Abstract
The current study used three different proteomic strategies, which differed by their extent of intact protein separation, to examine the proteome of a pluripotent mouse embryonic stem cell line, R1. Proteins from whole-cell lysates were subjected either to 2-D-LC, or 1-DE, or were unfractionated prior to enzymatic digestion and subsequent analysis by MS. The results yielded 1895 identified non-redundant proteins and, for 128 of these, the specific isoform could be determined based on detection of an isoform-specific peptide. When compared with two previously published proteomic studies that used the same cell line, the current study reveals 612 new proteins.
Keywords: 1-D gel electrophoresis, 2-D chromatography, Cell biology, Embryonic stem cell, Isoforms, Shotgun proteomics
The realization of stem cell therapy depends, in part, on understanding and manipulating mechanisms necessary for the maintenance of pluripotency as well as differentiation into specific cell types. Knowing the genes and proteins that play essential roles in these processes is an important part of understanding stem cell biology and developing viable therapies. Therefore, studies that characterize the proteome of pluripotent cells will benefit the stem cell community. Toward that end, the current study used three different proteomic strategies, which differed by their extent of intact protein separation, to examine the proteome of a pluripotent mouse embryonic stem (ES) cell line, R1. This work complements previous proteomic studies of the R1 proteome by our lab [1], which used 2-DE, and Graumann et al. [2], which used subcellular fractionation followed by 1-DE for protein separation and isoelectric focusing for peptide separation.
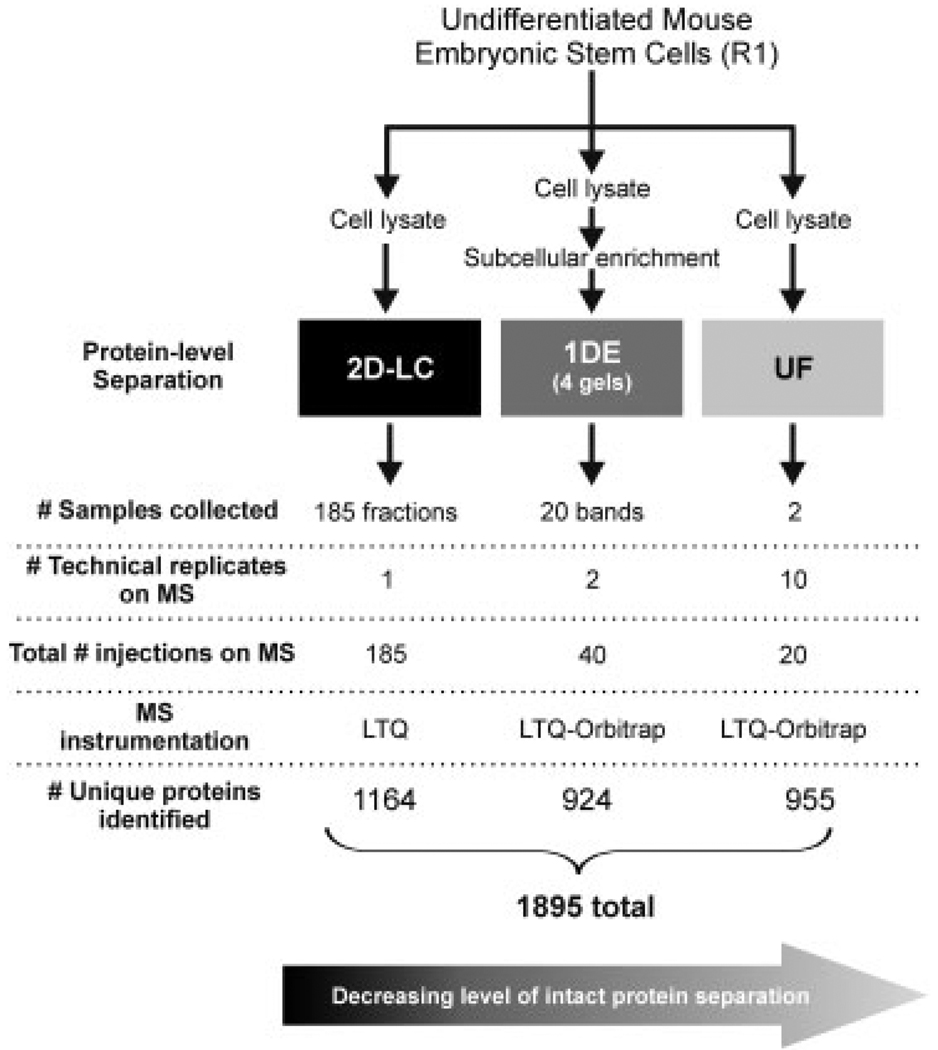
Pluripotent mouse ES cells (R1 cell line) were cultivated as described [3] and were passaged off feeder layers five times before lysis [1]. Under these conditions, ES cells contained mRNA transcripts to Oct4, sex-determining region Y-box 2, Nanog, Zfp42, and either weak or no expression of transcript markers of differentiation (Brachyury, CoupTF) (data not shown). Protein from whole-cell lysates were subjected either to 2-D-LC; (separation by pI and hydrophobicity), 1-DE (separation by molecular mass), or were unfractionated (UF; i.e. shotgun approach) prior to enzymatic digestion and subsequent analysis by MS (Fig. 1). Detailed methods are provided in the Supporting Information. Peptides from the 1-DE (n = 20 bands) and UF samples (n = 2 replicates) were analyzed on an Agilent 1200 nanoLC system (Agilent, Santa Clara, CA, USA) connected to an LTQ-Orbitrap mass spectrometer (Thermo). 