Abstract
Autism spectrum disorders are a group of highly heritable neurodevelopmental disorders with a complex genetic etiology. The International Molecular Genetic Study of Autism Consortium previously identified linkage loci on chromosomes 7 and 2, termed AUTS1 and AUTS5, respectively. In this study, we performed a high-density association analysis in AUTS1 and AUTS5, testing more than 3000 single nucleotide polymorphisms (SNPs) in all known genes in each region, as well as SNPs in non-genic highly conserved sequences. SNP genotype data were also used to investigate copy number variation within these regions. The study sample consisted of 127 and 126 families, showing linkage to the AUTS1 and AUTS5 regions, respectively, and 188 gender-matched controls. Further investigation of the strongest association results was conducted in an independent European family sample containing 390 affected individuals. Association and copy number variant analysis highlighted several genes that warrant further investigation, including IMMP2L and DOCK4 on chromosome 7. Evidence for the involvement of DOCK4 in autism susceptibility was supported by independent replication of association at rs2217262 and the finding of a deletion segregating in a sib-pair family.
Keywords: autistic disorder, disease susceptibility, single nucleotide polymorphisms, linkage disequilibrium, chromosome 7, chromosome 2
Introduction
Autism (OMIM: %209850) is a complex neurodevelopmental disorder characterized by impairments in reciprocal social interaction, difficulties in verbal and nonverbal communication, stereotyped behaviors and interests, and an onset in the first 3 years of life. Autism belongs to the group of pervasive developmental disorders (PDD), also known as autism spectrum disorders (ASDs), which also include Asperger syndrome and pervasive developmental disorder—not otherwise specified (PDD-NOS). The estimated population prevalence of core autism is around 15–20 in 10 000, with a male/female sex ratio of approximately 4:1. When all ASD subtypes are combined the prevalence is several times higher, reaching 116 in 10 000.1, 2, 3
Several lines of evidence indicate that genetic factors are important in susceptibility to idiopathic autism. Twin studies show a concordance of 60–92% for monozygotic (MZ) twins and 0–10% for dizygotic (DZ) twins, depending on phenotypic definitions, and the sibling recurrence risk is 25–60 times higher than the population prevalence.4 Furthermore, relatives of affected probands show a higher incidence of milder cognitive or behavioral features, consistent with the hypothesis of a ‘spectrum' of severity.5
Autism spectrum disorders exhibit wide clinical variability and a high degree of genetic heterogeneity. A variety of chromosomal abnormalities are found in a small proportion of affected individuals (6–7%), most frequently in syndromic cases with dysmorphic features and cognitive impairment.6 The autism phenotype is also associated with known genetic conditions such as the Fragile X syndrome and tuberous sclerosis. Recently, rare ASD-causing mutations were reported in a number of genes, including NLGN3, NLGN4,7 NRXN1,8 SHANK39 and NHE9.10
In recent years, the development of DNA microarray technologies has revealed that submicroscopic deletions and duplications of DNA, known as copy number variants (CNVs), may be significant in autism susceptibility.11, 12, 13, 14 Recent surveys identified a higher rate of de novo CNVs in autism pedigrees compared to controls, with the increased rate becoming more exaggerated in singleton than in multiplex families.10, 12, 13 Nevertheless, it remains difficult to interpret the significance of the numerous CNVs identified in ASDs, to distinguish those that influence susceptibility from normal polymorphic variation and to understand how they might interact with other genetic and non-genetic factors.
Although individually rare, highly penetrant abnormalities, such as microdeletions/microduplications or point mutations, may have a significant function in ASDs. It is also likely that genetic susceptibility may also result from the combined action of several common genetic variants. Common variation in several candidate genes has been implicated in autism (MET, CNTNAP2, SLC6A4, RELN, GABRB3),15 but in most cases consistent replication has not been achieved.
Because the strong genetic component in ASDs was clearly demonstrated over a decade ago, a large number of molecular genetic studies have searched for susceptibility genes, following the general approach of a genome-wide linkage scan using affected sibling/relative pair families. The International Molecular Genetic Study of Autism Consortium (IMGSAC) identified the first autism linkage locus on chromosome 7q21–q32 (designated autism susceptibility locus 1, AUTS1) with a multipoint maximum LOD score (MLS) of 2.53 in 87 families.16 This result was confirmed in follow-up studies conducted by the IMGSAC using additional families and markers.17, 18 Another linkage susceptibility locus (AUTS5) was identified by IMGSAC on chromosome 2q24–q33 with an MLS of 3.74 in 152 affected sibling pairs.17
Replication of linkage signals in independent studies has proven difficult for ASDs. To date, 13 whole-genome linkage scan for ASDs have been published,15 and no single locus has been consistently confirmed in all studies. This finding is likely to result from the small effect size attributable to individual genes, as well as from the clinical and genetic complexity of ASDs; differences in ascertainment and inclusion criteria may have been additional factors. However, AUTS1 is one of the few identified loci that has been supported by overlapping positive results in multiple multiplex collections,19, 20 and in meta-analyses.21, 22 Similarly, the chromosome 2q locus is supported by overlapping linkage findings in another two independent genome scans,23, 24 and by homozygosity mapping in consanguineous families.10 The largest genome scan published to date, carried out by the Autism Genome Project (AGP) using Affymetrix 10K single nucleotide polymorphism (SNP) arrays and 1181 multiplex families, also provided some support for both the chromosome 2q and 7q loci within the families of inferred European ancestry.8
Despite the support for linkage on chromosomes 2q and 7q, the candidate genomic intervals remain broad, each spanning approximately 40 Mb and containing approximately 200 known genes. Systematic screening and association studies of several positional candidate genes on chromosomes 2q and 7q have been conducted by the IMGSAC,25, 26, 27, 28, 29 but these studies have not led to the identification of confirmed autism susceptibility variants. Owing to the recent technological advances in high-density SNP genotyping and bioinformatic resources, we focused our efforts on performing a gene-based high-density SNP association study of the autism susceptibility loci on chromosomes 2q and 7q implicated by IMGSAC linkage studies. SNP genotype data were also used to investigate copy number variation within these regions. The genetic architecture of ASDs is likely to be extremely complex, with disease risk determined by both common variants of modest effect, as well as rare variants with a range of effect sizes. The strategy of focusing on linkage regions for fine-mapping studies by high-density association screens will prioritize genes containing penetrant rare variants, which would not be well identified through association analysis. However, we might expect that genes containing such variants also contain more common variants of lesser effect and thus are still natural candidates to follow-up through association studies.
Genotyping was conducted in two stages, based on HapMap Phase I and Phase II data, respectively. In total, 3002 SNPs were genotyped in each region, directly testing 173 genes on chromosome 2 and 270 genes on chromosome 7. The study sample consisted of 126 and 127 affected individuals and their parents, selected from 293 IMGSAC multiplex families based on identity-by-descent (IBD) sharing on chromosomes 2q and 7q, respectively, as well as 188 gender-matched controls. This study design, where the same probands are used for family-based and case–control analysis, should be more robust against the respective weaknesses of the case–control and TDT approaches (such as population structure and segregation distortion, respectively), and extract the maximum information from our sample.30 Moreover, by selecting families showing excess allele sharing in the region of interest, we are likely to increase the frequency of the disease-associated alleles in the case sample, thereby increasing the power of association studies.31 Power calculations were performed over a range of risk allele frequencies and odds ratios (OR), confirming that the strategy of selecting families for increased IBD sharing outperformed a strategy in which families are selected at random, given fixed genotyping resources (see Supplementary Information).
Our study thus represents a deep exploration of SNP and copy number variation within genic regions of the two autism linkage loci on chromosomes 2q and 7q and pinpoints several genes that need further investigation.
Materials and methods
Study populations
The chromosome 2 primary sample included 126 independent autism families, for 371 individuals (119 parent–parent–child trios and 7 single parent–child pairs). The chromosome 7 primary sample included 127 independent autism families (117 parent–parent–child trios and 10 single parent–child pairs). All families were Caucasian (Table 1). The assessment methods and diagnostic criteria used by the IMGSAC have been described in detail previously.17 Diagnosis was based on the Autism Diagnostic Interview—Revised (ADI-R) and the Autism Diagnostic Observation Schedule (ADOS) and clinical evaluation. Karyotypes were obtained on all affected individuals when possible, and gross karyotypic abnormalities were excluded in at least one affected individual per family in 93% of families and in both affected individuals in 83% of families.
