Abstract
A number of studies have evaluated two functional polymorphisms on p53 Arg72Pro and GSTP1 Ile105Val, in relation to esophageal cancer susceptibility. However, the results remain conflicting rather than conclusive. This meta-analysis on 2919 cases and 4074 controls for p53 Arg72Pro and 1885 cases and 2194 controls for GSTP1 Ile105Val from 13 published case-control studies showed that no significant general main effects for GSTP1 Ile105Val on esophageal cancer risk. However, we found that the p53 Arg72Pro was associated with an increased risk of esophageal cancer ((Pro/Arg +Pro/Pro) versus Arg/Arg: OR=1.20, 95%CI=1.06-1.36) without any between-study heterogeneity.
In the stratified analysis by ethnicity, we found that the increased esophageal cancer risk associated with p53 Arg72Pro polymorphism was more evident in Asian group ((Pro/Arg +Pro/Pro) versus Arg/Arg: OR=1.35, 95%CI=1.14-1.60, P=0.09 for heterogeneity test), although we still failed to find any significant association between GSTP1 Ile105Val polymorphism and esophageal cancer risk in different ethnicity. These results suggest that p53 Arg72Pro polymorphism, but not GSTP1 Ile105Val, may contribute to esophageal cancer development, especially in Asian. Additional well-designed large studies were required for the validation of this association.
Keywords: p53, GSTP1, polymorphism, esophageal cancer, meta-analysis
Introduction
Esophageal cancer, the sixth most common cause of cancer-related death in the world, occurs with increased frequency in specific regions. 1 Survival rates for esophageal cancer are poor; 75% of patients die within 1 year after diagnosis, and the 5-year survival rate is only 5-10% .1 The development of esophageal cancer is a multifactorial process associated with a variety of risk factors. Cumulative evidence suggests that tobacco smoking, heavy alcohol drinking, micronutrient deficiency, and dietary carcinogen exposure may cause the disease.2-5 However, even in the at-risk population, only a portion of exposed individuals develop the cancer in their life span, indicating that there may be important genetic basis rendering such individuals susceptible to the disease.
The tumor-suppressor gene p53 is important for cellular growth control once the DNA is subject to damage or mutation and arrests the cell cycle in the G1 phase to allow DNA repair or apoptosis.6 Its mutation is widely detected in all types of cancer, including esophageal cancer.6,7 It is now clear that disruption of p53 pathway, such as through inactivating p53 mutations, is associated with the formation and progression of malignancies. For example, it has been shown that >50% of human tumors have inactivating p53 mutations.8
Glutathione S-transferase P1 (GSTP1) is quantitatively the most important GST isoform in normal esophageal epithelium.9 GSTP1 expression, GSTP1 mRNA levels, glutathione content and GST enzyme activities are all reduced in BE (Barrett esophagus) compared with normal esophageal epithelium.9-13 Because accumulating evidence indicates p53 and GSTP1 play central role in cancer formation and progression, one may reason that functional single nucleotide polymorphisms in these genes might render the carrier susceptible to cancer, including esophageal cancer.
It was reported that the p53 gene is polymorphic and among its single nucleotide polymorphisms, a G>C change at codon 72 (rs1042522) results in Arg>Pro amino acid substitution.14 Although both variants are morphologically wild-type, the Pro/Pro genotype is less effective in suppressing cellular transformation.15 Several studies have reported that the p53 codon 72 polymorphism may be associated with tumor susceptibility to a variety of cancers recently.16-18 The GSTP1 gene displays a polymorphism, an A>G change at codon 105, resulting in an Ile-to-Val substitution (rs1695), which alters the enzymatic activity of the protein.18 This has been suggested as a putative high-risk genotype in various cancers.19 Therefore, it's reasonable to hypothesize that the p53 Arg72Pro and GSTP1 Ile105Val polymorphisms may functionally related to the risk of esophageal cancer.
A number of molecular epidemiology studies have been conducted to examine the association between p53 Arg72Pro, GSTP1 Ile105Val polymorphisms and esophageal cancer susceptibility 19-33, but the results remain inconsistent. To estimate the overall risk of p53 Arg72Pro, GSTP1 Ile105Val associated with esophageal cancer risk and to quantify the potential between-study heterogeneity, we conducted a meta-analysis on 13 published case-control studies with 2919 cases and 4074 controls for p53 Arg72Pro and 1885 cases and 2194 controls for GSTP1 Ile105Val.
Materials and Methods
Identification and Eligibility of Relevant Studies. We attempted to include all the case-control studies published to date on the association between p53 Arg72Pro, GSTP1 Ile105Val polymorphisms and esophageal cancer risk. Eligible studies were identified by searching the electronic literature PubMed for relevant reports (last search update February 2010, using the search terms “p53”, “polymorphisms” and “esophageal cancer”; “GSTP1”, “polymorphisms” and “esophageal cancer”). Additional studies were identified by hands-on searches from references of original studies or review articles on this topic. If studies had partly overlapped subjects, only the one with a larger and/or latest sample size was selected for the analysis.
