whether C-terminal peptides of TFPI possess antimicrobial activity, we investigated the effects of defined regions of TFPI previously reported to be generated by proteolytic action (plasmin and thrombin), as well as the peptide LIKTKRKRKKQRVKIAY (LIK17) comprising the C-terminal heparin-binding epitope of TFPI (see Fig. 1A for sequences). The results showed that the peptides were indeed antimicrobial in RDAs against Gram-negative E. coli and P. aeruginosa, Gram-positive B. subtilis and S. aureus, as well as the fungi C. albicans and C. parapsilosis (Fig. 1B). It is of note that the peptides displayed activities comparable to that of the “classic” AMP human cathelicidin LL-37 (Fig. 1B). In contrast, the control peptides GGL27(S), having the central K/R residues replaced by S, and the peptide DSE25, from the N terminus of TFPI (see “Experimental Procedures” for sequences), yielded no antimicrobial effects against the above microbes (supplemental Fig. 1). The antibacterial results above were further substantiated by matrix-free viable count assays. The results from these dose-response experiments utilizing E. coli confirmed that particularly GGL27 and LIK17 displayed significant antibacterial activity (Fig. 1C).
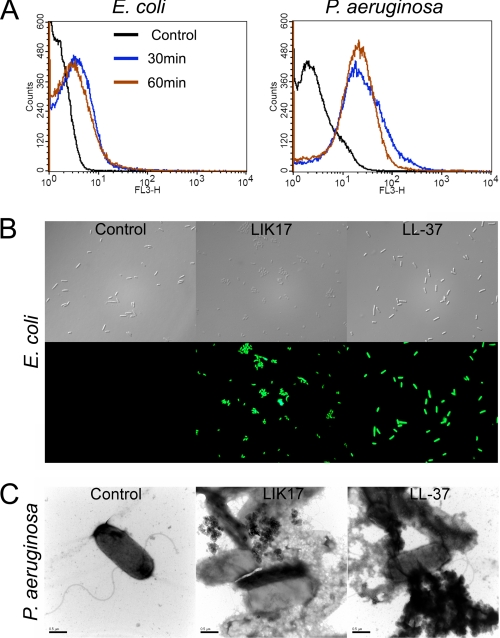
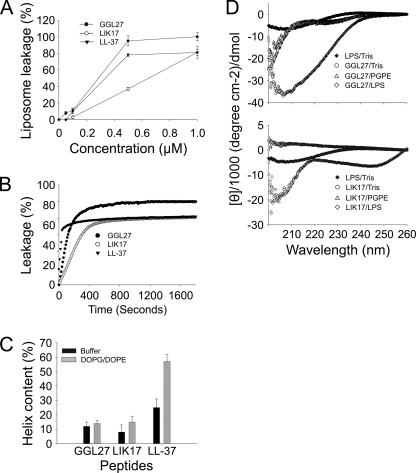
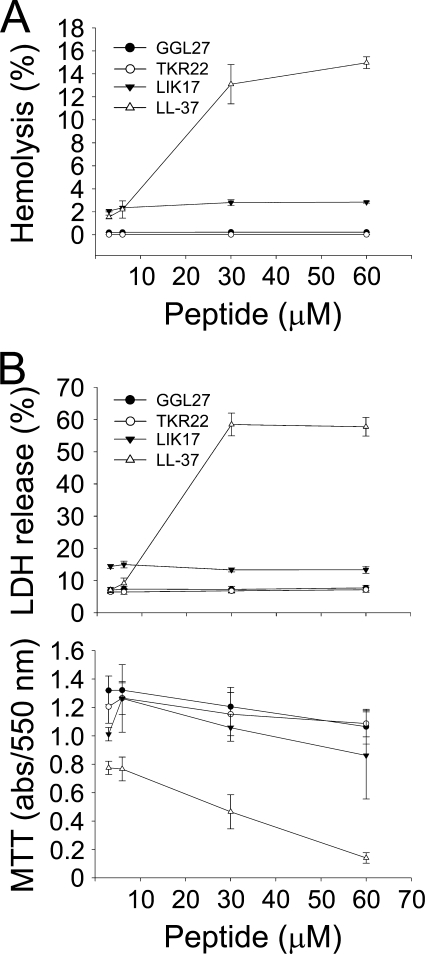
FACS analyses showed that plasmin-generated GGL27 avidly bound to E. coli and P. aeruginosa in human plasma (Fig. 2A). Studies employing the impermeant probe FITC showed that LIK17 (Fig. 2B) and GGL27 (supplemental Fig. 2) permeabilized bacterial membranes of E. coli similarly to those seen after treatment with LL-37 (30, 31) (Fig. 2B). Electron microscopy utilizing P. aeruginosa demonstrated extensive membrane damage, with cell envelopes of P. aeruginosa devoid of their cytoplasmic contents, and intracellular material found extracellularly (Fig. 2C). Again, similar findings were obtained with LL-37. These data suggest that the TFPI-derived peptides act on bacterial membranes; however, they do not demonstrate the exact mechanistic events following peptide addition to bacteria, because secondary metabolic effects on bacteria also may trigger bacterial death and membrane destabilization. Therefore, a liposome model was employed to study membrane permeabilization of LIK17 and GGL27. The peptides caused CF release (Fig. 3A), thus indicating a direct effect on lipid membranes. Kinetic analysis showed that ∼80% of the maximal release occurred within 5–10 min, comparable to results obtained with LL-37 (Fig. 3B). It is noteworthy that LIK17 and GGL27 displayed no significant conformational changes associated with binding to liposomes (Fig. 3C) and only relatively minor ones together with E. coli LPS (Fig. 3D), the latter originating from peptide and/or LPS, contrasting to LL-37, which showed a significant increase in helicity on liposome binding. AMPs that kill bacteria may also exhibit hemolytic and membrane-permeabilizing activities against eukaryotic cells. The results showed, however, that there was no hemolytic activity of the TFPI-derived peptides at doses of 3–60 μm (Fig. 4A). This contrasted to LL-37, which readily permeabilized erythrocytes at doses >6 μm. Likewise, the TFPI-derived peptides did not permeabilize HaCaT cells, nor did they display any significant toxicity at the concentrations studied (Fig. 4B).
FIGURE 2.
Effects on bacterial membranes. A, binding of TFPI to bacterial surfaces. The indicated bacteria (1–2 × 109 cfu/ml) were incubated with 3 μm 5-carboxytetramethylrhodamine-labeled GGL27 peptide in 500 μl of human plasma, and samples were analyzed by FACS. B, permeabilizing effects of peptides on E. coli. Bacteria were incubated with the indicated peptides, and permeabilization was assessed using the impermeant probe FITC. C, electron microscopy analysis. P. aeruginosa bacteria were incubated for 2 h at 37 °C with 30 μm of LIK17 and LL-37 and visualized by negative staining. Scale bar, 1 μm. Control, buffer control.