Table 1. Description of samples.
| Autism sample | Controls | ||||||
|---|---|---|---|---|---|---|---|
| Total affected | Sex (M/F) | Family type | Country of origin | Total | Sex (M/F) | Country | |
| IMGSAC chr. 2 | 126 | 103:23 | PPC 119, PC 7 | 73 UK, 25 USA, 16 Netherlands, 8 Germany, 3 France, 1 Greece | 188 | 154:34 | UK |
| IMGSAC chr. 7 | 127 | 101:26 | PPC 117, PC 10 | 66 UK, 28 USA, 13 Netherlands, 9 France, 7 Germany, 3 Denmark, 1 Greece | 188 | 148:40 | UK |
| IMGSAC replication | 294 | 236:58 | PPC 213, PC 47, C 34 | 129 UK, 85 Italy, 32 Germany, 31 Netherlands, 10 Denmark, 7 France | 180 | 144:36 | 133 UK, 47 Italy |
| ND | 96 | 85:11 | PPC 82, PC 14 | North of the Netherlands | |||
| ND-all | 204 | 175:29 | PPC 165, PC 39 | North of the Netherlands | |||
Abbreviations: C, single case; F, female; IMGSAC, International Molecular Genetic Study of Autism Consortium; M, male; ND, Northern Dutch; PC, parent–child pairs; PPC, parent–parent–child trios.
Trios for the primary sample were selected from the 293 multiplex families in the IMGSAC multiplex collection (using one affected sib per family) based on IBD sharing on chromosomes 2q and 7q, respectively. Calculation of IBD states was based on microsatellite marker data available from our genome scan18 and fine-mapping studies (unpublished data). Ranked Z-scores were calculated for each family using Merlin32 at the linkage peak position (D2S2302-D2S2310 and D7S2430-D7S684 for chromosomes 2 and 7, respectively).
Two main sample collections were used for replication (Table 1): (1) ‘IMGSAC replication' (IMGSAC-R) sample: 260 parent-affected child trios or pairs and 34 single cases and (2) ‘Northern Dutch' sample (ND): 96 singleton families from the north of the Netherlands, including 82 parent–parent–child trios and 14 parent–child pairs. Both replication sample collections fulfilled diagnostic criteria for Case ‘Type 1' or ‘Type 2' as defined by IMGSAC17 (meet ADI-R criteria or one point below threshold on one behavioral domain, meet ADOS/ADOS-G criteria for autism or PDD, performance IQ>35). An extended Northern Dutch sample (ND-all; Table 1) was available, including 108 cases that did not meet stringent criteria for one of the following reasons: (1) met ADI-R criteria but failed to meet ADOS criteria or did not undergo ADOS evaluation, (2) met ADI-R and ADOS criteria but had an IQ score <35, (3) did not meet full criteria for ASD on the ADI-R.
The most significant SNPs from the chromosome 2 locus were also tested in a collection of 358 multiplex families (‘Mount Sinai' sample), which have been previously described.23, 33 Similarly, three SNPs from two of the most strongly associated genes in the case–control and family-based analysis on chromosome 7 were genotyped in 62 Caucasian families selected for IBD sharing from a sample of 222 families showing linkage to the same region of chromosome 719 (‘University of Washington' sample).
Controls used in the primary experiment included 188 DNA samples from UK random blood donors from the ECACC HRC panels,34 sex-matched with the autism case sample. The additional set of 180 controls genotyped in the replication phase included 92 DNAs from ECACC HRC panels, 41 random donors from the UK and 47 random donors from Italy.
The study was reviewed by the relevant local ethics committees.
Genotyping
Single nucleotide polymorphisms for the primary analysis were genotyped using the GoldenGate assay (Illumina, San Diego, CA, USA) on an Illumina BeadStation according to the manufacturer's instructions. BeadArrays were scanned using the BeadArray Reader at 532 and 647 nm. BeadStudio genotyping module (version 3.2.23) was used to generate genotypes.
Genotyping was conducted in two parallel stages for both chromosomal loci. A total of 3072 SNPs were genotyped in each stage using two custom 1536-plex Illumina arrays, one for each chromosome. The regions of interest ranged from 94.246 to 136.661 Mb on chromosome 7 and from 152.305 to 191.605 Mb on chromosome 2 (NCBI Build 36). These intervals were defined using the approximate 1-LOD drop of the linkage peaks on the two chromosomes, based on IMGSAC microsatellite marker data.18
In the first stage of this study, we evaluated the patterns of linkage disequilibrium (LD) and the distribution of haplotype blocks in the CEU genotype data from the HapMap project release 13 (HapMap Phase I data). Genic regions were defined by NCBI Build 34, by merging all RefSeq and UCSC Known Genes, including all exonic, intronic and 3′ UTR sequences, as well as 5 kb upstream of the 5′ end. A total of 1496 tag SNPs on both chromosome 2q and 7q were identified using HaploView35 and the Gabriel algorithm for block definition from LD blocks overlapping all genic regions.
In the second stage of genotyping, we took advantage of the higher-density HapMap Phase II data to better represent genetic variation in regions of lower LD not previously captured by the HapMap Phase I data. We also used the latest genome annotation (NCBI Build 36) to investigate novel genes and ensure comprehensive coverage of all intragenic and putative regulatory regions on both chromosomes. We identified ‘non-genic' evolutionary conserved regions from PhastCons elements.36 We downloaded SNP genotype data from the CEU population from HapMap release 22, and selected all SNPs in all genic regions and in the top 5% of non-genic PhastCons elements. We also selected all nonsynonymous SNPs with minor allele frequency (MAF) ⩾0.05. We then used the Tagger program from HaploView35 (version 4) to select a second set of 1516 tag SNPs for each chromosomal region. Parameters used for Tagger were r2⩾0.75 (chromosome 2) and r2⩾0.63 (chromosome 7), minimum MAF of 5%, aggressive tagging and force including SNPs already genotyped in stage 1. We estimated that our two sets of SNPs were able to tag 96 and 85% of intragenic HapMap SNP variation (MAF>0.05) with r2>0.8 on chromosomes 2 and 7, respectively.
Genotypes for 212 SNPs (99 on chromosome 2 and 113 on chromosome 7), previously generated by the AGP using the Affymetrix 10K version 2 SNP array,8 were available on the IMGSAC family sample and were also included in the family-based association analysis.
A total of 50 genome-wide unlinked SNPs were genotyped for detection of population stratification,37 and 10 chromosome X SNP were also included to estimate levels of mistyping. In addition, for regions of high LD, where tagging SNPs captured the most genetic variation, extra SNPs were chosen in case of genotyping failure.
Replication SNP genotyping
Single nucleotide polymorphisms for replication were genotyped using a combination of the Mass Extend iPLEX Gold (Sequenom, San Diego, CA, USA) and TaqMan platforms. A 100% genotyping concordance was observed for two replicate DNA samples genotyped in each experiment. Twenty-five genome-wide SNPs were also genotyped in the IMGSAC-R sample to test for population stratification.
Statistical analysis
Association analysis
We evaluated evidence of association using both ‘frequentist' and Bayesian statistical approaches.
Primary association analysis of the 5880 SNPs (including the 212 SNPs available from the AGP linkage study8) successfully genotyped in the IMGSAC data set at the two loci was carried out using the PLINK package.38 To extend the amount of information captured by single-marker tests, an additional set of two-marker haplotype tags was devised using the ‘aggressive' option of the Tagger program39 implemented in HaploView.35 In total, 3526 tests (2959 single-marker tests and 567 haplotype tags) were performed for the chromosome 2 study, and 3380 tests (2921 single-marker tests and 459 haplotype tags) for the chromosome 7 study.