Data Extraction. Two investigators independently extracted data and reached a consensus on all of the items. Data extracted from these articles included the first author's name, year of publication, country of origin, ethnicity, number of cases and controls, genotype frequencies for cases and controls.
Meta-Analysis. The risk of esophageal cancer associated with p53 Arg72Pro, GSTP1 Ile105Val polymorphisms were estimated for each study by odds ratio (OR) with 95% confidence intervals (95%CI). For all studies, we evaluated the risk of the variant genotypes (Pro/Pro, Val/ Val), compared with the wild-type genotype (Arg/Arg, Ile/ Ile). Then we calculated the ORs of the polymorphisms, using both dominant and recessive genetic models of the variant 72Pro and 105Val alleles. In addition, we conducted stratification analysis by ethnicity. The χ2-based Q statistic test was used for the assessment of heterogeneity, and it was considered significant for P < 0.05. We used the fixed-effects model and the random-effects model based on the Mantel-Haenszel method and the DerSimonian and Laird method, respectively, to combine values from each of the studies. When the effects were assumed to be homogenous, the fixed-effects model was then used; otherwise, the random-effects model was more appropriate. We also computed the power of the selected studies by using the DSTPLAN4.2 software, in order to assess the probability of detecting an association between RANTES polymorphisms and asthma at the 0.05 level of significance, assuming a genotypic risk of 2.0 and 1.5. The Egger's test and inverted funnel plots were utilized to provide diagnosis of publication bias (Linear regression analysis, ref.34 All analysis was done by using the Statistical Analysis System software (v.9.1.3, SAS Institute, Cary, NC) and Review Manage (v.4.2). All the P values were two-sided.
Results
The selected study characteristics are listed in Table 1 and Table 2. All studies indicated that the distributions of two polymorphism's genotypes in the controls were both consistent with Hardy-Weinberg equilibrium except for one study 26for p53 Arg72Pro, and one studies 21 for GSTP1 Ile105Val. Considering the representation of samples, which may directly result in untruthful effect, we excluded these studies 21,26 with a departure from Hardy-Weinberg equilibrium from our analysis. As a result, 6 case-control studies (2919 cases and 4074 controls) for p53 Arg72Pro and 9 studies (1885 cases and 2194 controls) for GSTP1 Ile105Val were available for this meta-analysis. The minor Pro allele (for p53 Arg72Pro) and Val allele (for GSTP1 Ile105Val) frequency (MAF) were 0.44 and 0.20 for Asian studies, while around 0.60 and 0.32 for Mix and Caucasus populations, respectively.
Table 1.
Characteristics of published studies on p53Arg72Pro included in the meta-analysis
| Author (ref*) | Year | Origin | Ethnicity | SNPsite | Sample size(case/control) | HWE | MAF in controls | Genotypic ORs& | Power (%) † | |
|---|---|---|---|---|---|---|---|---|---|---|
| homozygotes/heterozygotes | OR>1.5 | OR>2.0 | ||||||||
| Lee JM22 | 2000 | China(Taiwan) | Asian | p53 Arg72Pro | 90/254 | 0.427 | 0.40 | 2.56/1.86 | 37.5 | 80.2 |
| Vos M23 | 2003 | South Afican | African | p53 Arg73Pro | 73/115 | 0.216 | 0.41 | 0.44/0.96 | 27.0 | 63.5 |
| Hong Y24 | 2005 | China | Asian | p53 Arg74Pro | 758/1420 | 0.105 | 0.44 | 1.77/0.99 | 99.4 | 100.0 |
| Cai L25 | 2006 | China | Asian | p53 Arg75Pro | 204/389 | 0.107 | 0.47 | 2.25/1.43 | 64.8 | 97.7 |
| Yang W26 | 2008 | China | Asian | p53 Arg76Pro | 435/550 | 0.000 | 0.32 | 0.39/0.07 | 86.0 | 100.0 |
| Liu G27 | 2009 | United States | Caucasian | p53 Arg77Pro | 302/453 | 0.066 | 0.26 | 1.05/01.18 | 70.6 | 99.2 |
| Canova C19 | 2009 | European | Caucasian | p53 Arg78Pro | 1492/1443 | 0.660 | 0.73 | 1.00/0.95 | 99.6 | 100.0 |
* The ref was referred to the reference numbers in this study.
& data from the same source, so selected by the latest sample size.
# NA: Not available.
& Genotypic odds ratios for homozygotes and heterozygotes.
† Power was calculated by the DSTPLAN4.2 software with MAF in controls as the frequency of risk factor, OR was selected 1.5 and 2.0 as the relative risk and а=0.05 as the significance.
Table 2.
