Abstract
Protein kinase A (PKA) has been suggested as a regulator of stage differentiation in Trypanosoma cruzi. Using a yeast two-hybrid system we have begun to characterize the downstream substrates of T. cruzi PKA. We identified several members of the trans-sialidase super family by this approach. Immunoprecitation demonstrated that a TcPKAc monoclonal antibody was able to pull-down proteins recognized by trans-sialidase antibodies as well as a SA85-1.1 antibody and vice versa. An in vitro phosphorylation assay demonstrated that PKA phosphorylated the recombinant protein of an active trans-sialidase. In addition, a phospho-(Ser/Thr) PKA substrate antibody detected bands on immunoblot analysis of trans-sialidase antibody precipitated proteins from parasite lysate and the media of L6E9 myoblasts infected with trypomastigotes as well as from a SA85-1.1 antibody precipitated proteins from parasite lysate. Immunofluorescence analysis suggested that some TcPKAc localizes to the plasma membrane surface of trypomastigotes. The identified trans-sialidases have PKA consensus phosphorylation sites located near the endoplasmic reticulum retention motif in the N-terminal. These data support that PKA phosphorylates trans-sialidase super family members in vivo.
Keywords: Trypanosoma cruzi, Protein Kinase A, Trans-sialidase
1. INTRODUCTION
According to the World Health Organization more than 15 million people have been infected by T. cruzi, which causes Chagas disease, a chronic debilitating disease in Central and South America. This disease is also recognized to be an opportunistic infection in immune compromised patients with AIDS [1–7]. This protozoan parasite has a complex life cycle, but the molecular mechanisms involved in these various stagespecific transformations remain ill defined. The available therapeutic agents for T. cruzi are highly toxic, and there is no effective treatment for chronic Chagas' disease. Improvement in our understanding of the basic biology of T. cruzi may provide new and novel therapeutic approaches for this parasitic infection.
Trypanosoma cruzi lacks the synthetic machinery to produce the monosaccharide sialic acid. It can, however, incorporate sialic acid derived from the host into its surface by expressing a trans-sialidase, which catalyzes the transfer of sialic acid from host glycoconjugates to mucin-like molecules located on the parasite surface membrane [8]. This enables the parasite to adhere and invade host cells. Analysis of the T. cruzi genome reveals that T. cruzi has hundreds of genes encoding trans-sialidase, trans-sialidase-like proteins and mucin core proteins (http://tcruzidb.org/tcruzidb/). The trans-sialidase super-family consists of active, inactive enzymes and trans-sialidase-like proteins, which are essential both in host invasion and in immuno-evasion and range from 85 kDa to 200 kDa in size, mainly depending on variable numbers of C-terminal 12 amino acids repeats (so called SAPA repeats) [8]. Trans-sialidase super-family proteins are much higher in abundance on the surface membrane of bloodstream trypomastigotes and metacyclic trypomastigotes than on epimastigotes.
In other microorganisms such as yeast and Dictyostelium discoideum, signal transduction pathways mediated by cyclic AMP (cAMP) and PKA play a key role in the processes of differentiation. It is probable that similar regulatory pathways also exist in T. cruzi and are involved in the various developmental transformations that occur during its life cycle [9–10]. Evidence exists for a role for the activation of PKA in metacyclogenesis; cAMP levels, for example, have been reported to be elevated in metacyclic trypomastigotes in contrast to log phase epimastigotes [11–12].
We have previously reported the molecular cloning and characterization of both PKAc and PKAr in T. cruzi [13–14]; recently, several PKA downstream interacting proteins or substrates in this organism were identified by our laboratory group [10]. In this paper we provide evidence that TcPKAc interacts and phosphorylates members of the trans-sialidase super-family. Immunofluorescence analysis demonstrates that some of the expressed TcPKAc resides on the membrane surface of trypomastigotes. In silico analysis reveals that all identified trans-sialidases possess endoplasmic reticulum (ER) retention motifs (RXR). PKA consensus phosphorylation sites are found near these ER retention motifs in the N-terminal region of these proteins and may be important in the trafficking of members of the trans-sialidase family. It is possible that such post-translational modification(s) of these proteins by PKA play a role in invasion and/or differentiation.