Standard TDT from PLINK was used for family-based analysis, and the Cochran–Armitage trend test (1 degree of freedom) for the case–control analysis. Haplotype-based tests were calculated using PLINK.
Bayesian logistic regression analysis was performed using the GENEBPM algorithm,40, 41 again using both a case–control and family-based approach (see Supplementary Methods). The logistic regression model allowed for additive and dominance effects of unobserved causal variants, a main effect of gender as well as for parent-of-origin effects in the family-based analysis. GENEBPM analyses were performed using a sliding window of five SNPs across each chromosomal region. For comparison with frequentist single-SNP analyses, the GENEBPM algorithm was also applied to each SNP in turn (that is, single-SNP ‘haplotypes').
Replication analysis
Association analysis of the IMGSAC-R and ND replication data sets was carried out using the UNPHASED package,42 given the presence of a higher proportion of families with missing parents (24%) (Table 1). UNPHASED implements maximum-likelihood-based association analysis for nuclear families and unrelated subjects allowing for missing genotypes and uncertain haplotype phase. In the presence of missing data it has only minor loss of robustness to population stratification and is more powerful than standard TDT.42
Analysis of the combined primary and replication cohorts was also carried out using UNPHASED, again using both a case–control approach and a family-based approach. Only the IMGSAC and IMGSAC-R data sets were combined for the population-based meta-analysis, because appropriate controls were not available for the ND population.
Copy number variation
We used transmission patterns of SNP genotypes within parent–offspring families to detect Mendelian errors consistent with the presence of a deletion. In addition, the clustering of all SNP genotypes was visually examined to identify abnormal clustering patterns or outlying samples that might point to CNVs associated with the autism phenotype. Sequencing was carried out to confirm the presence of microdeletions, CNVs or secondary SNPs.
After exclusion of whole-genome amplified samples, data from both GoldenGate arrays were combined for each region, no-calls were deleted, and run on QuantiSNP version 1.0.43 CNV validation and screening was carried out by multiplex PCR and quantitative multiplex PCR of short fluorescent fragments (QMPSF).44 Positive results were confirmed in a second independent QMPSF assay.
The distal breakpoint of the deletion detected in family 15-0084 was better defined by quantitative PCR (qPCR) of DOCK4 exons 52, 37, 31, 14 and 7, with GAPDH as a reference.
Additional information is available as Supplementary Methods.
Results
Genotyping
A total of 6004 SNPs—3002 in each chromosome region—were genotyped using the Illumina GoldenGate technology. After quality control procedures, we excluded 336 markers for one or more of the following reasons: MAF<0.05, more than 1 Mendelian error, genotyping rate <90%, poor clustering and deviations from Hardy–Weinberg equilibrium (P<0.001) in the control population.
For the 5668 (94%) SNPs that passed quality control, the genotyping efficiency exceeded 99.7%, with an estimated error rate from duplicate SNPs and from heterozygote calls of X chromosome SNPs in males in the order of 2–5 × 10−4. In summary, 2860 SNPs from the chromosome 2q23.3–q32.3 region were successfully genotyped in 559 DNA samples including 126 affected individuals, 245 parents and 188 gender-matched controls from the ECACC collection; 2808 SNPs from the chromosome 7q21.3–q33 region were successfully genotyped in 559 DNA samples including 127 affected individuals, 244 parents and 188 ECACC gender-matched controls. In addition, our family-based analysis included genotypes from 212 SNPs (99 on chromosome 2 and 113 on chromosome 7), which were generated by the AGP using the Affymetrix 10K version 2 SNP array.8
There was no significant difference in the pattern of LD between our sample and the HapMap CEU sample, indicating that the LD structure in the HapMap CEU data can be readily applied to our autism sample. SNPs were selected to capture efficiently the large majority of the currently known variation in all intragenic regions and highly conserved non-genic elements (see Supplementary Methods).
Population stratification
The presence of stratification in a population-based association study that is not suitably accounted for in case–control analysis can lead to an increase in the false-positive error rate. Furthermore, haplotype analyses in family-based association studies are not robust to population stratification if random mating is assumed among parents in the haplotype estimation step.
We tested for population structure in our primary IMGSAC sample using Structure45, 46 software, and testing 50 unlinked genome-wide SNPs. Comparing the fit of the admixture model for K=1, 2 and 3 strata, we found strongest support for a model of no stratification (K=1) in both of the following groups of individuals: (1) probands, controls and HapMap CEU founders; and (2) parents and HapMap CEU founders. Similarly, no evidence of stratification was detected in the combined IMGSAC primary and IMGSAC-R sample, using 25 unlinked genome-wide SNP markers. These results reassure us that no strong population stratification is present in our IMGSAC primary and IMGSAC-R sample.
Association analysis
The results of the case–control (Cochran–Armitage trend test) and family-based analysis (TDT) are shown in Figure 1 and summarized in Table 2.
Figure 1.
Graphical representation of chromosome 2 and 7 association results. −Log10 P-values are plotted against the chromosome position. (a) P-values obtained for single markers (Cochran–Armitage trend test) and 2-SNP haplotype case–control association (PLINK). (b) P-values for single-marker TDT and 2-SNP haplotype TDT.
Table 2. Summary of primary association results.
| Family-based | Case Control | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP/haplotype | Chr. | Position | Gene | Risk allele | P-value | LogBF | P-value | LogBF |
| rs1427395 | 2 | 153 442 168 | PhastConsa | T | 0.0016 | 0.78 | 0.0133 | 0.66 |
| rs3769357 | 2 | 157 101 520 | GPD2 | A | 0.0031 | 1.04 | ||
| rs6437129, rs6709528_CC | 2 | 158 669 000 | UPP2 | CC | 9.72E-04 | 1.00 | ||
| rs6437133 | 2 | 158 672 533 | UPP2 | C | 0.0045 | 0.65 | ||
| rs6709528 | 2 | 158 678 671 | UPP2 | T | 8.00E-04 | 1.09 | ||
| rs12620556 | 2 | 158 905 017 | LOC130940 | A | 0.0041 | 0.45 | 0.0153 | 0.55 |
| rs764660 | 2 | 165 921 543 | SCN2A | C | 0.0047 | 0.68 | ||
| rs1020626 | 2 | 166 106 880 | FAM130A2 | T | 3.80E-04 | 1.92 | ||