Characteristics of published studies on GSTP1I le105Val included in the meta-analysis
| Author (ref*) | Year | Origin | Ethnicity | SNPsite | Sample size(case/control) | HWE | MAF in controls | Genotypic ORs& | Power (%) † | |
|---|---|---|---|---|---|---|---|---|---|---|
| homozygotes/heterozygotes | OR>1.5 | OR>2.0 | ||||||||
| Lin DX&28 | 1998 | China | Asian | GSTP1 Ile105Val | 42/36 | 0.359 | 0.24 | 0.25/0.83 | 12.3 | 28.9 |
| Morita S29 | 1998 | Japan | Asian | GSTP1 Ile106Val | 66/164 | 0.412 | 0.16 | 0.26/0.19 | 19.2 | 49.2 |
| van Lieshout EM30 | 1999 | The Netherlands | Caucasian | GSTP1 Ile107Val | 34/247 | 0.739 | 0.23 | 3.65/3.44 | 16.4 | 40.7 |
| Tan W&31 | 2000 | China | Asian | GSTP1 Ile108Val | 150/150 | 0.616 | 0.22 | 1.47/0.89 | 33.5 | 77.1 |
| Lee JM22 | 2000 | China(Taiwan) | Asian | GSTP1 Ile109Val | 90/254 | NA# | NA# | NA#/ NA# | NA# | NA# |
| Casson AG21 | 2003 | Canada | Caucasian | GSTP1 Ile110Val | 45/45 | 0.019 | 0.29 | 0.78/2.51 | 14.6 | 35.1 |
| Roth MJ32 | 2004 | China | Asian | GSTP1 Ile111Val | 131/454 | 0.057 | 0.22 | 0.79/0.88 | 43.0 | 88.2 |
| Casson AG20 | 2006 | Canada | Caucasian | GSTP1 Ile112Val | 56/95 | 0.834 | 0.35 | 2.22/1.36 | 21.7 | 52.7 |
| Cai L25 | 2006 | China | Asian | GSTP1 Ile113Val | 204/393 | 0.872 | 0.18 | 0.46/0.93 | 48.4 | 92.6 |
| Murphy SJ33 | 2007 | Irish | Caucasian | GSTP1 Ile114Val | 207/223 | 0.201 | 0.36 | 0.99/0.93 | 54.0 | 94.4 |
| Canova C19 | 2009 | European | Caucasian | GSTP1 Ile115Val | 1471/1405 | 0.330 | 0.32 | 0.97/1.13 | 99.9 | 100.0 |
* The ref was referred to the reference numbers in this study.
& data from the same source, so selected by the latest sample size.
# NA: Not available.
&Genotypic odds ratios for homozygotes and heterozygotes.
† Power was calculated by the DSTPLAN4.2 software with MAF in controls as the frequency of risk factor, OR was selected 1.5 and 2.0 as the relative risk and а=0.05 as the significance.
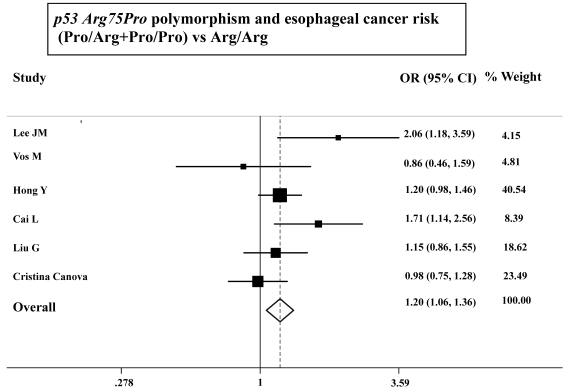
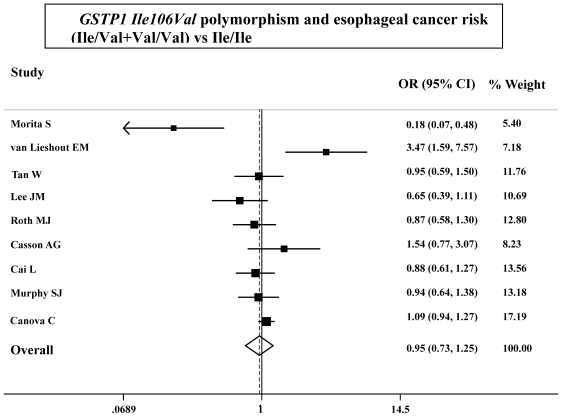
As shown in Table 3, the variant homozygote (Pro/Pro) for p53 Arg72Pro was associated with a significantly increased risk of esophageal cancer (Pro/Pro versus Arg/Arg: OR=1.43, 95%CI=1.23-1.68; P = 0.10 for heterogeneity test) compared with wild-type homozygote (Arg/Arg). We also found significant main effects in the dominant genetic model ((Pro/Arg +Pro/Pro) versus Arg/Arg: OR=1.20, 95%CI=1.06-1.36; P = 0.08 for heterogeneity test; Table 3 and Figure 1). However, we failed to find any significant main effects for GSTP1 Ile105Val on esophageal cancer risk in different genetic models tested (Table 3 and Figure 2).