2. MATERIALS AND METHODS
2.1. Cell Culture
Trypanosoma cruzi epimastigotes (HO 3/15, Brazil, Tulahuen and CL Brener) were grown at 26°C in liver infusion tryptose broth supplemented with 10% FCS (Life Technologies, Gaithersburg, MD). Trypomastigotes were obtained by growth in L6E9 myoblast cultures. The trypomastigotes were harvested 5–8 days post-inoculation, depending on the strain. Prior to harvesting the trypomastigotes, the cell cultures were washed with medium (Dulbecco’s Modified Eagle medium without serum) in order to remove small numbers of extracellular amastigotes that are frequently present. For the detection of trans-sialidase in L6E9 myoblast culture medium, the media were passed through a 0.45 µm filter to remove the parasites and cellular debris.
2.2. Yeast –two hybrid screening and confirming the interaction
Methods of construction of GAL4 AD library Yeast-two hybrid screening were previously described [10]. The bait construct (using BD the binding domain of GAL4) was produced by ligating the full length ORF of TcPKAc (13, Genbank accession number AY055783) containing EcoR1 and Pst1 restriction sites into a pBD-plasmid to generate pBDTcPKAc. Large scale transformation of the bait construct pBD- TcPKAc with the T. cruzi AD-plasmid library was carried out using YRG-2 yeast competent cells under a high stringency screen according to the manufacturer’s protocol (Stratagene, La Jolla, CA). Information from prey gene sequences was analyzed using BLAST (Genbank, NCI).
Among the identified genes, several candidates turned out to be members of the trans-sialidase super family. The entire ORFs of these genes were then amplified by RT-PCR using total RNA from CL Brener strain trypomastigotes and primers containing appropriate restriction sites (see table 1A). Briefly, cDNA was generated using the SuperScript™ First-Strand Synthesis System for RT-PCR according to manufacturer’s protocol (Invitrogen, Carlsbad, CA). 3ug of total RNA was converted into cDNA by incubating with 50U of Superscript II RT (reverse transcriptase), Oligo(dT), dNTP and appropriate buffer at 42 °C for 50 min. RNase H was then added to degrade the remaining RNA and the cDNA was stored as 2ul aliquot in each PCR tube at −80°C until used. PCR reactions were performed by adding 2ul of this cDNA and 3ul of primer mixes (150 pmole of each primer) into 45 ul of PCR Supermix High Fidelity solution (Invitrogen, Carlsbad, CA). PCR was performed using the conditions of: Initialization at 95 °C for 5 min; followed by 35 cycles of denaturation at 95°C for 30 second; annealing (primer dependence, see table 1A) and extension at 72°C for 3 min. A final elongation at 72°C for 10 min followed the 35 cycles and the mixtures were stored at 4°C until use. The amplicons were purified and digested with appropriate restriction enzymes (see table 1A), ligated into pAD-Gal4 vectors and sequenced to verify the correct genes.
Table 1.