| rs10930170 | 2 | 166 107 713 | FAM130A2 | G | 0.0020 | 1.40 | ||
| rs829957 | 2 | 169 367 080 | NOSTRIN | T | 0.0116 | 0.59 | 9.03E-05 | 2.47 |
| rs6433093 | 2 | 169 367 190 | NOSTRIN | A | 5.59E-04 | 1.94 | ||
| rs7583629 | 2 | 169 381 125 | NOSTRIN | A | 0.0027 | 1.09 | 3.22E-05 | 2.92 |
| rs482435 | 2 | 169 384 291 | NOSTRIN | C | 0.0084 | 0.78 | 1.39E-04 | 2.54 |
| rs2098802 | 2 | 170 760 429 | MYO3B | G | 0.0042 | 1.02 | ||
| rs6738892 | 2 | 170 768 975 | MYO3B | A | 0.0035 | 1.13 | ||
| rs13007575 | 2 | 174 386 625 | PhastConsa | A | 0.0077 | 0.81 | 0.0026 | 1.19 |
| rs6717587 | 2 | 175 865 296 | PhastConsa | A | 0.0015 | 1.40 | ||
| rs1434087 | 2 | 178 912 043 | OSBPL6 | T | 0.0010 | 1.56 | ||
| rs7590028 | 2 | 180 257 688 | ZNF533 | T | 0.0010 | 1.80 | ||
| rs11885327 | 2 | 180 276 318 | ZNF533 | C | 8.00E-04 | 1.56 | 0.0271 | 0.56 |
| rs11885327, rs1964081_TG | 2 | 180 287 992 | ZNF533 | TG | 0.0019 | 1.54 | ||
| rs2008230, rs1964081_GG | 2 | 180 288 449 | ZNF533 | GG | 0.0030 | 1.04 | ||
| rs881737, rs1964081_GG | 2 | 180 294 093 | ZNF533 | GG | 0.0016 | 1.75 | ||
| rs1964081 | 2 | 180 299 666 | ZNF533 | A | 0.0014 | 1.61 | ||
| rs2126424, rs1139_CG | 2 | 180 312 034 | ZNF533 | CG | 0.0073 | 1.18 | 0.0038 | 1.11 |
| rs1139 | 2 | 180 318 326 | ZNF533 | G | 0.0067 | 1.73 | 0.0049 | 1.26 |
| rs415994 | 2 | 183 266 932 | 5′ of DNAJC10 | C | 0.0027 | 1.54 | ||
| rs3755248 | 2 | 188 078 477 | TFPI | T | 0.0036 | 0.90 | ||
| rs7573488 | 2 | 188 106 325 | TFPI | G | 0.0046 | 0.91 | ||
| rs3811608 | 2 | 191 043 302 | FLJ20160 | T | 0.0023 | 0.81 | ||
| rs6757698 | 2 | 191 071 741 | FLJ20160 | C | 0.0012 | 0.31 | ||
| rs12538145 | 7 | 95 636 377 | SLC25A13 | C | 0.0039 | 0.52 | 0.0168 | 0.55 |
| rs2307355 | 7 | 99 531 488 | MCM7 | A | 0.0040 | 0.88 | ||
| rs11768465 | 7 | 100 036 322 | FBX024 | C | 0.0184 | 0.57 | 0.0031 | 1.21 |
| rs875659 | 7 | 101 696 376 | CUX1 | C | 2.00E-04 | 1.82 | ||
| rs3819479 | 7 | 103 184 318 | RELN | T | 0.0028 | 1.02 | ||
| rs6976167 | 7 | 103 848 209 | LHFPL3 | T | 0.0026 | 1.16 | ||
| rs12666599 | 7 | 103 905 157 | LHFPL3 | T | 0.0038 | 0.89 | ||
| rs4730037 | 7 | 104 129 973 | LHFPL3 | C | 0.0032 | 2.07 | ||
| rs176481 | 7 | 105 515 161 | SYPL1 | T | 0.0385 | 8.19E-04 | 1.92 | |
| rs9690688 | 7 | 107 507 398 | LAMB4 | T | 0.0047 | 0.75 | ||
| rs6951925 | 7 | 108 588 860 | NT_007933.689b | G | 0.0029 | 1.20 | ||
| rs1464895 | 7 | 110 111 977 | IMMP2L | A | 0.0049 | 0.95 | ||
| rs2030781 | 7 | 110 149 994 | IMMP2L | C | 0.011c | 1.57 | 0.0037 | 1.15 |
| rs12537269 | 7 | 110 184 783 | IMMP2L | A | 1.20E-04 | 2.85 | ||
| rs10500002 | 7 | 110 229 091 | IMMP2L | T | 0.0012 | 1.58 | ||
| rs1528039 | 7 | 110 230 008 | IMMP2L | C | 6.28E-04 | 1.77 | ||
| rs12531640 | 7 | 110 266 771 | IMMP2L | T | 0.0021 | 1.27 | ||
| rs2217262 | 7 | 111 583 613 | DOCK4 | A | 0.0143 | 0.41 | 0.0042 | 1.02 |
| rs989613 | 7 | 113 233 792 | NT_007933.632b | G | 0.0049 | 0.53 | ||
| rs7807053 | 7 | 120 137 743 | KCND2 | A | 0.0022 | 1.18 | ||
| rs41620 | 7 | 120 213 054 | 3′ of TSPAN12 | A | 0.0046 | 1.07 | ||
| rs2525720 | 7 | 120 392 266 | ING3 | A | 0.0049 | 1.25 | ||
| rs538558 | 7 | 121 724 673 | 3′ of FEZF1 | A | 0.0065 | 0.98 | ||
| rs11978485 | 7 | 122 480 367 | 3′ of SLC13A1 | G | 0.0295 | 0.98 | 0.0039 | 1.23 |
| rs6962740 | 7 | 128 614 047 | 5′ of SMO | G | 3.39E-04 | 2.14 | ||
| rs4110091 | 7 | 128 719 985 | AHCYL2 | T | 0.0012 | 1.56 | ||
| rs2030974 | 7 | 129 693 119 | 5′ of CPA2 | C | 0.0197 | 0.56 | 0.0032 | 1.17 |
| rs2171493 | 7 | 129 693 383 | 5′ of CPA2 | C | 0.0412 | 0.39 | 0.0038 | 1.18 |
| rs13226219 | 7 | 129 806 727 | 5′ of CPA1 | T | 0.0032 | 1.26 | ||
| rs1863009 | 7 | 130 649 715 | AK054623 | T | 9.20E-04 | 1.60 | ||
| rs7787173 | 7 | 131 107 683 | NT_007933.1017b | A | 6.00E-04 | 1.12 | ||
| rs4731863 | 7 | 131 674 323 | PLXNA4 | T | 1.00E-04 | 2.02 | 0.0321 | 0.45 |
Only SNPs showing P<0.005 in either family-based or case–control analysis are reported. P-values >0.05 are not shown. P-values<0.001 are in bold. The reported risk allele is consistent in the two approaches.
PhastCons, highly conserved region.
Predicted genes, reference sequence annotation changed from Build 34.
Parent-specific TDT.
Chromosome 2 association results
Three SNPs in the NOSTRIN gene provided the strongest association in the case–control analysis (rs7583629, P=3.2 × 10−5; rs829957, P=9.0 × 10−5; rs482435, P=1.4 × 10−4), followed by rs1020626 (P=3.8 × 10−4) in the FAM130A2 gene.
For the TDT analysis the strongest results came from SNPs in the ZNF533 gene (rs11885327, P=8.0 × 10−4; rs1964081, P=1.4 × 10−3), and an SNP in the UPP2 gene (rs6709528, P=8.0 × 10−4).
Single-marker logistic regression analysis provided a similar ranking of results. In the case–control analysis the most strongly associated SNP rs7583629 in NOSTRIN provided a log10 Bayes factor (logBF) of 2.9, whereas in the family-based analysis the top signal was for rs1139 in the ZNF533 gene (logBF=1.7). GENEBPM multimarker analysis using 5-SNP sliding windows (Supplementary Figure S1) showed increased evidence in favor of association for the NOSTRIN locus (logBF=3.2) in the case–control analysis, but did not identify additional interesting signals. Family-based multimarker analysis revealed an additional association signal with a haplotype spanning 75 kb in the METTL8 gene (logBF=2.3).
Chromosome 7 association results
The strongest signal for the case–control (trend test) analysis was from IMMP2L (rs12537269, P=1.2 × 10−4; rs1528039, 6.3 × 10−4) and just upstream of SMO (rs6962740, P=3.4 × 10−4). The TDT test implicated Plexin A4 (PLXNA4; rs4731863, P=1.0 × 10−4) and cut-like homeobox 1 isoform b (CUX1; rs875659, P=2.0 × 10−4).
Single-marker logistic regression analysis provided a similar ranking of results in the case–control analysis with rs12537269 (logBF=2.9) in IMMP2L showing the most significance. In the family-based analysis, the most significant result was seen for rs4730037 in LHFPL3 (logBF=2.1), closely followed by rs4731863 in PLXNA4 (logBF=2.0). Moreover, GENEBPM revealed a parent-of-origin effect in the IMMP2L locus, with increased risk for causal variants inherited from the father compared to those inherited from the mother. For this reason we investigated SNPs in IMMP2L by parent-specific TDT, which revealed a P-value of 0.01 for rs2030781, with a transmitted/untransmitted allele ratio of 31:14 for paternal transmissions (Table 2).
GENEBPM multimarker analysis using 5-SNP sliding windows showed increased evidence of association for the IMMP2L locus in the case–control analysis (logBF=2.9) and for PLXNA4 (logBF=2.9) in the family-based analysis, but did not identify additional interesting signals (Supplementary Figure S1).