Table 3.
Summary ORs of p53 and GSTP1 polymorphisms and esophageal cancer risk
| Comparison | No. of Cases | No. of Controls | OR | 95%CI | P* |
|---|---|---|---|---|---|
| p53 Arg75Pro | |||||
| Pro/Arg vs Arg/Arg | 1761 | 2850 | 1.09 | 0.95-1.24 | 0.25 |
| Pro/Pro vs Arg/Arg | 1720 | 2263 | 1.43 | 1.23-1.68 | 0.06 |
| Pro/Pro vs (Arg/Arg+Pro/Arg) | 2919 | 4074 | 1.31 | 0.95-1.80 | 0.00 |
| (Pro/Arg +Pro/Pro) vs Arg/Arg | 2919 | 4074 | 1.20 | 1.06-1.36 | 0.08 |
| GSTP1 Ile106Val | |||||
| Ile/Val vs Ile/Ile | 1687 | 1917 | 0.99 | 0.74-1.32 | 0.00 |
| Val/Val vs Ile/Ile | 1063 | 1295 | 1.00 | 0,81-1.23 | 0.28 |
| Val/Val vs (Ile/Ile+Ile/Val) | 1885 | 2194 | 0.95 | 0.79-1.17 | 0.57 |
| (Ile/Val+Val/Val) vs Ile/Ile | 1885 | 2194 | 0.95 | 0.73-1.25 | 0.00 |
* Test for heterogeneity. Fixed-effects model was used when P value for heterogeneity test > 0.05; otherwise, random-effects model was used.
Figure 1.
ORs (log scale) of esophageal cancer associated with p53 Arg75Pro for the Pro/Arg+Pro/Pro genotypes, compared with the Arg/Arg genotype.
Figure 2.
ORs (log scale) of esophageal cancer associated with GSTP1 Ile106Val for the Ile/Val+Val/Val genotypes, compared with the Ile/Ile genotype.
We further performed stratified analysis according to ethnicity (Asian and Mixed/ Caucasian group). As shown in the Table 4, we found that the increased esophageal cancer risk associated with p53 Arg72Pro polymorphism was more evident in Asian ((Pro/Arg +Pro/Pro) versus Arg/Arg: OR=1.35, 95%CI=1.14-1.60, P=0.09 for heterogeneity test). Unfortunately, we still failed to find any significant association between GSTP1 Ile105Val polymorphism and esophageal cancer risk in different ethnicity.
Table 4.
Association between esophageal cancer risk and the p53, GSTP1 polymorphisms, stratified by ethnicity.
| SNP site | Studies of available& | No. of Cases | No. of Controls | OR# | 95%CI | P* |
|---|---|---|---|---|---|---|
| p53 Arg72Pro | ||||||
| Asian | 22,24,25 | 1052 | 2063 | 1.35 | 1.14-1.60 | 0.09 |
| Mix | 19,23,27 | 1868 | 2011 | 1.04 | 0.86-1.25 | 0.60 |
| GSTP1 Ile105Val | ||||||
| Asian | 22,25,29,31,32 | 641 | 1415 | 0.99 | 0.66-1.49 | 0.00 |
| Caucasian | 19,20,30,33 | 1768 | 1970 | 1.06 | 0.86-1.31 | 0.02 |
# The OR was obtained in dominant genetic model.
* Test for heterogeneity. Fixed-effects model was used when P value for heterogeneity test > 0.05; otherwise, random-effects model was used.
& Studies of available was referred to the reference resource of the stratified variable, which data was available.
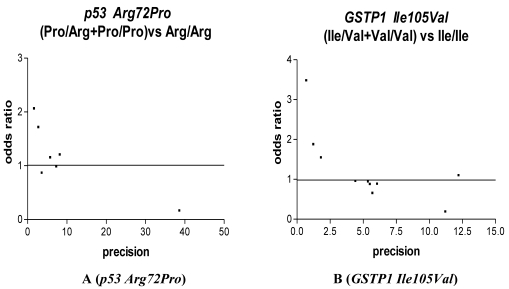
We used Funnel plot and Egger's test to access the publication bias of literatures. As shown in Fig. 3 A, the shape of the funnel plots seemed nonsymmetrical in the dominant genetic model for the p53 Arg72Pro, suggesting that there was significant publication bias. Egger's test was used to provide statistical evidence. As a result, the publication bias was observed slightly for p53 Arg72Pro (t=4.55, P = 0.01) but was disappeared (t=1.35, P = 0.25) when we excluded the study 26 departure from Hardy-Weinberg equilibrium. No publication bias was observed for GSTP1 Ile105Val (t=1.13, P = 0.29), we also excluded the study 21 departure from Hardy-Weinberg equilibrium and still did not found any publication bias for 28C/G (t=0.90, p=0.39).