| Table 1 A. PCR primers and annealing temperatures for trans-sialidase and TcPKAc genes | ||
|---|---|---|
| Trans-sialidase genes and TcPKAc gene |
PCR primers | Annealing Temperature |
| Tc00.1047053508563.20 | Forward- (Nhe1) CTAGCTAGC ATGCTCTCACGTGTTGCTGC Reverse- (Xbal1) GATCTCTAGA TCACGCAGTCGCAAAGCCCC |
55°C |
| Tc00.104753509187.10 | Forward- (Nhe1) CTAGCTAGCATGTCCCGGCGTGTGTTTACT Reverse- (Xbal1) GATCTCTAGATCACGCAGCCGCAAACCC |
55°C |
| Tc00.1047053507839.40 | Forward- (Xho1) CCGCTCGAG ATGCTCTCACGTGTTGCTG Reverse- (Sal1) ACGCGTCGAC TCACGCAGCCGCAAAGCCC |
66°C |
| Tc00.104753510111.20 | Forward- (Xho1) CCGCTCGAG ATGCTCTCACGTGTTGCTG Reverse- (Sal1) ACGCGTCGAC TCACGCAGTCGCAAGGCCC |
66°C |
| TcPKAc (Genbank, AY055783) | Forward- (EcoR1) CGGAATTCATGTCAGGGGAGGCAAATTATG Reverse- (Pst1) AACTGCAGTTAGAACCCAATAAACTCCGCCTG |
55°C |
| Table 1B. In silico analysis of PKA phosphorylation site. | |
|---|---|
| Each of trans-sialidase protein sequences was analyzed against PROSITE patterns and profiles (http://us.expasy.org/prosite). All of these proteins contain typical PKA phosphorylation sites. The numbers indicate the location of the sites in the various protein sequences. | |
| Trans-sialidase genes | cAMP- dependent protein kinase phosphorylation site |
| Tc00.1047053508563.20 | 113 – 116: KKgT; 120 – 123: RRdS; 465 – 468: RRvS; 624 – 627: KKlT |
| Tc00.104753509187.10 | 17 – 20: RRvT; 635 – 638: KKiS |
| Tc00.1047053507839.40 | 17 – 20: RRvT; 239 – 242: KRiS; 633 – 636: KKaS |
| Tc00.104753510111.20 | 17 – 20: RRvT; 468 – 471: RRgS; 649 – 652: KKrS; 699 – 702: KKpS |
| Tc00.1047053508285.60(SA1.1) | 17 – 20: RRvT; 119 – 122: RRdS; 466 – 469: RRdS; 624 – 627: KKpT |
| TcTS 6 11/2 | 12 – 15: KRqS |
| Table 1C. Myristoylation sites exist in TcPKAc | |
|---|---|
| The PROSITE program was used to identify N-myristoylation sequences in both TcPKAc isoforms | |
| TcPKAc | Myristoylation site |
| Protein kinase A catalytic subunit isoform-1 (Tc00.1047053508461.310) |
306 – 311: GLlqTD; 317 – 322: GTlkGG |
| Protein kinase A catalytic subunit isoform- 2 (Tc00.1047053508461.280) |
25 – 30: GstlGA; 250 – 255: GLlqTD; 261 – 266: GTlkGG |
A SA85-1.1 cDNA insert, a member of gp85 trans-sialidase super family [15], was excised by BamHI and XhoI from Bluescript SK− and ligated into the pAD-Gal4 vector. In addition, the DNA insert coding for TCTS 611/2, an active trans-sialidase lacking SAPA repeats [16], in a pTrcHisA vector (kind gift from Drs. Laura Ratier and Alberto C. C. Frasch) was derived by digesting the construct with EcoR1 and ligating the TCTS 611/2 gene into a pAD-Gal4 vector.
Transformation using these pAD-Gal4 constructs and the TcPKAc bait construct was performed under the highest stringent condition. Constructs, pADwt with pBDwt, were used as positive controls and pADwt with pLaminC as a negative control for these experiments.
2.3. The generation of a T. cruzi trans-sialidase antibody
To generate a polyclonal antibody against active trans-sialidase, TCTS 611/2, recombinant protein [16] was used to immunize mice with Freund’s adjuvant (Sigma, St. Louis, MO) at a 1:1 dilution with boosting at 4 and 12 weeks. The specificity of this antiserum was confirmed by immunoblot.
2.4. Immunoprecipitation and immunoblot
To confirm yeast two hybrid interactions with a complementary method, co-immunoprecitation (co-IP) was performed using Triton X-100 protein extracts from trypomastigotes and a trans-sialidase polyclonal antibody (anti-TCTS 6 11/2), a sialidase antibody (Immunogen: native sialidase from Clostridium perfringens; Biogenesis UK), a T. cruzi SA85-1.1poly-clonal antibody, and a TcPKAc mAb [14–15]. The commercial sialidase antibody was first tested and verified for its ability to recognize the trans-sialidases of T. cruzi, such as the recombinant protein of TCTS 6 11/2 and the Gal4-AD T. cruzi trans-sialidase fusion proteins by immunoblot before other uses (Fig. 2A and 2B).