2-D-LC samples (n = 185 fractions) were analyzed on an Agilent 1100 nano-LC system connected to an LTQ mass spectrometer (Thermo). All MS acquisition details are provided in the Supporting Information.
Figure 1.
Experimental workflow. Listed are the number of fractions the technique resulted in (# samples collected), number of times each digested fraction was analyzed independently by mass spectrometry (# technical replicates), multiplying # samples × # replicates = total number samples run on MS, MS instrumentation used to analyze that particular sample, and # unique proteins identified by that platform which were identified by 2 or more unique peptide sequences with a protein probability > 0.9
Raw MS data were searched against the International Protein Index (IPI) Mouse v3.47 database [4] (55 298 entries; 8/26/08) using Sorcerer 2™-SEQUEST® (Sage-N Research, Milpitas, CA, USA) with post search analysis performed using the Trans-Proteome Pipeline (TPP), implementing PeptideProphet [5] and ProteinProphet [6] algorithms. Database search parameters are provided in the Supporting Information. The ProteinProphet interact-prot.xml result files were input into ProteinCenter (Proxeon Bioinformatics, Odense, Denmark) and filtered to display proteins with protein probability scores p>0.9 (corresponding to false discovery rate ~1.0%), which were identified by two or more unique peptides. To remove redundancy in protein identifications, proteins were grouped according to “indistinguishable proteins,” which resulted in 1895 protein groups. For the final protein database, isoform notation is provided only when a peptide with an amino acid sequence that is unique to a specific protein isoform was identified. Membrane topology predictions were based on TMAP [7], which is integrated into ProteinCenter. All proteins identified are provided in Supporting Information Table S2 and detailed information regarding the data set can be found in Supporting Information Table S3 and the PRIDE database [8] (www.ebi.ac.uk/pride), accession numbers 11364 – 11379 (inclusive). The data were converted using PRIDE Converter [9] (http://code.google.com/p/pride-converter).