Figure 3.
Funnel plot analysis to detect publication bias in esophageal cancer. Each point represents a separate study for the indicated association. For each study, the OR is plotted on a logarithmic scale against the precision (the reciprocal of the SE).
Discussion
The GSTP1 gene, which encodes the GST π isoenzyme, is the most important form in the esophagus.35 It can eliminate DNA oxidative products of thymidine or uracil propenal.36 The 105Val form shows altered affinity and enzymatic activity for some substrates.37-39 However, our analysis results showed there was no significant relations between GSTP1 Ile105Val polymorphism and esophageal cancer, but this conclusion was consistent with Hiyama T et al' s review.40 These findings suggest that the GSTP1 Ile105Val genotype alone does not show any association with the susceptibility to esophageal cancer, even when stratified by subgroup. This finding is perhaps not surprising, because the functional evidence to support the role of GSTP1 Ile105Val as an esophageal cancer risk factor is not strong. Although GSTP1 may encode the GST π isoenzyme in the esophagus, positive effect for esophageal cancer frequently has been detected in those who had some environment exposures such as smoke cigarettes, alcohol drinkers or low level of dietary selenium intake. Therefore, it is reasonable to hypothesize that the GSTP1 Ile105Val polymorphism may be at best a modifier for esophageal cancer by interactive with some lifestyle and dietary habits, but is not a significant independent susceptibility factor.
The p53 tumor suppressor pathway is well-known to be crucial in maintaining genomic integrity and preventing cells from oncogenic transformation. When a cell is exposed to genotoxic stress such as DNA damage and oncogene activation, the p53 protein accumulates rapidly through posttranscriptional mechanisms and is also activated as a transcriptional factor, which leads to cell cycle arrest for DNA repair or apoptotic cell death 41. Both mice and humans harboring germ line inactivating mutations in one p53 allele are highly susceptible to cancer: they develop cancer very early in life and at very high frequencies. 42,43
The functional impact of this p53 polymorphism has been reported and the Arg/Arg genotype seems to induce apoptosis with faster kinetics and to suppress transformation more efficiently than the Pro/Pro genotype.15 It was shown that p53 Pro/Pro exhibits a lower ability to induce apoptosis in vitro than p53Arg/Arg.15 In a pilot study, Zhang et al.44 showed that subjects carrying the p53 72Pro/Pro genotype had a >2-fold increased risk for developing esophageal cancer. These results are consistent with our present meta-analysis study. Thus, it is reasonable to hypothesize that the Arg72Pro polymorphism with reduced activity of p53 may play more important role in esophageal cancer risk.
In the present meta-analysis on the association between p53 Arg72Pro, GSTP1 Ile105Val polymorphisms and risk of esophageal cancer, we found that variant 72Pro of alleles p53 Arg72Pro could significantly increase the risk of esophageal cancer, although the association were not significantly evident in most studies individually. However, we failed to find any significant association between GSTP1 Ile105Val and esophageal cancer risk. In stratified analysis, we further observed that the association between p53 Arg72Pro and risk of esophageal cancer was remained significant in Asian population. The different effect of p53 Arg72Pro polymorphism between ethnicity may result from different genetic background and environmental exposures, which may contribute to the frequency of ethnic difference.
It is worth emphasizing that several environment exposures are regarded as risk factors of esophageal cancer, especially tobacco smoking, which is an established etiologic factor for esophageal cancer 3,45, and exposure to smoke causes genotoxic stress including DNA damage or avoids potential saturation of enzyme activity.46,47 Several data provided some support for one hypothesis that there may be existed significant interaction between p53 Arg72Pro or GSTP1 Ile105Val polymorphism and smoking, though there were not enough report support us to make meta-analysis in current research. Studies with a larger sample size, especially including smoking or another environment factors will be helpful to confirm the findings.
Although there have been consistent findings that the p53 codon 72 Pro/Pro genotype was associated with increased esophageal SCC risk 40, it is worth mentioning that there are 2 main forms of esophageal cancer histologically, squamous cell carcinoma (SCC) and adenocarcinoma, and each has distinct etiologic and pathologic characteristics. Squamous cell is cancer located in epithelial cell of the mouth throat or lungs and adenocarcinoma is composed of cells of glandular tissue. Over the past 5 decades, many changes in the prevalence of esophageal cancer have occurred. Prior to this, SCC comprised more than 95% of esophageal malignancies 48. In our meta-analysis, we had wanted to analysis the association between these two gene polymorphisms and risk of esophageal cancer according to the different pathological type, but most of the included research were majored on SCC, so we failed to conduct related stratified analysis. More molecular epidemiological studies on adenocarcinoma are needed to further elucidate the real association of the p53 Arg72Pro and GSTP1 Ile105Val polymorphism with esophageal carcinogenesis.