FIGURE 3.
Structure and effects on liposomes. A, effects of the indicated peptides on liposome leakage. The membrane-permeabilizing effect was recorded by measuring fluorescence release of carboxyfluorescein from PA (negatively charged) liposomes. The experiments were performed in 10 mm Tris-buffer. Values represents mean of triplicate samples. B, kinetics of CF release from liposomes. 1 μm peptides was used. C, helical content of the TFPI-derived C-terminal LIK16 and GGL27 peptides in the presence of negatively charged liposomes (PA). LIK17and GGL27 structures were largely unaffected by the addition of liposomes. D, CD spectra of LIK17 and GGL27 in Tris buffer and in the presence of LPS. For control, CD spectra for buffer and LPS alone are also presented.
FIGURE 4.
Activities on eukaryotic cells. A, hemolytic effects of the indicated peptides. The cells were incubated with different concentrations of the peptides, 2% Triton X-100 (Sigma-Aldrich) served as positive control. The absorbance of hemoglobin release was measured at λ 540 nm and is expressed as % of Triton X-100-induced hemolysis (note the scale of the y axis). Effects of LL-37 are shown for comparison. In B, upper panel, HaCaT keratinocytes were subjected to the indicated TFPI-peptides as well as LL-37. Cell-permeabilizing effects were measured by the LDH-based TOX-7 kit. LDH release from the cells was monitored at λ 490 nm and was plotted as % of total LDH release. Lower panel: the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used to measure viability of HaCaT keratinocytes in the presence of the indicated peptides (at 60 μm). In the assay, MTT is modified into a dye, blue formazan, by enzymes associated with metabolic activity. The absorbance of the dye was measured at λ 550 nm.
Because the activity of many antimicrobial peptides is abrogated by the presence of physiological salt as well as presence of “biomatrices” such as plasma or serum (32, 33), we also investigated the influence of salt and human plasma on peptide activity. As demonstrated in Fig. 5A, the bactericidal activity of all three TFPI-derived peptides (GGL27, TKR22, and LIK17), particularly against E. coli, was not only retained in the presence of human plasma, but significantly enhanced. It should here be noted that killing of E. coli by C-terminal TFPI peptides was recently shown to be mediated via the classic pathway and linked to formation of the MAC (34). Consequently, heat inactivation of plasma abolished the potentiation, findings compatible with previous results showing that the presence of an intact complement system promotes the killing of this microbe. Additionally, it should be noted that the TFPI-derived peptides also mediated killing in heat-inactivated plasma, although at higher concentrations (Fig. 5A), compatible with a direct peptide-mediated antimicrobial effect, as demonstrated in Figs. 2 and 3. Interestingly, in contrast to the findings with E. coli, killing of P. aeruginosa was not enhanced in human plasma when compared with physiological buffer. Furthermore, the potentiating effect of native plasma, when compared with heat-inactivated plasma, was less significant (notably for GGL27 and LIK17). The above findings were generalized using a panel of E. coli and P. aeruginosa isolates (Fig. 5B). In addition, kinetic studies demonstrated that the bacterial killing by the peptides occurred within 5–20 min indicating a fast direct action compatible with many antimicrobial peptides (supplemental Fig. 3).