Analysis of the LD landscape across the AUTS1 region, using both HapMap (CEU) and data from the 127 probands used in our primary sample, indicated that the six associated SNPs in IMMP2L (Table 2) are all within a single block of LD, and thus likely to be indexing the same effect. In contrast, the modest association seen in the first intron of the neighboring DOCK4 gene was in a separate block of LD.
Replication
We attempted replication of 56 SNPs (28 on each chromosome) that attained the most significant association results in primary case–control and TDT analyses (Table 3; Supplementary Table S1). The replication population consisted of the IMGSAC-R and the ND collections, including 390 affected individuals (see Table 1; Materials and methods for a description of samples). Family-based analysis of the replication sample showed significant overtransmission of the common allele of SNP rs2217262 in the DOCK4 gene (P=9.2 × 10−4, OR=2.28, confidence interval 1.37–3.77) (Table 3; Supplementary Table S1). This result remains significant after Bonferroni correction for multiple testing (28 SNPs tested on chromosome 7, P=0.026). The trend toward association of rs2217262 (P=0.029) was also seen in the extended ND sample, which included additional subjects fulfilling broader diagnostic criteria (ND-all, 204 affected subjects; Table 1).
Table 3. Family-based analysis of replication samples using UNPHASED.
| IMGSAC-R (294 affected subjects) | ND (96 affected subjects) | IMGSAC-R+ND (390 affected subjects) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr. | Gene | Alleles | Risk allele | P-value | Ca-Freq | Co-Freq | P-value | Ca-Freq | Co-Freq | P-value | Ca-Freq | Co-Freq |
| rs1427395 | 2 | PhastConsa | A/T | T | 0.3634 | 0.564 | 0.532 | 0.0216 | 0.500 | 0.387 | 0.0505 | 0.547 | 0.496 |
| rs6437133 | 2 | UPP2 | C/T | C | 0.1630 | 0.543 | 0.500 | 0.0395 | 0.482* | 0.599* | 0.7106 | 0.529 | 0.519 |
| rs12620556 | 2 | LOC130940 | A/G | A | 0.8454 | 0.899 | 0.905 | 0.0235 | 0.905* | 0.965* | 0.2247 | 0.901 | 0.920 |
| rs13007575 | 2 | PhastConsa | A/G | A | 0.1337 | 0.921 | 0.946 | 0.0063 | 0.958 | 0.886 | 0.8726 | 0.931 | 0.929 |
| rs1434087 | 2 | OSBPL6 | C/T | T | 0.0399 | 0.928 | 0.890 | 0.7597 | 0.916 | 0.925 | 0.0988 | 0.925 | 0.898 |
| rs11768465 | 7 | FBX024 | C/T | C | 0.3915 | 0.785 | 0.765 | 0.0184 | 0.761* | 0.863* | 0.6561 | 0.779 | 0.790 |
| rs1464895 | 7 | IMMP2L | A/G | A | 0.4854 | 0.161 | 0.145 | 0.0042 | 0.120* | 0.235* | 0.3043 | 0.150 | 0.170 |
| rs12537269 | 7 | IMMP2L | A/G | A | 0.0485 | 0.262 | 0.210 | 0.7737 | 0.255 | 0.243 | 0.0667 | 0.260 | 0.220 |
| rs2217262 | 7 | DOCK4 | A/C | A | 0.0272 | 0.955 | 0.924 | 0.0055 | 0.979 | 0.916 | 9.21E-04 | 0.962 | 0.921 |
| rs2171493 | 7 | 5′ of CPA2 | A/C | C | 0.0230 | 0.242* | 0.301* | 0.8506 | 0.216 | 0.224 | 0.0459 | 0.235* | 0.282* |
| rs4731863 | 7 | PLXNA4 | A/T | T | 0.1591 | 0.907 | 0.931 | 0.0987 | 0.891 | 0.938 | 0.0391 | 0.903* | 0.934* |
Abbreviations: Ca-Freq, frequency in affected offsprings; Co-Freq, frequency in untransmitted parental alleles; IMGSAC-R, International Molecular Genetic Study of Autism Consortium-replication; ND, Northern Dutch; SNP, single nucleotide polymorphism.
Only nominal P-values <0.05 are shown. Allele frequencies are reported for the risk allele detected in the primary association analysis. Flip-flop of associated allele is flagged by an asterisk.
PhastCons, highly conserved region.
The remaining SNPs did not show significant replication after correction for multiple testing, and no parent-of-origin effects were seen for rs2030781.
Finally, the 56 SNPs selected for replication were investigated in the combined primary and replication data sets; only 7 SNPs attained uncorrected significance of P<0.001 (Table 4). The DOCK4 SNP rs2217262 reached a nominal significance of P=5.23 × 10−5 in the family-based analysis of all cohorts (IMGSAC primary, IMGSAC-R and ND). In the case–control analysis of the combined IMGSAC collections (421 cases and 368 controls), rs12537269 in IMMP2L achieved the most significant result (P=7.3 × 10−5). Additional loci retaining association evidence in case–control meta-analysis were ZNF533 on chromosome 2, and TSPAN12, FEZF1 and SLC13A1 on chromosome 7.
Table 4. Combined analysis of primary and replication samples.
| All samples combineda Family-based analysis | IMGSAC samples combinedb Case–control analysis | ||||||
|---|---|---|---|---|---|---|---|
| Chr | SNP | Gene location | Risk allele | P-value (Ca-Co Freq) | OR (CI) | P-value (Ca-Co Freq) | OR (CI) |
| 2 | rs7590028 | ZNF533 intronic | T | 0.3227 | 6.56E-04 | 1.41 | |
| (0.52, 0.50) | (0.54, 0.45) | (1.16–1.72) | |||||
| 7 | rs2030781 | IMMP2L intronic | C | 0.08613 | 4.63E-04 | 1.53 | |
| (0.25, 0.22) | (0.27, 0.19) | (1.20–1.95) | |||||
| 7 | rs12537269 | IMMP2L intronic | A | 0.01047 | 7.26E-05 | 1.62 | |
| (0.27, 0.22) | (0.27, 0.19) | (1.27–2.06) | |||||
| 7 | rs2217262 | DOCK4 intronic | A | 5.23E-05 | 2.37 | 1.75E-03 | 2.08 |
| (0.96, 0.92) | (1.53–3.68) | (0.96, 0.92) | (1.31–3.32) | ||||
| 7 | rs41620 | 3′ of TSPAN12 | A | 0.08796 | 8.14E-04 | 1.48 | |
| (0.77, 0.74) | (0.78, 0.71) | (1.18–1.86) | |||||
| 7 | rs538558 | 3′ of FEZF1 | A | 0.4688 | 5.77E-04 | 1.45 | |
| (0.36, 0.34) | (0.37, 0.28) | (1.17–1.80) | |||||
| 7 | rs11978485 | 3′ of SLC13A1 | G | 0.04972 | 2.89E-04 | 1.59 | |
| (0.82, 0.79) | (0.84, 0.76) | (1.24–2.04) | |||||
Abbreviations: Ca-Co freq, risk allele frequency in affected offspring and in untransmitted parental alleles (family-based) or in control (case–control); IMGSAC, International Molecular Genetic Study of Autism Consortium; OR (CI), odds ratio and 95% confidence interval; SNP, single nucleotide polymorphism. Results generated by UNPHASED. Only SNPs with nominal P<0.001 are shown. P-values<0.001 are in bold.
IMGSAC primary sample, IMGSAC-R, ND (515–516 affected individuals).
IMGSAC primary sample, IMGSAC-R (420–421 cases, 368 controls).
Several SNPs in the most interesting genes from the primary analysis were also tested in two additional family collections, which had previously shown evidence of linkage to the chromosome 2q and 7q loci19, 23, 33 (Supplementary Table S1). Five SNPs in NOSTRIN, ZNF533 and OSBPL6 were tested in a sample of 358 multiplex families (‘Mount Sinai' cohort),23, 33 but no significant results were obtained. Of the 28, 3 AUTS1 replication SNPs in IMMP2L and CUX1 were genotyped in 62 Caucasian families selected for IBD sharing from 222 families showing linkage to the same region of chromosome 7 (‘University of Washington' sample),19 again with no evidence for association.