In conclusion, this meta-analysis of 13 case-control studies provided evidence that the p53 Arg72Pro polymorphism, but not the GSTP1 Ile105Val, was significantly associated with increased risk of esophageal cancer, especially in Asian. Further well-designed large studies, particularly referring to gene-gene and gene-environment interactions are warranted to confirm the real contribution of these polymorphisms to esophageal cancer susceptibility.
References
- 1.Xing D, Tan W, Lin D. Genetic polymorphisms and susceptibility to esophageal cancer among chinese population (review) Oncol Rep. 2003;10:1615–1623. [PubMed] [Google Scholar]
- 2.Franceschi S, Bidoli E, Negri E, Zambon P, Talamini R, Ruol A, Parpinel M, Levi F, Simonato L, La Vecchia C. Role of macronutrients, vitamins and minerals in the aetiology of squamous-cell carcinoma of the oesophagus. Int J Cancer. 2000;86:626–631. doi: 10.1002/(sici)1097-0215(20000601)86:5<626::aid-ijc4>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 3.Gao YT, McLaughlin JK, Blot WJ, Ji BT, Benichou J, Dai Q, Fraumeni JFJr. Risk factors for esophageal cancer in shanghai, china. I. Role of cigarette smoking and alcohol drinking. Int J Cancer. 1994;58:192–196. doi: 10.1002/ijc.2910580208. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y, Chen J, Ohshima H, Pignatelli B, Boreham J, Li J, Campbell TC, Peto R, Bartsch H. Geographic association between urinary excretion of n-nitroso compounds and oesophageal cancer mortality in china. Int J Cancer. 1993;54:713–719. doi: 10.1002/ijc.2910540502. [DOI] [PubMed] [Google Scholar]
- 5.Hu J, Nyren O, Wolk A, Bergstrom R, Yuen J, Adami HO, Guo L, Li H, Huang G, Xu X. et al. Risk factors for oesophageal cancer in northeast china. Int J Cancer. 1994;57:38–46. doi: 10.1002/ijc.2910570108. [DOI] [PubMed] [Google Scholar]
- 6.Hollstein M, Sidransky D, Vogelstein B, Harris CC. P53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 7.Bennett WP, Hollstein MC, Metcalf RA, Welsh JA, He A, Zhu SM, Kusters I, Resau JH, Trump BF, Lane DP. et al. P53 mutation and protein accumulation during multistage human esophageal carcinogenesis. Cancer Res. 1992;52:6092–6097. [PubMed] [Google Scholar]
- 8.Soussi T, Beroud C. Assessing tp53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 9.Peters WH, Roelofs HM, Hectors MP, Nagengast FM, Jansen JB. Glutathione and glutathione s-transferases in barrett's epithelium. Br J Cancer. 1993;67:1413–1417. doi: 10.1038/bjc.1993.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobbe SC, Scobie GC, Pohler E, Hayes JD, Kernohan NM, Dillon JF. Alteration of glutathione s-transferase levels in barrett's metaplasia compared to normal oesophageal epithelium. Eur J Gastroenterol Hepatol. 2003;15:41–47. doi: 10.1097/00042737-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Chandra RK, Bentz BG, Haines GK3rd, Robinson AM, Radosevich JA. Expression of glutathione s-transferase pi in benign mucosa, barrett's metaplasia, and adenocarcinoma of the esophagus. Head Neck. 2002;24:575–581. doi: 10.1002/hed.10093. [DOI] [PubMed] [Google Scholar]
- 12.Compton KR, Orringer MB, Beer DG. Induction of glutathione s-transferase-pi in barrett's metaplasia and barrett's adenocarcinoma cell lines. Mol Carcinog. 1999;24:128–136. doi: 10.1002/(sici)1098-2744(199902)24:2<128::aid-mc7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.van Lieshout EM, Tiemessen DM, Witteman BJ, Jansen JB, Peters WH. Low glutathione and glutathione s-transferase levels in barrett's esophagus as compared to normal esophageal epithelium. Jpn J Cancer Res. 1999;90:81–85. doi: 10.1111/j.1349-7006.1999.tb00669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koushik A, Platt RW, Franco EL. P53 codon 72 polymorphism and cervical neoplasia: A meta-analysis review. Cancer Epidemiol Biomarkers Prev. 2004;13:11–22. doi: 10.1158/1055-9965.epi-083-3. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Solari A, Wang X, Zhang Z, Xu Y, Wang L, Hu X, Guo J, Wei Q. P53 codon 72 polymorphism and risk of gastric cancer in a chinese population. Oncol Rep. 2004;11:1115–1120. [PubMed] [Google Scholar]