FIGURE 5.
Activities of C-terminal TFPI-derived peptides. A, antibacterial effects of the TFPI-derived peptides and LL-37 against E. coli or P. aeruginosa in viable count assays. 2 × 106 cfu/ml bacteria were incubated in 50 μl with peptides at the indicated concentrations in 10 mm Tris, 0.15 m NaCl, pH 7.4 (buffer), or in 0.15 m NaCl, 10 mm Tris, pH 7.4, containing 20% human citrate plasma (CP), or in the same buffer but with heat-inactivated human citrate plasma (HCP) (n = 3, S.D. is indicated). B, the indicated bacterial isolates were subjected to GGL27 at 3 μm in buffer, native plasma (CP), or heat-inactivated plasma (HCP) for 2 h, and the number of cfu was determined.
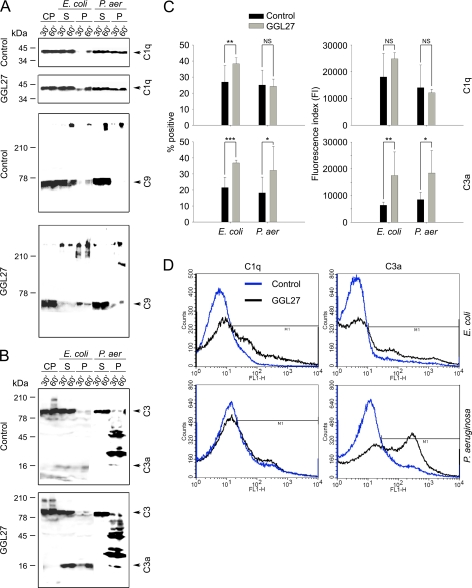
Western blot experiments (Fig. 6, A and B) and FACS analyses (Fig. 6, C and D), showed that GGL27 enhanced binding of C1q to E. coli resulting in increased formation of MAC (Fig. 6, A, C, and D). It also induced a significant generation of C3a (Fig. 6, B–D), an anaphylatoxin previously shown to exert antimicrobial effects under physiological conditions against various bacteria, mediated by bacterial binding and membrane lysis (25). Taken together, these observations provide a novel mechanism for bacterial killing, based on initial bacterial binding of GGL27 (Fig. 2A), followed by boosting of formation and bacterial binding of antimicrobial C3a (Fig. 6, B–D), which therefore should further add to the total antimicrobial effect induced by GGL27 (Fig. 5B). In contrast to these results, no significant C1q/MAC alterations were induced after subjecting P. aeruginosa to GGL27 (Fig. 6, A, C, and D). Considering C3a in relation to P. aeruginosa, the Western blots of bacteria-bound peptides identified minor C3a bands, as well as peptides of higher molecular weight, containing the C-terminal epitope of C3a. FACS analysis showed increased generation and binding of these C3a-containing fragments to the bacterial surface (Fig. 6, C and D).
FIGURE 6.
The C-terminal TFPI peptide GGL27 enhances C1q, C3a, and MAC binding to E. coli. A, E. coli ATCC 25922 and P. aeruginosa 15159 bacteria were washed, resuspended, and incubated with citrate plasma either alone or supplemented with GGL27 (at 3 μm) for 30 min or 1 h at 37 °C. The bacterial cells were collected and washed with PBS, and bound proteins and corresponding supernatants were subjected to Tris-Tricine SDS-PAGE under reducing conditions, followed by immunoblotting with antibodies recognizing C1q or C5b-9. CP, citrate plasma; S, supernatant or unbound bacteria; P, pellet or material bound to bacterial cells. B, as in A, but antibodies against the C3a were used (25). In C: left panel, comparison of the mean proportion of bacteria positive for C1q/C3a binding in citrate plasma (control, black columns) and in plasma supplemented with GGL27 (gray columns). Right panel, comparative degree of C1q and C3a binding to E. coli and P. aeruginosa strains, expressed as means of the fluorescence index (FI; proportion of bacteria positive for C1q/C3a multiplied by the mean intensity of C1q/C3a binding). (*, p < 0.05; **, p < 0.01; and ***, p < 0.001, t test). D, examples of flow cytometry histograms of C1q/C3a binding to E. coli and P. aeruginosa in citrate plasma and in plasma supplemented with GGL27.