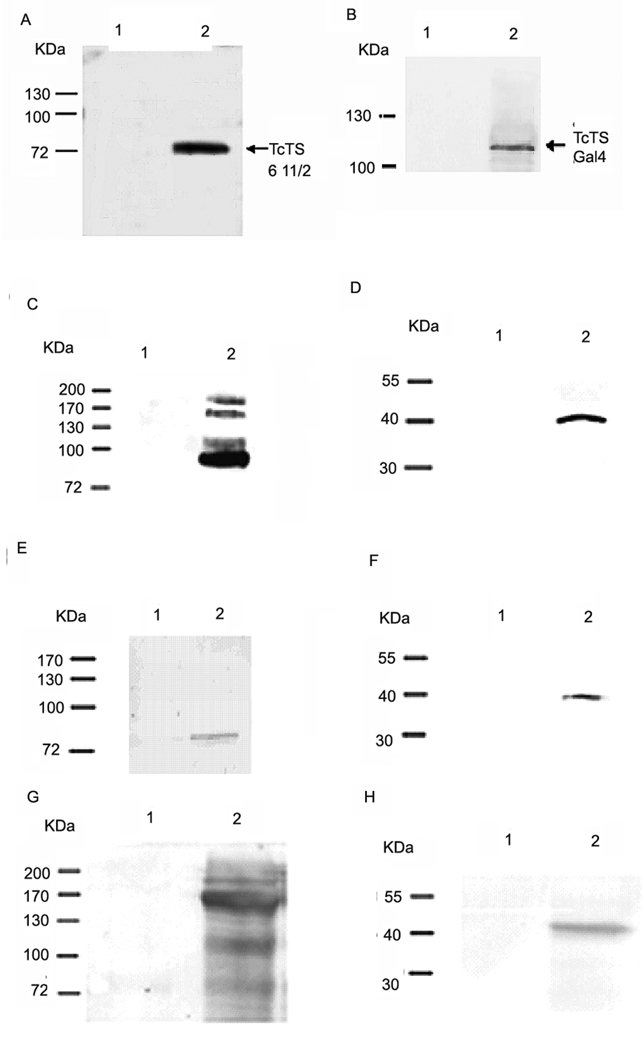
Fig. 2. Verification of a trans-sialidase antibody and Interactions of TcPKAc with the members of trans-sialidase super family.
A commercial anti- sialidases antibody (rabbit) was first tested and verified for its ability to recognize T. cruzi trans-sialidases recombinant proteins, including a purified active trans-sialidase and trans-sialidase fusion proteins in crude yeast lysates (primary antibody dilution 1:1000; secondary antibody dilution 1: 5000) (Fig.2A–B). Co-immunoprecitation was performed using Triton X-100 protein extracts from trypomastigotes and a trans-sialidase polyclonal antibody (anti-TCTS 6 11/2, mouse), a commercial sialidase antibody, a T. cruzi SA85-1.1polyclonal antibody (rabbit) and a TcPKAc mAb(antibody dilutions:1;100 for immunoprecipitation, 1:1000 for immunoblot)(Fig.2C–H).
A. The sialidase antibody binds to recombinant protein of TCTS 611/2.
Lane1, 100ug of BSA as negative control;
Lane 2, 100ng of recombinant protein of TCTS 611/2.
B. The sialidase antibody binds to fusion protein of trans-sialidase (Tc00.1047053509187.10) in yeast lysate.
Lane 1, 150 ug of wild type yeast lysate without transformation as negative control;
Lane 2, 150ug of yeast lysate with transformation of both bait and pAD- trans-sialidase (Tc00.104053509187.10) constructs. Antibody recognized the pAD- trans-sialidase protein.
C. Co-IP in Triton X −100 protein extracts from trypomastigotes with a TcPKAc mAb. Complex pulled down by a TcPKAc mAb contained proteins recognized by a commercial sialidase antibody on immunoblot.
Lane 1, Negative control using an unrelated mAb (anti-bag5);
Lane 2, Immunoprecipitation with TcPKAc mAb. There are four bands ranged from 85 kDa to 200 kDa, which reacted with this sialidase antibody.
D. Co-IP in Triton X −100 protein extracts from trypomastigotes with a commercial sialidase antibody. The sialidase antibody was able to pull down a protein, which reacted with a TcPKAc mAb on immunoblot.
Lane 1, negative control using pre-immune rabbit serum;
Lane 2, Immunoprecipitation with Trans-sialidase antibody. There is a 40-kDa TcPKAc protein band.