Copy number variation
A Mendelian error in one family for SNP rs7585982 pinpointed a potentially interesting deletion in the UPP2 gene on chromosome 2. The deletion boundaries were defined by sequence analysis of additional SNPs flanking rs7585982. Using long-range PCR followed by sequencing, we refined the deletion to 5897 bp of the UPP2 gene (158 681 612–158 687 508 bp; UCSC Build 36), removing two coding exons (exons 6 and 7) and predicted to cause a frameshift leading to a premature termination codon (Supplementary Figure S2A). This deletion was not present in the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation/), suggesting it could be an autism-specific CNV. We screened the same sample used for the SNP association experiment (126 cases and 188 controls) for the presence of this deletion using multiplex PCR (Supplementary Figure S2B). The frequency of the deletion was not significantly different between cases and controls (1.6 and 3.2%, respectively, P=0.2). To investigate if the deletion segregates with the ASD phenotype, we also screened 265 sib-pair families from the IMGSAC collection, including relatives of the 126 cases. Of these, we found 30 families with a parent carrying the deleted allele, and in only 13 families was it transmitted to affected children (in 5 families to both affected siblings and in 8 families to a single affected individual). These results suggest that the UPP2 deletion is not involved in autism susceptibility. The coding sequence of UPP2 was also sequenced in 47 unrelated subjects, including 12 probands carrying the deletion of exons 6 and 7; no novel coding variants were identified, except one silent change in exon 4 in only one individual.
By combining data from both SNP arrays for each candidate region, a sufficient SNP density was achieved to carry out copy number analysis on these samples using QuantiSNP.43 We detected 17 CNVs in seven regions of chromosome 7 and 6 CNVs in five regions of chromosome 2 (Supplementary Table S2). For the chromosome 7 analysis, an ∼800 kb duplication was detected in family 13-3023 that was transmitted from father to proband (Supplementary Figure S3). This duplication includes two genes: IMMP2L and DOCK4. Another duplication overlapping EMID2 and RABL5 was detected in three families where it was transmitted from mother to proband, whereas a smaller duplication containing only EMID2 was detected in a father, but not transmitted, and in one control. A third CNV in EXOC4 was detected as a nontransmitted loss in a father and as a gain (four copies) in a control.
On chromosome 2, five duplications and one deletion were detected in parents and a single control, but never transmitted to an affected child (Supplementary Table S2).
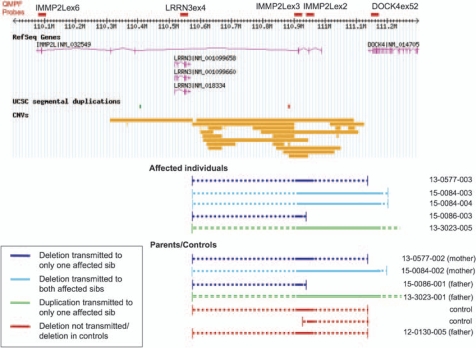
Most of the identified CNVs are well represented in the DGV, suggesting that they do not have a major function in autism susceptibility. However, the duplication involving IMMP2L and DOCK4 warranted further analysis, as it involved two adjacent genes showing possible SNP association with autism. Therefore we developed a QMPSF assay able to simultaneously test CNVs in exons 2, 3 and 6 of IMMP2L, exon 4 of LRRN3 and the last exon of DOCK4 (number 52). We validated the duplication in family 13-3023, identified by QuantiSNP, and verified that it is transmitted from the father to the affected son, but it was not transmitted to the other affected sib or to an unaffected sib. Screening of 475 UK controls and 285 IMGSAC multiplex families with 487 affected individuals was then carried out using the QMPSF assay, to check if CNVs in these genic regions segregated with the autism phenotype in families and/or have a higher frequency in cases than controls. We identified six additional deletions of different length, of which some were transmitted. One deletion disrupted exons 2 and 3 of IMMP2L and the last exon of DOCK4, and was transmitted from the mother to both affected sons, as well as to a daughter, who did not have an ASD. Both the carrier mother and daughter were reported to have dyslexia. qPCR indicated that the deletion distal breakpoint is located between exons 31 and 14 of DOCK4 (Supplementary Figure S4). Two smaller deletions were transmitted from the parent to only one of their affected children, one was found only in the father but not transmitted and the other two were found in controls. The relative length and position of the CNVs identified are depicted in Figure 2.
Figure 2.
Summary of IMMP2L and DOCK4 copy number variants (CNVs). Fragments tested by QMSPF are shown as red bars at the top. CNVs from the Database of Genomic Variants (DGV) are shown as orange bars. Deletions and duplications identified in affected individuals and in parents or controls are depicted at the bottom. Dashed and continuous lines indicate the maximum and minimum length of the CNVs, respectively. The distal breakpoint of the deletion in pedigree 15-0084 was defined by qPCR. The distal breakpoint of the duplication in pedigree 13-3023 was not defined precisely.
Discussion
Several linkage studies have suggested that chromosomes 2q and 7q may harbor one or more genes contributing to the risk for developing an ASD. Here, we have presented a comprehensive high-density SNP genotyping, association and CNV study covering the 2q23.3–q32.3 and 7q21.3–q33 chromosome regions. We have tested more than 3000 SNPs in each region, covering all known genes, as well as in highly conserved non-genic sequences.
The complementary case–control and family-based approach taken in our study allowed us to extract the maximum information from our sample, taking into consideration the advantages and disadvantages of the two different approaches. Case–control studies are more powerful compared to family-based approaches, but are sensitive to the presence of population stratification. Structure analysis using 50 genome-wide SNPs did not reveal strong population stratification, although we cannot exclude that undetected low levels may be present. Family-based approaches are more robust to confounding by population stratification and in addition they enable testing for parent-of-origin effects.
Although the strongest signals identified by the two approaches did not coincide, comparison of the results led us to pinpoint the most interesting loci supported by both methods, albeit with different strength. In addition, consistency of the results obtained by frequentist and Bayesian approaches suggested that our strongest signals are independent of the analysis method.
Primary association analysis of the chromosome 2 region identified the most interesting results in NOSTRIN, UPP2 and ZNF533. NOSTRIN encodes the nitric oxide synthase trafficker. Interestingly, the nitric oxide signaling pathway has been recently shown to be overrepresented in genes disrupted by CNVs in schizophrenia.47 However, the NOSTRIN association was stronger in the case–control analysis with only minor support from the TDT, and it was not confirmed in the replication sample or in the combined meta-analysis, suggesting that it might represent a false-positive result.
Similarly, the ZNF533 association was not replicated, however rs7590028 remained one of the strongest signals in case–control combined analysis of IMGSAC samples. ZNF533 encodes a protein containing four matrin-type zinc fingers and is highly conserved in evolution. Given its putative nuclear location, it is thought to act as a repressor of transcription, although no specific targets are currently known. ZNF533 is widely expressed in adult tissues, including brain. Expression of all isoforms in fetal brain was confirmed by reverse transcriptase–PCR (data not shown). Deletions including ZNF533 have been described in several patients with a neurological phenotype including mental retardation,48, 49 and other zinc-finger genes have also been implicated in mental retardation cases.50, 51, 52 The zinc-finger gene ZNF804A was recently identified as the strongest result in a genome-wide association study of schizophrenia and bipolar disorder,53 suggesting that they may act as transcription regulators in a wide range of human cognitive processes.