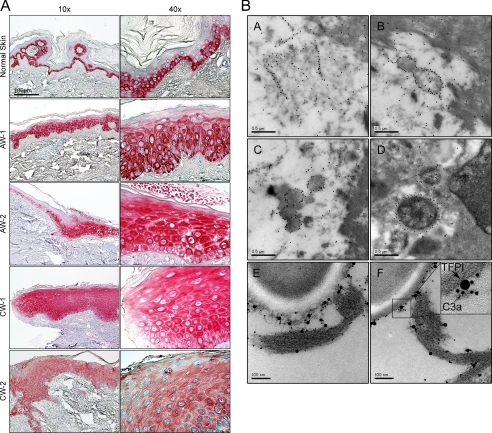
TFPI is mostly considered a plasma protein, but is also expressed by endothelial cells, monocytes, and macrophages (12). Immunohistochemistry analyses showed that TFPI was expressed in normal human skin, particularly in the basal epidermal layers (Fig. 7A). Furthermore, the molecule showed a ubiquitous expression at the wound edges of both acute wounds and chronic leg ulcers. Chronic leg ulcers are characterized by an excessive chronic inflammatory state, high levels of proteinases (such as plasmin), and frequent bacterial colonization with P. aeruginosa (35). Hence, we analyzed fibrin slough from such an ulcer, infected with P. aeruginosa. As demonstrated by electron microscopy and immunogold-labeled antibodies against the C-terminal part of TFPI, these peptide epitopes were found in association with bacterial membranes, as well as fibrin fibers. Importantly, using double staining, C3a was found to be particularly associated with these TFPI-peptides (Fig. 7B). Hence, these in vivo data are compatible with previous results (Fig. 6) on microbial binding of GGL27 and its relation to enhanced generation of antimicrobial C3a in plasma in vitro.
FIGURE 7.
Identification of TFPI in human skin and wounds. A, immunohistochemical identification of TFPI in normal skin, acute wound (AW), and in chronic venous leg ulcer tissue (chronic wounds, CW). Skin biopsies were taken from normal skin (n = 3), from acute wounds (AW-1 and AW-2, 5 and 8 days after wounding, respectively), and from the wound edges of patients with chronic venous ulcers (CW, n = 3). Representative sections are shown. In normal skin TFPI is detected mainly in the basal layers of epidermis. TFPI in AW and CW are ubiquitously found in all epidermal layers. Scale bar, 100 μm. B, TFPI-derived peptides are found in human wounds. Visualization of binding of C-terminal TFPI peptides to bacteria found in fibrin slough from a P. aeruginosa-infected chronic wound surface. The peptides bind to fibrin (A), bacteria (B and C), and bacteria inside a macrophage (D). In E and F, TFPI and C3a peptides were visualized by immunogold using gold-labeled antibodies of different sizes, specific for C3a (10 nm) and C termini of TFPI (20 nm), respectively. Evaluation of 50 bacterial profiles showed that ∼70% of TFPI-molecules were associated with C3a. See inset for exemplification.
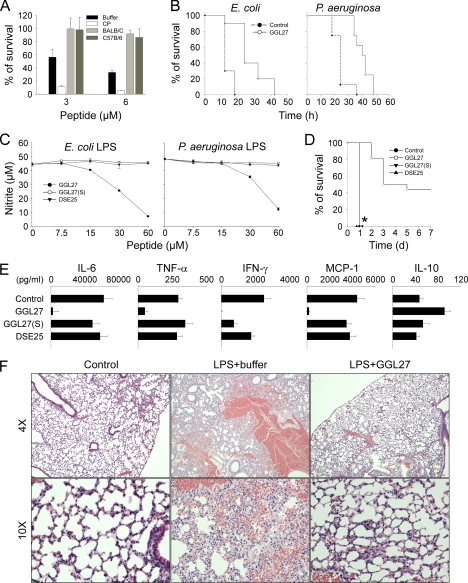
It is of note that the human TFPI peptides did not enhance bacterial killing in mouse plasma (Fig. 8A). Nevertheless, a prolongation of survival in the mouse infection models was observed (Fig. 8B). To delineate a possible mechanism underlying this observation, and considering the previously reported LPS-binding property of TFPI (16), we investigated whether GGL27 could exert anti-endotoxin effects in vitro and in vivo. The results showed indeed that the peptide inhibited LPS-mediated NO release from mouse-derived macrophages (RAW 264.7 cells) (Fig. 8C) and also significantly increased survival in a mouse model of LPS shock (Fig. 8D). Analyses of cytokines 20 h after LPS injection, showed significant reductions of pro-inflammatory IL-6, IFN-γ, TNF-α, and MCP-1, whereas anti-inflammatory IL-10 was increased (Fig. 8E). Furthermore, a marked reduction of inflammation and vascular leakage in the lungs of the GGL27-treated animals was observed (Fig. 8F). In contrast, the two control peptides DSE25 from the N-terminal region of TFPI, and the peptide GGL27(S), having the “core” K/R residues substituted with S, did neither block NO release in vitro nor significantly alter overall cytokine release, as well as survival in vivo (Fig. 8, C–E). These data demonstrate a specific anti-endotoxic role of GGL27, compatible with the observed improvement in the E. coli and P. aeruginosa mouse infection models. Furthermore, the abrogated anti-endotoxic activity of GGL27(S) also highlights the importance of the central cationic K/R residues in this molecule. Taken together, the results are indicative of differential and host-related effects of GGL27, a host-independent anti-endotoxic effect and a host-dependent and complement-based antimicrobial function, likely logical consequences of different structural prerequisites for LPS and complement interactions. In line with this reasoning, Fig. 9 illustrates the evolution of this region of TFPI and highlights the conservation of the cationic “core” but also significant sequence changes between mice and men in this C-terminal region of TFPI.