- 18.Lacerda LL, Serrano SV, Mathes A, Rey JA, Bello MJ, Casartelli C. An intronic variant in the tp53 gene in a brazilian woman with breast cancer. Cancer Genet Cytogenet. 2005;160:160–163. doi: 10.1016/j.cancergencyto.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Canova C, Hashibe M, Simonato L, Nelis M, Metspalu A, Lagiou P, Trichopoulos D, Ahrens W, Pigeot I, Merletti F, Richiardi L, Talamini R, Barzan L, Macfarlane GJ, Macfarlane TV, Holcatova I, Bencko V, Benhamou S, Bouchardy C, Kjaerheim K, Lowry R, Agudo A, Castellsague X, Conway DI, McKinney PA, Znaor A, McCartan BE, Healy CM, Marron M, Brennan P. Genetic associations of 115 polymorphisms with cancers of the upper aerodigestive tract across 10 european countries: The arcage project. Cancer Res. 2009;69:2956–2965. doi: 10.1158/0008-5472.CAN-08-2604. [DOI] [PubMed] [Google Scholar]
- 20.Casson AG, Zheng Z, Porter GA, Guernsey DL. Genetic polymorphisms of microsomal epoxide hydroxylase and glutathione s-transferases m1, t1 and p1, interactions with smoking, and risk for esophageal (barrett) adenocarcinoma. Cancer Detect Prev. 2006;30:423–431. doi: 10.1016/j.cdp.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Casson AG, Zheng Z, Chiasson D, MacDonald K, Riddell DC, Guernsey JR, Guernsey DL, McLaughlin J. Associations between genetic polymorphisms of phase i and ii metabolizing enzymes, p53 and susceptibility to esophageal adenocarcinoma. Cancer Detect Prev. 2003;27:139–146. doi: 10.1016/s0361-090x(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 22.Lee JM, Lee YC, Yang SY, Shi WL, Lee CJ, Luh SP, Chen CJ, Hsieh CY, Wu MT. Genetic polymorphisms of p53 and gstp1,but not nat2,are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 2000;89:458–464. doi: 10.1002/1097-0215(20000920)89:5<458::aid-ijc10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 23.Vos M, Adams CH, Victor TC, van Helden PD. Polymorphisms and mutations found in the regions flanking exons 5 to 8 of the tp53 gene in a population at high risk for esophageal cancer in south africa. Cancer Genet Cytogenet. 2003;140:23–30. doi: 10.1016/s0165-4608(02)00638-6. [DOI] [PubMed] [Google Scholar]
- 24.Hong Y, Miao X, Zhang X, Ding F, Luo A, Guo Y, Tan W, Liu Z, Lin D. The role of p53 and mdm2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res. 2005;65:9582–9587. doi: 10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- 25.Cai L, Mu LN, Lu H, Lu QY, You NC, Yu SZ, Le AD, Zhao J, Zhou XF, Marshall J, Heber D, Zhang ZF. Dietary selenium intake and genetic polymorphisms of the gstp1 and p53 genes on the risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:294–300. doi: 10.1158/1055-9965.EPI-05-0680. [DOI] [PubMed] [Google Scholar]
- 26.Yang W, Zhang Y, Tian X, Ning T, Ke Y. P53 codon 72 polymorphism and the risk of esophageal squamous cell carcinoma. Mol Carcinog. 2008;47:100–104. doi: 10.1002/mc.20368. [DOI] [PubMed] [Google Scholar]
- 27.Liu G, Cescon DW, Zhai R, Zhou W, Kulke MH, Ma C, Xu W, Su L, Asomaning K, Heist RS, Wain JC, Lynch TJ, Christiani DC. P53 arg72pro, mdm2 t309g and ccnd1 g870a polymorphisms are not associated with susceptibility to esophageal adenocarcinoma. Dis Esophagus. 2010;23(1):36–9. doi: 10.1111/j.1442-2050.2009.00960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin DX, Tang YM, Peng Q, Lu SX, Ambrosone CB, Kadlubar FF. Susceptibility to esophageal cancer and genetic polymorphisms in glutathione s-transferases t1, p1, and m1 and cytochrome p450 2e1. Cancer Epidemiol Biomarkers Prev. 1998;7:1013–1018. [PubMed] [Google Scholar]
- 29.Morita S, Yano M, Tsujinaka T, Ogawa A, Taniguchi M, Kaneko K, Shiozaki H, Doki Y, Inoue M, Monden M. Association between genetic polymorphisms of glutathione s-transferase p1 and n-acetyltransferase 2 and susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 1998;79:517–520. doi: 10.1002/(sici)1097-0215(19981023)79:5<517::aid-ijc12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 30.van Lieshout EM, Roelofs HM, Dekker S, Mulder CJ, Wobbes T, Jansen JB, Peters WH. Polymorphic expression of the glutathione s-transferase p1 gene and its susceptibility to barrett's esophagus and esophageal carcinoma. Cancer Res. 1999;59:586–589. [PubMed] [Google Scholar]