On chromosome 7, the most significant association result from the primary cohort was in the IMMP2L gene. Although SNPs in this gene failed to replicate in independent samples, the IMMP2L intronic SNP rs12537269 achieved the strongest result in the case–control meta-analysis of the IMGSAC sample (P=7.3 × 10−5). This gene encodes an inner mitochondrial membrane protease-like protein and is a plausible candidate for autism, because it was previously reported to be disrupted in an individual with Tourette syndrome, a complex neuropsychiatric disorder showing phenotypic overlap with ASDs.54 Moreover, IMMP2L contains a neuronal leucine-rich repeat gene (LRRN3) nested within its large third intron. The expression profile of LRRN3 also makes it an interesting candidate gene for autism, as it is most highly expressed in fetal brain. Studies in Drosophila demonstrate that many members of the LRR family provide an essential role in target recognition, axonal pathfinding and cell differentiation during neural development,55 and murine studies suggest these LRR proteins could have similar functions in mammalian neural development.56
The only SNP that achieved significant replication, after Bonferroni correction for multiple testing, is rs2217262 in the neighboring gene DOCK4, also a good autism candidate. This gene encodes a protein that activates Rac GTPase and is often deleted during tumor progression.57 A recent study in rats indicates that DOCK4 is predominantly expressed in the hippocampus as well as in the lung.58 This study further demonstrated that in cultured hippocampal neurons, DOCK4 is upregulated at the same time as dendrites start growing, and that knockdown of this gene by RNA interference results in impaired dendritic morphogenesis.
The association result for rs2217262 indicates that the common allele in the population is associated with increased risk for autism, or the minor allele is a ‘protective' variant. It has been shown that in presence of missing data, SNPs with a low MAF may show a bias in TDT, resulting in artificial overtransmission of the common allele.59 This problem is not likely to apply to rs2217262, as this association was supported also by case–control analysis.
Although only the rs2217262 association was confirmed by replication analysis, suggesting that the other results may represent false positives, this polymorphism (with MAF only about 5%) would not alone account for the linkage signal seen at AUTS1 in the IMGSAC sample. It is thus possible that multiple loci might contribute to the overall linkage seen for this region, and that the other significant SNPs from primary analysis may in reality be true signals but with lower OR, which our replication study was underpowered to detect. We do recognize that several limitations may have affected our replication sample. The primary sample was composed of trios selected from multiplex families based on IBD sharing, thereby more likely to be enriched for susceptibility alleles. By contrast, the replication population was a more heterogeneous sample, not preselected on linkage, and was mostly composed of singleton families. Power calculation suggested that our replication sample (IMGSAC-R and ND) should give us sufficient power to replicate the most significant primary results. However, the well-known ‘winners curse' theory also suggests that the effect sizes from the initial study may have been overestimated, thus requiring a much larger sample for replication. We did not detect presence of structure in the combined IMGSAC primary and IMGSAC-R samples, but it is possible that heterogenity may be present among the different samples used in this study (ND, Mount Sinai and University of Washington). This could have also contributed to the lack of replication, as could have gene–environment interactions, when different environmental exposures are present between population samples.
De novo and/or inherited CNVs are emerging as important causes of ASDs and other complex disorders.8, 11, 12, 13 Hence we exploited our dense SNP genotyping data to mine for structural variants. The most interesting discovery is the occurrence of deletions and duplications in four independent families in the IMMP2L/DOCK4 locus, given the coincident SNP association also seen for these genes. A maternal deletion was transmitted to both affected sons and the unaffected daughter in family 15-0084. In all other instances (two deletions and one duplication) the second affected sib did not inherit the CNV. Interestingly, the maternally segregating deletion extends to the 3′ end of the DOCK4 gene, whereas the non-segregating deletions or those identified in controls and in the DGV were limited to IMMP2L. Taken together, these data seem to suggest that a copy number loss of DOCK4 may influence susceptibility to ASDs, whereas duplications may not be damaging. The effect of DOCK4 deletions might be less penetrant in women because the mother and the unaffected daughter also carried the deletion. Larger studies will be needed to confirm this hypothesis.
The predominantly gene-based nature of our study represents a possible limitation, as we may have missed susceptibility alleles in intergenic regions. Recent findings from the ENCODE Consortium emphasize the importance of looking at noncoding sequence, as several functional elements in the genome seem to be in these regions.60 We attempted to minimize this limitation by including several SNPs in non-genic evolutionary conserved elements.
Our study also suggests that no common variants of large effect size are present within genic regions at AUTS1 and AUTS5 and highlights the importance of very large sample sizes for identification of robust associations and rare CNVs with sufficient power for statistical significance. Evidence from recent genome-wide association studies for various disorders clearly shows that effect sizes for loci contributing to complex traits are generally lower than those predicted a few years ago.61 Several whole-genome association and CNV studies for autism are currently in progress by large consortia, and it will be interesting to see if any of the genes highlighted by this study are also identified by these extensive studies.
It is possible that rare variants, both point mutations and CNVs, may account for a larger fraction of the overall genetic risk in complex psychiatric disorders than previously assumed. The present study was not designed to assess the contribution of rare sequence variants and our results do not preclude that the chromosome 2q and 7q linkage regions may harbor rare variation showing allelic heterogeneity across families, which may require resequencing to uncover.
The inconclusive findings identified with this study reflect the status of the field of autism genetics and suggest that classical approaches such as linkage and association analyses alone may not be sufficient to deal with the genetic and phenotypic heterogeneity seen in autism. One recent study of note used homozygosity mapping to uncover a number of large homozygous deletions in consanguineous pedigrees, highlighting the utility of this approach for heterogeneous disorders like autism.10 Another successful study found linkage to 15q13.3–q14 in a subset of families with IQ ⩾70, suggesting that the use of informative subphenotypes to define homogeneous sets of ASD families could be very important in detecting susceptibility loci involved in autism.62 Finally, another report indicated that level of somatic CNVs between MZ twins may be higher than expected.63 If confirmed, this finding could be a powerful tool for identification of autism susceptibility loci in MZ twins with a discordant phenotype. We believe a combination of these (and other) novel approaches, together with traditional methods will be required to uncover all the genes and biological pathways leading to autism.
In summary, the present high-density SNP association and CNV screen have provided evidence that variants in the IMMP2L/DOCK4 locus on chromosome 7 and in ZNF533 on chromosome 2 may increase susceptibility to ASDs. Association of the common allele of SNP rs2217262 in DOCK4 was supported by an independent replication, whereas the associations in IMMP2L and ZNF533 are not sufficiently significant in the context of multiple testing and warrant further studies.