FIGURE 8.
In vitro and in vivo effects of GGL27. A, 2 × 106 cfu/ml E. coli were incubated in 50 μl with GGL27 peptide at the indicated concentrations in 10 mm Tris, pH 7.4, buffer (Buffer), or in 0.15 m NaCl, 10 mm Tris, pH 7.4, containing 20% human citrate plasma (CP), or in the same buffer but with mice citrate plasma (Balb/c or C57B/6) (n = 3, ±S.D. is indicated). B, the GGL27 peptide prolongs survival in E. coli- and P. aeruginosa-infected mice. Mice were injected intraperitoneally with E. coli or P. aeruginosa bacteria. In the E. coli infection model GGL27 (200 μg) or buffer alone was injected intraperitoneally after 30 min (n = 10 in each group, p < 0.001, Kaplan-Meier Survival Analysis Log-Rank test). In the P. aeruginosa infection model GGL27 (500 μg) or buffer alone was injected subcutaneously after 1 h (n = 8 in each group, p < 0.001, Kaplan-Meier Survival Analysis Log-Rank test). C, GGL27 inhibits NO production. RAW264.7 mouse macrophages were stimulated with LPS from E. coli (left panel) or P. aeruginosa (right panel) in presence of GGL27 or the two control peptides GGL27(S) and DSE25 at the indicated concentrations. The difference between GGL27 and the control peptides is statistically significant (p < 0.01). D, GGL27 significantly increases survival in LPS-induced shock. Mice were injected with E. coli LPS followed by intraperitoneal administration of GGL27 (500 μg). Buffer and the peptides GGL27(S) and DSE25 (500 μg) served as controls. Survival was followed for 7 days (GGL27, n = 16; buffer, n = 15; GGL27(S), n = 8; DSE25, n = 8; the difference between GGL27 and buffer, or the control peptides, is significant, p < 0.01, Kaplan-Meier Survival Analysis Log-Rank test). *, indicates that the lines for GGL27(S), DSE25, and buffer are overlaid. E, GGL27 attenuates pro-inflammatory cytokines. In a separate experiment, mice were sacrificed 20 h after intraperitoneal injection of LPS followed by treatment as above with GGL27 (500 μg), buffer, or the control peptides GGL27(S) or DSE25, and the indicated cytokines were analyzed in blood (control, n = 8; GGL27, n = 12; GGL27(S), n = 7; DSE25; n = 8). In all cases, the difference between GGL27-treated animals and buffer was significant. p values for the respective cytokines are IL-6, 0.0023; TNF-α, 0.0018; IFN-γ, 0.0002; MCP-1, 0.0002; and IL-10, 0.0023. There was a significant difference between controls and GGL27(S) with respect to IFN-γ. F, lungs were analyzed 20 h after intraperitoneal LPS injection followed by treatment with GGL27 (500 μg) or buffer. Histochemical analysis shows marked attenuation of inflammatory changes in GGL27-treated lungs (a representative lung section is shown).
FIGURE 9.
Evolution of TFPI. The phylogenetic tree and sequence similarities show that TFPI in Homo sapiens, Pongo abelii, and Sus scrofa are closely related.
DISCUSSION
The key findings in this study are the identification of a dual antimicrobial activity of C-terminal TFPI peptides, based on direct and complement-mediated bactericidal effects, the demonstration of an anti-endotoxic effect, combined with the identification of TFPI in skin, its up-regulation during wounding, and presence in wounds.