E. Co-IP in Triton X −100 protein extracts from trypomastigotes with a TcPKAc mAb.
Complex pulled down by a TcPKAc mAb contained proteins recognized by a SA85-1.1 antibody on immunoblot.
Lane 1, Negative control using an unrelated mAb (anti-bag5);
Lane 2, Immunoprecipitation with TcPKAc mAb. There is an 85 kDa SA85-1.1 protein band.
F. Co-IP in Triton X −100 protein extracts from trypomastigotes with a SA85-1.1 antibody.
Precipitated complex by a SA85-1.1 antibody contained TcPKAc.
Lane 1, Negative control using pre-immune rabbit serum;
Lane 2, Immunoprecipitation with SA85-1.1 antibody. There is a 40-kDa TcPKAc protein band.
G. Co-IP in Triton X −100 protein extracts from trypomastigotes with a TcPKAc mAb. Complex pulled down by a TcPKAc mAb contained proteins recognized by a trans-sialidase antibody (anti-TCTS 6 11/2) on immunoblot.
Lane 1, Negative control using an unrelated mAb (anti-bag5);
Lane 2, Immunoprecipitation with TcPKAc mAb. There are bands with molecular weights from 85–200 kDa.
H. Co-IP in Triton X −100 protein extracts from trypomastigotes with a TCTS 6 11/2 antibody. Precipitated complex by a TCTS 6 11/2 antibody contained TcPKAc.
Lane 1, Negative control using pre-immune mouse serum;
Lane 2, Immunoprecipitation with Anti-TCTS 6 11/2;
To produce parasite lysates for co-immunoprecipitation, 6 ×109 CL Brener strain trypomastigotes derived from cell-culture were resuspended in Buffer A (20mM Tris-HCl pH 7.4, 2 mM EDTA, 1% Triton X-100 and one tablet of protease inhibitor cocktail tablet [Roche, Bavaria, Germany] per 5ml of buffer) and subjected to three freeze - thaw cycles, followed by centrifugation at 16000g for 30 minutes at 4°C. The supernatant was then used for immunoprecipitation followed by immunoblot. Briefly, 2mg Triton X-100 protein extracts from trypomastigotes was used for each precipitation. Either pre-immune sera or primary antiserum was added at optimal dilutions (1:100) and incubated at 4°C overnight with a rocking motion. The antibody–antigen complexes were isolated by incubating the reaction mixture with a slurry of protein A-Sepharose CL-4B (Sigma, St. Louis, MO), followed by centrifugation at 10,000×g and washing three times with Buffer A prior to immunoblot. For immunoblot, the protein A-Sepharose beads of each immunoprecipitation were resuspended in 30ul sample buffer, boiled for five minutes, briefly centrifuged and ran on a 10% SDS- polyacrylamide gel, transferred onto nitrocellulose membrane, blocked with 5% non-fat milk and detected with appropriate primary antibodies (1:1000) and secondary antibody conjugated with alkaline phosphatase (1:5000), visualized by using BCIP/NBT as substrate (Roche, Bavaria, Germany).
To detect PKA phosphorylation of trans-sialidase proteins in vivo, A PKA specific phosphorylation antibody, which recognizes the phosphorylated PKA consensus sequences RXXT and RRXS (Cell Signaling Technology, Inc. Danvers, MA) was used to examine proteins in immunoprecipitation complexes from Triton X-100 protein extracts by immunoblot (1:1000 dilutions). In order to examine whether the shed trans-sialidases were phosphorylated by PKA, co-IP was performed with the trans-sialidase antibodies and the PKA phosphorylated substrate antibody using trypomastigote lysate and L6E9 myoblast culture media heavily infected with trypomastigotes. Uninfected L6E9 myoblast culture media was used as a negative control. Briefly, the media were filtered to remove the parasites and cellular debris and 1.5ml of the media were used for each precipitation employing the same method detailed above.