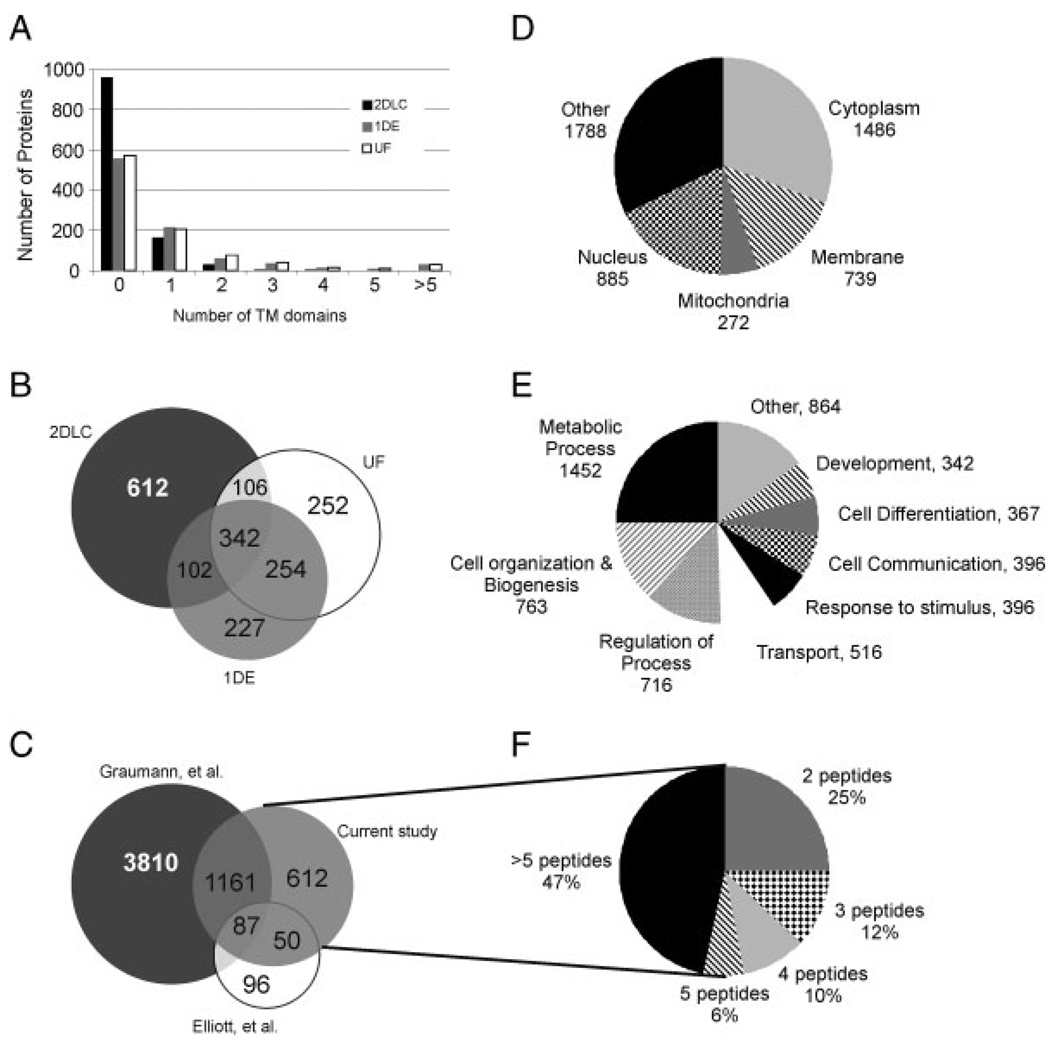
A total of 1895 non-redundant proteins were identified among all three strategies, with 1164 identified via 2-D-LC, 924 via 1-DE, and 955 via UF (Figs. 1 and 2B). A higher percentage of the proteins identified via 1-DE (40%) and UF (40%) compared to 2-D-LC (18%) were predicted or known transmembrane proteins based on TMAP (Fig. 2A). The gene ontology classifications for subcellular localization most represented in the complete data set include cytoplasm (29%) and nucleus (17%) (Fig. 2D) and the biological processes most represented include cell organization/biogenesis and regulatory proteins (Fig. 2E). Known ES protein markers such as sex-determining region Y-box 2 (Sox2), nestin (Nes), catenin-1 (Ctnna1), telomere-associated protein RIF1 (Rif1), E3 ubiquitin-protein ligase RING2 (Rnf2), undifferentiated embryonic cell transcription factor 1 (Utf1), and sal-like protein 4 (Sall4) were identified under the stringent conditions used for this database. The pluripotency markers Oct4 and Nanog were also identified, but by a single peptide (data not shown) and thus were not included in the final database. In total, 112 proteins known to be involved in the pluripotency regulatory network [10–13] or are part of a protein interaction network common to pluripotent cells (PluriNet [14]) were identified (Table 1 and Supporting Information Table S1).