The presently disclosed direct antibacterial action of C-terminal peptides of TFPI is in line with observations indicating that heparin-binding proteins, such as complement C3 (25), kininogen (36, 37), heparin-binding protein (38), heparin-binding epidermal growth factor and other growth factors (39), heparin/heparan sulfate-interacting protein (40), β2-glycoprotein (41), histidine-rich glycoprotein (42), and human thrombin (43) may, either as holoproteins or after fragmentation, exert antimicrobial activities in vitro and, in several cases, in vivo (36, 37, 42). In addition, peptide-mediated C3a generation, along with enhanced MAC formation, represents a mechanism by which a host defense peptide may selectively enhance microbial killing by generation of additional complement-derived AMPs. Recent evidence showing a significant cross-talk between the coagulation and complement systems (44) further adds biological relevance to the facilitated generation of C3a by the TFPI-peptides.
Considering the C-terminal region of TFPI, peptides derived from this region resemble other linear peptides of low helical content. For example, antimicrobial peptides derived from growth factors also display a low helical content in presence of membranes, reflecting their low content of features typical of classic helical peptides, such as regularly interspersed hydrophobic residues (39). Furthermore, studies on a kininogen- derived antimicrobial peptide, HKH20 (HKHGHGHGKHKNKGKKNGKH) (45) showed that the HKH20 peptide displays predominantly random coil conformation in buffer and at lipid bilayers, the interactions dominated by electrostatics (45). It is noteworthy that, like the TFPI-derived peptides, both HKH20 (36) and GKR22 (GKRKKKGKGLGKKRDPCLRKYK) (39), a peptide derived from heparin-binding growth factor, retain their antibacterial activity at physiological conditions. In relation to the above, it is interesting that recent studies indicate that certain low amphipathic peptides exhibit activity mainly by strong electrostatic interactions, e.g. leading to negative curvature strains, to membrane thinning, or to the formation of lipid domains, sometimes resulting in bacterial membrane fluidity changes affecting biological function (46).
As previously mentioned, many antimicrobial peptides, including LL-37 (31), C3a (25), thrombin- and kininogen-derived peptides (36, 37, 43), are released during proteolysis. Considering TFPI, enzymes such as plasmin and thrombin release distinct C-terminal fragments of relevance for physiological events, such as wounding. Therefore, TFPI-derived peptides generated in fibrin clots by thrombin (20) may contribute to antimicrobial activity during wound healing. Furthermore, subsequent proteolysis by plasmin (generating GGL27) (21) may further add to the spectrum of host defense peptides released. As mentioned above, it is of note that TFPI, in addition to its synthesis in microvascular endothelial cells and subsequent occurrence in the endothelium, plasma, and platelets, is also produced by the liver, and found in monocytes and macrophages (12). Production of TFPI has also been demonstrated in capillaries, megakaryocytes, several cell lines, as well as neoplastic cells. This evidence combined with a marked up-regulation of TFPI during wounding of human skin, as well as its occurrence in fibrin and on bacteria, suggests that high local levels of endogenous TFPI peptides may occur in vivo. The finding that the peptides were particularly active in human plasma against E. coli, and to a lesser extent against P. aeruginosa, is particularly relevant and interesting, in the light of the ubiquitous occurrence of P. aeruginosa in chronic leg ulcers, contrasting to the very sparse E. coli colonization in these wounds (35, 47). Furthermore, based on the demonstration that bacterial omptins (such as OmpT from E. coli) may release GGL27 fragments from TFPI, it was recently proposed that TFPI has evolved sensitivity to omptin-mediated proteolytic inactivation to potentiate procoagulant responses to E. coli infection in certain conditions (48). Our data further substantiate this hypothesis by adding another host-response mechanism to E. coli infection: OmpT-mediated generation of the host defense peptide GGL27.
Simultaneous control of inflammation and coagulation plays a key role in maintaining homeostasis, and it is notable that trials utilizing recombinant TFPI indicate that the protein protects from E. coli-induced severe sepsis (49), and furthermore, TFPI is also under evaluation in a phase III clinical trial involving patients with severe community-acquired pneumonia. In this context, it is interesting to note that the here observed anti-endotoxic effect of GGL27 in vitro and in vivo may explain, at least partly, the previously observed protective effects of TFPI during E. coli sepsis (49). The current findings showing direct and indirect, MAC- and C3a-mediated, antimicrobial effects of the C-terminal epitope of TFPI, GGL27, may have implications for future attempts in designing and developing peptide-based therapeutics combating severe infection. Finally, the finding that the TFPI-peptide did not significantly enhance bacterial killing in mouse plasma ex vivo as well as increase total survival during bacterial sepsis (although a prolonged survival was observed), illustrates that mouse models might be disadvantageous when it comes to assessing certain aspects of the physiological roles of human C-terminal TFPI peptides, such as those involving complement-mediated bacterial clearance. Interestingly, whereas the TFPI-α form (having the C-terminal cationic sequence) is the predominant form in humans, mice appear to largely produce TFPI-β (a form lacking the C-terminal sequence) (50), an observation compatible with the abovementioned evolution of unique C-terminal and OmpT-releasable peptides in humans.