2.5. Recombinant protein expression and purification
We have previously published these methods [10]. Briefly, to examine whether PKAc can phosphorylate active trans-sialidase in vitro, the clone TCTS 611/2 in pTrcHisA was introduced into E. coli BL21 (DE3) (Invitrogen, Carlsbad, CA) by transformation. One colony was inoculated into 50 ml of LB broth with 100 µg ampicillin/ml. The bacteria were allowed to grow to a density of 0.6–0.8 Abs600 and were then induced to express the recombinant protein by adding 0.5 mM isopropylthiogalactoside (IPTG) (Sigma, St. Louis, MO). Cells were then harvested and stored at −80°C until needed. Protein purification was performed using the His Trap FF crude Kit (GE Healthcare, Fairfield, CT). Purified proteins were analyzed by SDS-polyacrylamide gel electrophoresis under reducing conditions and stained with Coomassie Blue R250. The eluate containing the recombinant proteins were dialyzed against 2 liters of Tris- buffer (10mM Tris-HCL pH 7.4) at 4°C for 24 hrs and protein concentration was determined using a Nanodrop ND1000 spectrophotometer (NanoDrop Technologies, Inc. Wilmington, DE) prior to in vitro phosphorylation assays.
2.6. In vitro phosphorylation
We have found that TcPKAc can phosphorylate a PKA specific Kemptide LRRASLG, (the bold letters are a typical PKA consensus sequence), is inhibited by PKI (containing RRNA as a pseudosubstrate), which is a classic characteristic of PKA, and that the catalytic domain of TcPKAc is similar to that of bovine PKA [13]. TcPKAc is, therefore, a typical PKA family member. Bovine PKAc was used for the in vitro phosphorylation assay as it is routinely used as the reagent for assays of eukaryotic PKA substrates including protozoa [17–18]. Phosphorylation of 10 µg of recombinant protein 611/2 (active trans-sialidase) by 4 units PKAc (purified from Bovine heart, Sigma, St. Louis, MO) was measured in a final volume of 20 ul in a kinase buffer containing 25 mM Tris (pH 7.4), 10mM MgCI2, 10 µCi [γ-32P] ATP (3000 Ci/mmol; GE-Health Care, Fairfield, CT) in the presence or absence of PKI (10 µg). A negative control was performed using the same reagents without the recombinant protein. The reactions were incubated in 30°C for 30 min and then 4 ul of 6x SDS sample buffer (60% Glycerol, 300 mM Tris (pH 6.8), 12 mM EDTA, 12% SDS 864 mM 2-mercaptoethanol and 0.05% bromophenol blue) was added to each tube, which was heated at 100°C for 5 min and then analyzed by SDS-polyacrylamide gel electrophoresis under reducing conditions. Coomassie Blue R250 staining was used to confirm protein-loading equivalency, followed by gel drying and autoradiography.
2.7. Immunofluorescence analysis (IFA)
The parasites were treated in two ways after fixation, with Triton X-100 for permeabilization of the membrane or without Triton X-100 for non-permeabilized parasites. Briefly, trypomastigotes (Tulahuen strain) were fixed with 4% paraformaldehyde, adhered to poly-L-lysine coverslips, permeabilized for 5 min with Dulbecco's phosphate-buffered saline (PBS), 0.3% Triton X-100, (non-permeabilized parasite slides were not treated with Triton X-100), blocked for 1 h with PBS, 3% bovine serum albumin, 1% fish gelatin, 5% goat serum, 50 mM NH4Cl, and incubated with primary antibody (anti-TcPKAc, mAb) for 1 h at 37°C, washed three times in PBS and then secondary antibody (goat anti-mouse IgG Fluoresceiin linked, 1:5000) for 45 min at 37°C. Coverslips were then washed three times in PBS, incubated with 40,6-diamidino-2-phenylindole-2 HCl (DAPI) solution (5 ug/ml, Molecular Probes) to allow visualization of nuclei and kinetoplasts. Subsequently, 200 µl of DABCO solution (2.5% of DABCO (1,4-diazabicyclo [2.2.2] octane (Sigma, St. Louis, MO) and 20% of glycerol in PBS) was placed on the slides. The slides were examined using a Fluorescence microscope (Nikon, Model: HB-10101AF, using a 100X oil lens) with Fluoresceiin (Nikon filter cube: UV2B) and Blue filter (Nikon filter cube: B2A) sets. Either an unrelated mAb (anti-bag5 of Toxoplasma gondii) or a secondary antibody was used as negative control.