Figure 2.
Analysis of current dataset. (A) Chart illustrating distribution of the number of proteins with transmembrane domains identified via each proteomic strategy. Venn diagrams illustrating the overlap in the proteins identified via each proteomic strategy in the current study (B) and the overlap of proteins identified among three studies which examined the proteome of R1 mouse embryonic stem cells [1, 2] (C). Pie charts illustrating the distribution of the gene ontology designations for cellular component (D) and biological process (E). (F) Pie chart illustrating the number of peptides per protein for the 612 proteins identified in the current study but not in Elliott et al. [1] or Graumann et al. [2].
Table 1.
Pluripotency network proteins
| Uniprot | Gene | Protein | Role [citation] | 2-D-LC | 1-DE | UF |
|---|---|---|---|---|---|---|
| P07356 | Anxa2 | Annexin A2 | PluriNet [14] | X | X | |
| P28352 | Apex1 | DNA-(apurinic or apyrimidinic site) lyase | PluriNet [14] | X | X | X |
| P24860 | Ccnb1 | G2/mitotic-specific cyclin-B1 | PluriNet [14] | X | ||
| P00375 | Dhfr | Dihydrofolate reductase | PluriNet [14] | X | ||
| P00493 | Hprt1 | Hypoxanthine-guanine phosphoribosyl- transferase |
PluriNet [14] | X | X | X |
| Q64433 | Hspe1 | 10 kDa heat shock protein, mitochondrial | PluriNet [14] | X | X | X |
| Q922D8 | Mthfd1 | C-1-tetrahydrofolate synthase, cytoplasmic | PluriNet [14] | X | X | |
| P50580 | Pa2g4 | Proliferation-associated protein 2G4 | PluriNet [14] | X | X | X |
| Q3TF18 | Parp1 | Putative uncharacterized protein | PluriNet [14] | X | X | X |
| P62962 | Pfn1 | Profilin-1 | PluriNet [14] | X | X | X |
| Q3TUQ5 | Pnn | Pinin | PluriNet [14] | X | ||
| Q9CR16 | Ppid | 40 kDa peptidyl-prolyl cis-trans isomerase | PluriNet [14] | X | X | X |
| Q8CIG8 | Prmt5 | Protein arginine N-methyltransferase 5 | PluriNet [14] | X | X | |
| Q8R323 | Rfc3 | Replication factor C subunit 3 | PluriNet [14] | X | ||
| Q8C671 | Sfrs2 | Putative uncharacterized protein | PluriNet [14] | X | X | |
| Q62189 | Snrpa | U1 small nuclear ribonucleoprotein A | PluriNet [14] | X | X | X |
| P54227 | Stmn1 | Stathmin | Interacts with Zfp281 [12] |
X | X | X |
| P70460 | Vasp | Vasodilator-stimulated phosphoprotein | PluriNet [14] | X | ||
| P27641 | Xrcc5 | ATP-dependent DNA helicase 2 subunit 2 | PluriNet [14] | X | ||
| Q99LI5 | Zfp281 | Zinc finger protein 281 | Interacts with Nanog, pluripotency transcriptional network, protein interaction network [10, 11] |
X |
Proteins identified in the current study, but not in previous proteomic studies of R1 cells, which have been experimentally linked to the pluripotency network in human and/or mouse ES cells, are markers of pluripotency, or are part of the protein–protein network shared by pluripotent cells. Listed are the Uniprot accession number, gene, protein name, role and corresponding citation, and by which method it was identified in the current study.