- 31.Tan W, Song N, Wang GQ, Liu Q, Tang HJ, Kadlubar FF, Lin DX. Impact of genetic polymorphisms in cytochrome p450 2e1 and glutathione s-transferases m1, t1, and p1 on susceptibility to esophageal cancer among high-risk individuals in china. Cancer Epidemiol Biomarkers Prev. 2000;9:551–556. [PubMed] [Google Scholar]
- 32.Roth MJ, Abnet CC, Johnson LL, Mark SD, Dong ZW, Taylor PR, Dawsey SM, Qiao YL. Polymorphic variation of cyp1a1 is associated with the risk of gastric cardia cancer: A prospective case-cohort study of cytochrome p-450 1a1 and gst enzymes. Cancer Causes Control. 2004;15:1077–1083. doi: 10.1007/s10552-004-2233-3. [DOI] [PubMed] [Google Scholar]
- 33.Murphy SJ, Hughes AE, Patterson CC, Anderson LA, Watson RG, Johnston BT, Comber H, McGuigan J, Reynolds JV, Murray LJ. A population-based association study of snps of gstp1, mnsod, gpx2 and barrett's esophagus and esophageal adenocarcinoma. Carcinogenesis. 2007;28:1323–1328. doi: 10.1093/carcin/bgm007. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JM, Wu MT, Lee YC, Yang SY, Chen JS, Hsu HH, Huang PM, Kuo SW, Lee CJ, Chen CJ. Association of gstp1 polymorphism and survival for esophageal cancer. Clin Cancer Res. 2005;11:4749–4753. doi: 10.1158/1078-0432.CCR-04-2333. [DOI] [PubMed] [Google Scholar]
- 36.Berhane K, Widersten M, Engstrom A, Kozarich JW, Mannervik B. Detoxication of base propenals and other alpha, beta-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc Natl Acad Sci U S A. 1994;91:1480–1484. doi: 10.1073/pnas.91.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundberg K, Johansson AS, Stenberg G, Widersten M, Seidel A, Mannervik B, Jernstrom B. Differences in the catalytic efficiencies of allelic variants of glutathione transferase p1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis. 1998;19:433–436. doi: 10.1093/carcin/19.3.433. [DOI] [PubMed] [Google Scholar]
- 38.Zimniak P, Nanduri B, Pikula S, Bandorowicz-Pikula J, Singhal SS, Srivastava SK, Awasthi S, Awasthi YC. Naturally occurring human glutathione s-transferase gstp1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem. 1994;224:893–899. doi: 10.1111/j.1432-1033.1994.00893.x. [DOI] [PubMed] [Google Scholar]
- 39.Hu X, Xia H, Srivastava SK, Herzog C, Awasthi YC, Ji X, Zimniak P, Singh SV. Activity of four allelic forms of glutathione s-transferase hgstp1-1 for diol epoxides of polycyclic aromatic hydrocarbons. Biochem Biophys Res Commun. 1997;238:397–402. doi: 10.1006/bbrc.1997.7311. [DOI] [PubMed] [Google Scholar]
- 40.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121:1643–1658. doi: 10.1002/ijc.23044. [DOI] [PubMed] [Google Scholar]
- 41.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DEJr, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 42.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CAJr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 43.Malkin D, Li FP, Strong LC, Fraumeni JFJr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA. et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Xing D, He Z, Lin D. [p53 gene codon 72 polymorphism and susceptibility to esophageal squamous cell carcinoma in a chinese population] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:10–13. [PubMed] [Google Scholar]
- 45.Engel LS, Chow WH, Vaughan TL, Gammon MD, Risch HA, Stanford JL, Schoenberg JB, Mayne ST, Dubrow R, Rotterdam H, West AB, Blaser M, Blot WJ, Gail MH, Fraumeni JFJr. Population attributable risks of esophageal and gastric cancers. J Natl Cancer Inst. 2003;95:1404–1413. doi: 10.1093/jnci/djg047. [DOI] [PubMed] [Google Scholar]
- 46.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: A review. Mutat Res. 2004;567:447–474. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Malats N, Camus-Radon AM, Nyberg F, Ahrens W, Constantinescu V, Mukeria A, Benhamou S, Batura-Gabryel H, Bruske-Hohlfeld I, Simonato L, Menezes A, Lea S, Lang M, Boffetta P. Lung cancer risk in nonsmokers and gstm1 and gstt1 genetic polymorphism. Cancer Epidemiol Biomarkers Prev. 2000;9:827–833. [PubMed] [Google Scholar]
- 48.Kuwano H, Kato H, Miyazaki T, Fukuchi M, Masuda N, Nakajima M, Fukai Y, Sohda M, Kimura H, Faried A. Genetic alterations in esophageal cancer. Surg Today. 2005;35:7–18. doi: 10.1007/s00595-004-2885-3. [DOI] [PubMed] [Google Scholar]