Supplementary Material
Acknowledgments
We thank Maria Baumgarten, Mina Davoudi, and Lotta Wahlberg for expert technical assistance. The help of Rita Wallén, Department of Cell and Organism Biology, with electron microscopy is gratefully acknowledged.
This work was supported by grants from the Swedish Research Council (projects 521-2009-3378, 7480, and 621-2003-4022), the Royal Physiographic Society in Lund, the Welander-Finsen, Crafoord, Österlund, and Kock Foundations, the Marianne and Marcus Wallenberg Foundation, XImmune AB, and The Swedish Government Funds for Clinical Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- TFPI
- tissue factor pathway inhibitor
- C3
- complement factor 3
- CF
- carboxyfluorescein
- RDA
- radial diffusion assay
- LDH
- lactate dehydrogenase
- TSB
- Trypticase soy broth
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- TF
- tissue factor
- FX
- factor X
- MAC
- membrane attack complex.
REFERENCES
- 1.Yount N. Y., Bayer A. S., Xiong Y. Q., Yeaman M. R. (2006) Biopolymers 84, 435–458 [DOI] [PubMed] [Google Scholar]
- 2.Zelezetsky I., Tossi A. (2006) Biochim. Biophys. Acta 1758, 1436–1449 [DOI] [PubMed] [Google Scholar]
- 3.Tossi A., Sandri L. (2002) Curr. Pharm. Des. 8, 743–761 [DOI] [PubMed] [Google Scholar]
- 4.Powers J. P., Hancock R. E. (2003) Peptides 24, 1681–1691 [DOI] [PubMed] [Google Scholar]
- 5.Bulet P., Stöcklin R., Menin L. (2004) Immunol. Rev. 198, 169–184 [DOI] [PubMed] [Google Scholar]
- 6.Dürr U. H., Sudheendra U. S., Ramamoorthy A. (2006) Biochim Biophys Acta 1758, 1408–1425 [DOI] [PubMed] [Google Scholar]
- 7.Brogden K. A. (2005) Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 8.Lohner K., Blondelle S. E. (2005) Comb. Chem. High Throughput Screen. 8, 241–256 [DOI] [PubMed] [Google Scholar]
- 9.Zanetti M. (2004) J. Leukoc. Biol. 75, 39–48 [DOI] [PubMed] [Google Scholar]
- 10.Elsbach P. (2003) J. Clin. Invest. 111, 1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganz T. (2003) Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 12.Lwaleed B. A., Bass P. S. (2006) J. Pathol. 208, 327–339 [DOI] [PubMed] [Google Scholar]
- 13.Girard T. J., Warren L. A., Novotny W. F., Likert K. M., Brown S. G., Miletich J. P., Broze G. J., Jr. (1989) Nature 338, 518–520 [DOI] [PubMed] [Google Scholar]
- 14.Mine S., Yamazaki T., Miyata T., Hara S., Kato H. (2002) Biochemistry 41, 78–85 [DOI] [PubMed] [Google Scholar]
- 15.Crawley J. T., Lane D. A. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 233–242 [DOI] [PubMed] [Google Scholar]
- 16.Park C. T., Creasey A. A., Wright S. D. (1997) Blood 89, 4268–4274 [PubMed] [Google Scholar]
- 17.Hembrough T. A., Ruiz J. F., Swerdlow B. M., Swartz G. M., Hammers H. J., Zhang L., Plum S. M., Williams M. S., Strickland D. K., Pribluda V. S. (2004) Blood 103, 3374–3380 [DOI] [PubMed] [Google Scholar]
- 18.Wesselschmidt R., Likert K., Girard T., Wun T. C., Broze G. J., Jr. (1992) Blood 79, 2004–2010 [PubMed] [Google Scholar]
- 19.Ettelaie C., Adam J. M., James N. J., Oke A. O., Harrison J. A., Bunce T. D., Bruckdorfer K. R. (1999) FEBS Lett. 463, 341–344 [DOI] [PubMed] [Google Scholar]
- 20.Ohkura N., Enjyoji K., Kamikubo Y., Kato H. (1997) Blood 90, 1883–1892 [PubMed] [Google Scholar]
- 21.Li A., Wun T. C. (1998) Thromb. Haemost. 80, 423–427 [PubMed] [Google Scholar]
- 22.Cunningham A. C., Hasty K. A., Enghild J. J., Mast A. E. (2002) Biochem. J. 367, 451–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson E., Rydengård V., Sonesson A., Mörgelin M., Björck L., Schmidtchen A. (2004) Eur. J. Biochem. 271, 1219–1226 [DOI] [PubMed] [Google Scholar]
- 24.Lehrer R. I., Rosenman M., Harwig S. S., Jackson R., Eisenhauer P. (1991) J. Immunol. Methods 137, 167–173 [DOI] [PubMed] [Google Scholar]