Acknowledgments
We thank all the families who have participated in the study and the professionals who made this study possible. We also thank John Broxholme for bioinformatics support, Joseph Trakalo and Chris Allan at the WTCHG core genomics facility for Illumina and Sequenom genotyping, respectively. We especially thank Professor Giovanni Romeo at the Medical Genetics Unit, S Orsola-Malpighi Hospital, University of Bologna for his generous provision of laboratory space and equipment to EM, EB and CT. The CPEA (Collaborative Program of Excellence in Autism) thank Jeffery Munson, and Raphael Bernier and Annette Estes. This work was funded by the Nancy Lurie Marks Family Foundation; the Simons Foundation; the EC Sixth FP AUTISM MOLGEN, Telethon-Italy; the Korczak Foundation for Autism and Related Disorders; the Netherlands Organization for Scientific Research (NWO). The IMGSAC was funded by UK Medical Research Council, Wellcome Trust, BIOMED 2 (CT-97-2759), EC Fifth Framework (QLG2-CT-1999-0094), Deutsche Forschungsgemeinschaft, Fondation France Telecom, Conseil Regional Midi-Pyrenees, Danish Medical Research Council, Sofiefonden, Beatrice Surovell Haskells Fund for Child Mental Health Research of Copenhagen, Danish Natural Science Research Council (9802210) and National Institutes of Health (U19 HD35482, MO1 RR06022, K05 MH01196, K02 MH01389). AJ Bailey is the Cheryl and Reece Scott Professor of Psychiatry. AP Monaco is a Wellcome Trust principal research fellow.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry. 2005;162:1133–1141. doi: 10.1176/appi.ajp.162.6.1133. [DOI] [PubMed] [Google Scholar]
- Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J Clin Psychiatry. 2005;66 (Suppl 10:3–8. [PubMed] [Google Scholar]
- Baird G, Simonoff E, Pickles A, Chandler S, Loucas T, Meldrum D, et al. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) Lancet. 2006;368:210–215. doi: 10.1016/S0140-6736(06)69041-7. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Bolton P, Macdonald H, Pickles A, Rios P, Goode S, Crowson M, et al. A case–control family history study of autism. J Child Psychol Psychiatr. 1994;35:877–900. doi: 10.1111/j.1469-7610.1994.tb02300.x. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L.Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism Mol Psychiatry 200611118–28. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMGSAC A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Molec Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- IMGSAC A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet. 2001;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Barnby G, Bonora E, Sykes N, Bacchelli E, Blasi F, et al. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J Med Genet. 2005;42:132–137. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg GD, Dawson G, Sung YJ, Estes A, Munson J, Rosenthal E, et al. Evidence for multiple loci from a genome scan of autism kindreds Mol Psychiatry 2006111049–1060.979. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Wolpert CM, Menold MM, Bass MP, Hauser ER, Donnelly SL, et al. Chromosome 7 and autistic disorder (AD) Am J Hum Genet. 1998;63:A16. [Google Scholar]
- Trikalinos TA, Karvouni A, Zintzaras E, Ylisaukko-oja T, Peltonen L, Jarvela I, et al. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry. 2006;11:29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES. Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol Psychiatry. 2002;7:56–66. doi: 10.1038/sj.mp.4000922. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, et al. Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet. 2001;68:1514–1520. doi: 10.1086/320588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Raiford KL, Wolpert CM, Cope HA, Ravan SA, Ashley-Koch AA, et al. Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Hum Genet. 2002;70:1058–1061. doi: 10.1086/339765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora E, Bacchelli E, Levy ER, Blasi F, Marlow A, Monaco AP, et al. Mutation screening and imprinting analysis of four candidate genes for autism in the 7q32 region. Mol Psychiatry. 2002;7:289–301. doi: 10.1038/sj.mp.4001004. [DOI] [PubMed] [Google Scholar]
- Bonora E, Beyer KS, Lamb JA, Parr JR, Klauck SM, Benner A, et al. Analysis of reelin as a candidate gene for autism. Mol Psychiatry. 2003;8:885–892. doi: 10.1038/sj.mp.4001310. [DOI] [PubMed] [Google Scholar]
- Bonora E, Lamb JA, Barnby G, Sykes N, Moberly T, Beyer KS, et al. Mutation screening and association analysis of six candidate genes for autism on chromosome 7q. Eur J Hum Genet. 2005;13:198–207. doi: 10.1038/sj.ejhg.5201315. [DOI] [PubMed] [Google Scholar]
- Bacchelli E, Blasi F, Biondolillo M, Lamb JA, Bonora E, Barnby G, et al. Screening of nine candidate genes for autism on chromosome 2q reveals rare nonsynonymous variants in the cAMP-GEFII gene. Mol Psychiatry. 2003;8:916–924. doi: 10.1038/sj.mp.4001340. [DOI] [PubMed] [Google Scholar]
- Blasi F, Bacchelli E, Carone S, Toma C, Monaco AP, Bailey AJ, et al. SLC25A12 and CMYA3 gene variants are not associated with autism in the IMGSAC multiplex family sample. Eur J Hum Genet. 2006;14:123–126. doi: 10.1038/sj.ejhg.5201444. [DOI] [PubMed] [Google Scholar]
- Ackerman H, Usen S, Jallow M, Sisay-Joof F, Pinder M, Kwiatkowski DP. A comparison of case–control and family-based association methods: the example of sickle-cell and malaria. Ann Hum Genet. 2005;69:559–565. doi: 10.1111/j.1529-8817.2005.00180.x. [DOI] [PubMed] [Google Scholar]
- Fingerlin TE, Boehnke M, Abecasis GR. Increasing the power and efficiency of disease-marker case–control association studies through use of allele-sharing information. Am J Hum Genet. 2004;74:432–443. doi: 10.1086/381652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Ramoz N, Cai G, Reichert JG, Silverman JM, Buxbaum JD. An analysis of candidate autism loci on chromosome 2q24–q33: evidence for association to the STK39 gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1152–1158. doi: 10.1002/ajmg.b.30739. [DOI] [PubMed] [Google Scholar]
- http://www.hpacultures.org.uk/collections/ecacc.jsp .
- Barrett JC, Fry B, Maller J, Daly MJ. HaploView: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, Silva G, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Morris AP. Direct analysis of unphased SNP genotype data in population-based association studies via Bayesian partition modelling of haplotypes. Genet Epidemiol. 2005;29:91–107. doi: 10.1002/gepi.20080. [DOI] [PubMed] [Google Scholar]
- Morris AP. A flexible Bayesian framework for modeling haplotype association with disease, allowing for dominance effects of the underlying causative variants. Am J Hum Genet. 2006;79:679–694. doi: 10.1086/508264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugier-Veber P, Goldenberg A, Drouin-Garraud V, de La Rochebrochard C, Layet V, Drouot N, et al. Simple detection of genomic microdeletions and microduplications using QMPSF in patients with idiopathic mental retardation. Eur J Hum Genet. 2006;14:1009–1017. doi: 10.1038/sj.ejhg.5201661. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Mencarelli MA, Caselli R, Pescucci C, Hayek G, Zappella M, Renieri A, et al. Clinical and molecular characterization of a patient with a 2q31.2–32.3 deletion identified by array-CGH. Am J Med Genet A. 2007;143A:858–865. doi: 10.1002/ajmg.a.31602. [DOI] [PubMed] [Google Scholar]
- Monfort S, Rosello M, Orellana C, Oltra S, Blesa D, Kok K, et al. Detection of known and novel genomic rearrangements by array based comparative genomic hybridisation: deletion of ZNF533 and duplication of CHARGE syndrome genes. J Med Genet. 2008;45:432–437. doi: 10.1136/jmg.2008.057596. [DOI] [PubMed] [Google Scholar]
- Shoichet SA, Hoffmann K, Menzel C, Trautmann U, Moser B, Hoeltzenbein M, et al. Mutations in the ZNF41 gene are associated with cognitive deficits: identification of a new candidate for X-linked mental retardation. Am J Hum Genet. 2003;73:1341–1354. doi: 10.1086/380309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Yntema HG, Oudakker AR, Banning MJ, Kalscheuer VM, Chelly J, et al. Zinc finger 81 (ZNF81) mutations associated with X-linked mental retardation. J Med Genet. 2004;41:394–399. doi: 10.1136/jmg.2003.016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg D, Yntema HG, Banning MJ, Oudakker AR, Firth HV, Willatt L, et al. ZNF674: a new Kruppel-associated box-containing zinc-finger gene involved in nonsyndromic X-linked mental retardation. Am J Hum Genet. 2006;78:265–278. doi: 10.1086/500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008. [DOI] [PubMed]
- Petek E, Windpassinger C, Vincent JB, Cheung J, Boright AP, Scherer SW, et al. Disruption of a novel gene (IMMP2L) by a breakpoint in 7q31 associated with Tourette syndrome. Am J Hum Genet. 2001;68:848–858. doi: 10.1086/319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye R, Stevens A, Perry RL, Jacobs JR. Repellent signaling by Slit requires the leucine-rich repeats. J Neurosci. 2001;21:4290–4298. doi: 10.1523/JNEUROSCI.21-12-04290.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukamachi K, Matsuoka Y, Ohno H, Hamaguchi T, Tsuda H. Neuronal leucine-rich repeat protein-3 amplifies MAPK activation by epidermal growth factor through a carboxyl-terminal region containing endocytosis motifs. J Biol Chem. 2002;277:43549–43552. doi: 10.1074/jbc.C200502200. [DOI] [PubMed] [Google Scholar]
- Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- Ueda S, Fujimoto S, Hiramoto K, Negishi M, Katoh H. Dock4 regulates dendritic development in hippocampal neurons. J Neurosci Res. 2008;86:3052–3061. doi: 10.1002/jnr.21763. [DOI] [PubMed] [Google Scholar]
- Mitchell AA, Cutler DJ, Chakravarti A. Undetected genotyping errors cause apparent overtransmission of common alleles in the transmission/disequilibrium test. Am J Hum Genet. 2003;72:598–610. doi: 10.1086/368203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WTCCC Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Paterson AD, Szatmari P, Autism Genome Project Consortium Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biol Psychiatry. 2008;64:561–570. doi: 10.1016/j.biopsych.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, de Ståhl TD, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.