Three hundred and forty-two proteins were common among all three proteomic strategies used in this study (Fig. 2B). Comparing the sequence coverage of these 342 proteins among each strategy revealed that for 160 proteins (47%) the 1-DE provided the highest sequence coverage, for 113 (33%) the 2-D-LC provided highest coverage, and for 69 (20%) the UF provided the highest coverage. Though it was expected that the more extensive fractionation provided by the 2-D-LC would have resulted in the highest sequence coverage, the 1-DE provided the highest sequence coverage regardless of protein length (Supporting Information Fig. S2). However, it is noted that the 2-D-LC fractions were analyzed using an LTQ and it is predicted that had they been analyzed on the LTQ-Orbitrap, the sequence coverage would have been higher as is our experience in other studies (unpublished data). The median sequence coverage for all 1895 proteins identified were 15, 18 and 12%, respectively, for 2-D-LC, 1-DE, and UF.
It has been suggested that maintaining protein integrity during sample fractionation will facilitate the identification of protein isoforms [15, 16]. Even though the sequence coverage achieved was very similar for the 342 proteins observed in all three approaches, we independently manually examined this subset to determine whether there were any proteins for which the specific isoform could be determined based on the criteria that a peptide corresponding to an amino acid sequence unique to the isoform was identified by MS. This analysis was facilitated using ProteinCenter, which visually maps the identified peptides to all protein isoforms contained within the database. Of the 1895 proteins identified, the specific isoform could be determined for 128 proteins. Of these, 96 could be determined by 2-D-LC, 38 by 1-DE, and 25 by UF. The method by which each protein isoform was determined is listed in Supporting Information Table S2. For 17 proteins that were observed by multiple methods, the isoform could only be differentiated in the 2-D-LC analysis but not by other methods. The increased number of isoforms determined via 2-D-LC is consistent with the hypothesis that more protein fractionation can lead to a more complete characterization of the protein. The determination of protein isoform can be important from a biological perspective as the isoform of a protein can affect its localization and function. For example, protein isoforms found in the current study, which have been found to be specifically involved with functional changes in the differentiation of stem cells include cell division control protein 42 homolog (CDC42), staufen (RNA binding protein) homolog 1 (Stau1), pyruvate kinase isozymes M1/M2 (Pkm2), and 2-oxoglutarate dehydrogenase E1 component, mitochondrial (Ogdh) [17]. Specifically, the current study identified the isoform M2 of pyruvate kinase isozyme type M2, which promotes proliferation, is regulated by fructose-1,6-bisphosphate (FBP) and is present only during embryonic development, whereas the M1 isoform is found in adult heart, skeletal muscle, and brain and is not regulated by FBP [17–19].
The current data were compared with other proteomic studies of undifferentiated R1 cells by importing the protein accession numbers reported by our lab (Elliott et al. [1]) as well as Graumann et al. [2] into ProteinCenter. Clustering the proteins to remove redundancy resulted in a total of 5826 protein groups collectively for the 3 studies, with only 87 proteins common to all three studies (Fig. 2C). The overlap is limited among all three data sets by the relatively smaller data size contained in the 2-DE study of Elliott et al. (218 proteins) [1]. The overlap between the larger data set contained in Graumann et al. [20] and this current study is 1161 proteins. Of the 612 proteins found in the current study but not in the other studies, 75% were identified by 3 or more peptides, which allows us to have high confidence in these identifications (Fig. 2F). Also, of the 612 proteins found only in the current study, 18 are part of the protein interaction network common among pluripotent cells (PluriNet [14]) and 2 are experimentally linked to the pluripotency regulatory network [10, 12] (Table 1).