- 25.Nordahl E. A., Rydengård V., Nyberg P., Nitsche D. P., Mörgelin M., Malmsten M., Björck L., Schmidtchen A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16879–16884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlemalm E., Villiger W., Hobot J. A., Acetarin J. D., Kellenberger E. (1985) J. Microsc. 140, 55–63 [DOI] [PubMed] [Google Scholar]
- 27.Pollock J. S., Förstermann U., Mitchell J. A., Warner T. D., Schmidt H. H., Nakane M., Murad F. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10480–10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenfield N., Fasman G. D. (1969) Biochemistry 8, 4108–4116 [DOI] [PubMed] [Google Scholar]
- 29.Sjögren H., Ulvenlund S. (2005) Biophys. Chem. 116, 11–21 [DOI] [PubMed] [Google Scholar]
- 30.Tossi A., Sandri L., Giangaspero A. (2000) Biopolymers 55, 4–30 [DOI] [PubMed] [Google Scholar]
- 31.Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 32.Ganz T. (2001) Semin. Respir. Infect. 16, 4–10 [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Agerberth B., Löthgren A., Almstedt A., Johansson J. (1998) J. Biol. Chem. 273, 33115–33118 [DOI] [PubMed] [Google Scholar]
- 34.Schirm S., Liu X., Jennings L. L., Jedrzejewski P., Dai Y., Hardy S. (2009) J. Infect. Dis. 199, 1807–1815 [DOI] [PubMed] [Google Scholar]
- 35.Lundqvist K., Herwald H., Sonesson A., Schmidtchen A. (2004) Thromb. Haemost. 92, 281–287 [DOI] [PubMed] [Google Scholar]
- 36.Nordahl E. A., Rydengård V., Mörgelin M., Schmidtchen A. (2005) J. Biol. Chem. 280, 34832–34839 [DOI] [PubMed] [Google Scholar]
- 37.Frick I. M., Akesson P., Herwald H., Mörgelin M., Malmsten M., Nägler D. K., Björck L. (2006) EMBO J. 25, 5569–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira H. A. (1995) J. Leukoc. Biol. 57, 805–812 [DOI] [PubMed] [Google Scholar]
- 39.Malmsten M., Davoudi M., Walse B., Rydengård V., Pasupuleti M., Mörgelin M., Schmidtchen A. (2007) Growth Factors 25, 60–70 [DOI] [PubMed] [Google Scholar]
- 40.Meyer-Hoffert U., Hornef M., Henriques-Normark B., Normark S., Andersson M., Pütsep K. (2008) FASEB J. 22, 2427–2434 [DOI] [PubMed] [Google Scholar]
- 41.Nilsson M., Wasylik S., Mörgelin M., Olin A. I., Meijers J. C., Derksen R. H., de Groot P. G., Herwald H. (2008) Mol Microbiol 67, 482–492 [DOI] [PubMed] [Google Scholar]
- 42.Rydengård V., Shannon O., Lundqvist K., Kacprzyk L., Chalupka A., Olsson A. K., Mörgelin M., Jahnen-Dechent W., Malmsten M., Schmidtchen A. (2008) PLoS Pathog. 4, e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papareddy P., Rydengard V., Pasupuleti M., Walse B., Morgelin M., Chalupka A., Malmsten M., Schmidtchen A.PLoS Pathog 6, e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amara U., Rittirsch D., Flierl M., Bruckner U., Klos A., Gebhard F., Lambris J. D., Huber-Lang M. (2008) Adv. Exp. Med. Biol. 632, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ringstad L., Andersson Nordahl E., Schmidtchen A., Malmsten M. (2007) Biophys J 92, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto N., Tamura A. (2010) Peptides 31, 794–805 [DOI] [PubMed] [Google Scholar]
- 47.Schmidtchen A., Wolff H., Hansson C. (2001) Acta Derm. Venereol. 81, 406–409 [DOI] [PubMed] [Google Scholar]
- 48.Yun T. H., Cott J. E., Tapping R. I., Slauch J. M., Morrissey J. H. (2009) Blood 113, 1139–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Creasey A. A., Chang A. C., Feigen L., Wün T. C., Taylor F. B., Jr., Hinshaw L. B. (1993) J. Clin. Invest. 91, 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maroney S. A., Ellery P. E., Mast A. E. (2010) Thromb. Res. 125, Suppl. 1, S52–S56 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.