In addition, we performed IFA on trypsinized trypomastigotes to confirm surface localization of identified proteins. Trypsinized trypomastigotes were generated using a published method [19]. Briefly, washed trypomastigote forms (108) were resuspended in 1 ml medium 199 containing 150 µg trypsin (Sigma) and then incubated at 37°C for 20 min. The parasites were then washed with PBS three times and fixed on coverslips using the techniques described above for IFA. Trypsinized parasites (non-permeabilized) were used for IFA with TcPKAc mAb using the same conditions described above. SA85-1.1 at 1:50 dilution, an antibody which recognizes some trans-sialidase superfamily surface antigens, was used as a control antibody to examine the effect of trypsin treatment on a known family of surface protein in trypomastigotes.
2.8. In silico analysis of PKA phosphorylation site and N-myristoylation site
For every trans-sialidase interacting with TcPKAc, the protein sequence was analyzed against PROSITE patterns and profiles (http://us.expasy.org/prosite) to examine their PKA consensus phosphorylation sites. Since N-myristoylation sites in PKAc play a role in plasma membrane localization [20], the PROSITE program was used to detect whether N-myristoylation sequences exist in both TcPKAc isoforms.
3. RESULTS
3.1. Trans-sialidase family members interact with PKAc
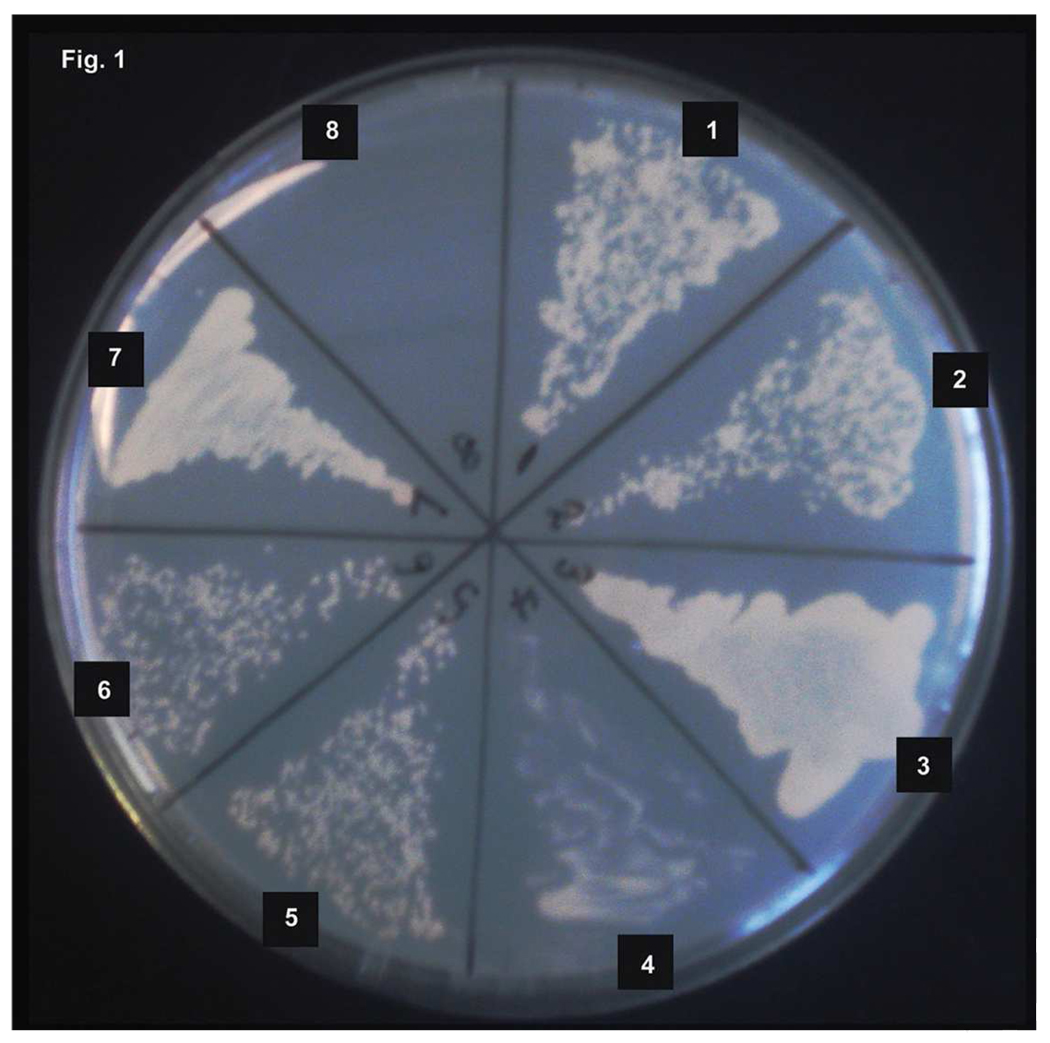
In order to find the downstream substrates of PKA in T. cruzi, we performed a yeast two-hybrid screen. Several trans-sialidase genes and gp85 super family members were among the identified candidates. Using the highest stringency for selection, the entire ORFs of four genes and a partial SA85-1.1 gene were demonstrated to interact with TcPKAc (Fig. 1). Using this method, however, we did not identify any candidate trans-sialidase with variable SAPA repeats in the C-terminus (expected molecular weight between 120–200kDa). We reasoned that the SAPA repeats might interfere with the translocation of these proteins into the yeast nuclei in a yeast two-hybrid system. To test this hypothesis, an active trans-sialidase clone (TCTS 6 11/2) lacking SAPA repeats was subcloned into the pAD-Gal4 vector and transformed with pBD- TcPKAc into YRG-2 yeast competent cells under the highest stringency selection condition. This test yielded positive colonies, indicating that this active