In summary, the current data set adds new information to the growing knowledge of the pluripotent stem cell proteome by identifying proteins known to be important for the maintenance of pluripotency, proteins not previously identified via proteomic approaches, and the identification of specific protein isoforms. Overall, this data set should be a useful reference for future studies of stem cells.
Supplementary Material
Acknowledgments
This research was supported by funding from the Intramural Research Program of the NIH, National Institute on Aging (K. R. B.), NIH Pathway to Independence Award K99-L094708-01 (R. L. G.), the NHLBI Proteomics Innovation Contract N01-HV-28180 (J. E. V.), NIH-R01-HL085434 (J. E. V.), and AHA Grant-in-Aid #09GRNT2500002 (J. E. V.). The authors thank the Technical Implementation and Coordination Core at JHMI for their technical assistance as well as Rui Wang and Juan Vizcaino at EBI for their assistance in uploading the data to PRIDE.
Abbreviations
- ES
embryonic stem
- UF
unfractionated
Footnotes
Dataset information was uploaded to the PRIDE database, accession numbers 11364–11379.
The authors have declared no conflict of interest.
References
- 1.Elliott ST, Crider DG, Garnham CP, Boheler KR, Van Eyk JE. Two-dimensional gel electrophoresis database of murine R1 embryonic stem cells. Proteomics. 2004;4:3813–3832. doi: 10.1002/pmic.200300820. [DOI] [PubMed] [Google Scholar]
- 2.Graumann J, Hubner NC, Kim JB, Ko K, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Tarasova Y, Riordon D, Tarasov K, Boheler K. In: Derivation and Manipulation of Embryonic Stem Cells: A Practical Approach. Evans ENaM., editor. Oxford: IRL Press; 2006. pp. 130–168. [Google Scholar]
- 4.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 5.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 6.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 7.Persson B, Argos P. Prediction of membrane protein topology utilizing multiple sequence alignments. J. Protein Chem. 1997;16:453–457. doi: 10.1023/a:1026353225758. [DOI] [PubMed] [Google Scholar]
- 8.Vizcaíno JA, Côté R, Reisinger F, Foster JM, et al. A guide to the Proteomics Identifications Databse proteomics data repository. Proteomics. 2009;9:4276–4283. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barsnes H, Vizcaino JA, Eidhammer I, Martens L. PRIDE Converter: making proteomics data-sharing easy. Nat. Biotechnol. 2009;27:598–599. doi: 10.1038/nbt0709-598. [DOI] [PubMed] [Google Scholar]
- 10.Orkin SH, Wang J, Kim J, Chu J, et al. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb. Symp. Quant. Biol. 2008;73:195–202. doi: 10.1101/sqb.2008.72.001. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Rao S, Chu J, Shen X, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 12.Wang ZX, Teh CH, Chan CM, Chu C, et al. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells. 2008;26:2791–2799. doi: 10.1634/stemcells.2008-0443. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller FJ, Laurent LC, Kostka D, Ulitsky I, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vegvari A, Rezeli M, Welinder C, Malm J, et al. Identification of prostate-specific antigen (PSA) isoforms in complex biological samples utilizing complementary platforms. J. Proteomics. 2010;73:1137–1147. doi: 10.1016/j.jprot.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnott D, Gawinowicz MA, Kowalak JA, Lane WS, et al. ABRF-PRG04: differentiation of protein isoforms. J. Biomol. Tech. 2007;18:124–134. [PMC free article] [PubMed] [Google Scholar]
- 17.Salomonis N, Nelson B, Vranizan K, Pico AR, et al. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput. Biol. 2009;5:e1000553. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Biol. 2008;40:1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Graumann J, Hubner NC, Kim JB, Ko K, et al. SILAC-labeling and proteome quantitation of mouse embryonic stem cells to a depth of 5111 proteins. Mol. Cell. Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.