trans-sialidase clone interacts with TcPKAc and suggests that the lack of trans-sialidases containing SAPA repeats in our yeast-two hybrid screen was most likely a consequence of the screening methodology rather than a lack of PKA interactions with these proteins. As demonstrated in Figure 1, we identified six genes of the trans-sialidase super- family which interacted with TcPKAc by this technique: (1) trans-sialidase Tc00. 1047053508563.20; (2) trans-sialidase Tc00.1047053509187.10; (3) trans-sialidase-like protein SA85-1.1 Genbank accession number X53545; (4) trans-sialidase Tc00.1047053507839.40; (5) trans-sialidase Tc00.104753510111.20; and (6) trans-sialidase active clone lacking SAPA repeats TCTS 611/2 [16].
Fig. 1. T. cruzi trans-sialidase proteins interact with TcPKAc.
Using a yeast two-hybrid system six members of the trans-sialidase super family target plasmids interacting with pBD- TcPKAc were obtained. Four ORFs of these genes and two partial genes were subcloned into a pAD-Gal4 vector. Each pAD-Gal4 construct was co-transformed into yeast with pBD-TcPKAc and cultured in high stringency condition (-Leu, -Trp, -His)
(1) trans-sialidase Tc00. 1047053508563.20;
(2) trans-sialidase Tc00.1047053509187.10;
(3) trans-sialidase-like protein SA85-1.1 Genbank accession number X53545;
(4) trans-sialidase Tc00.1047053507839.40;
(5) trans-sialidase Tc00.104753510111.20;
(6) trans-sialidase active clone lacking SAPA repeats TCTS 611/2.
(7) Positive control (pADwt + pBDwt);
(8) Negative control (pADwt + pLaminC).
The T. cruzi genome contains over a hundred trans-sialidases, however universal antibodies to this T. cruzi family have not been described. It is probable that an antibody that recognizes a common conservative motif SXDXGXTW can serve this purpose. The trans-sialidase gene products of T. cruzi have a significant degree of structural and biochemical similarity to the sialidases (also named neuraminidase) found in bacteria [21]. In both T. cruzi and C. perfringens, sialidases contain multiple copies of the conserved motif, SXDXGXTW. We evaluated a commercially available C. perfringens, sialidase antiserum for its ability to recognize recombinant T. cruzi trans-sialidases, including a purified active trans-sialidase and several trans-sialidase fusion proteins in crude yeast lysates. Our experiment demonstrated that this antibody could recognize trans-sialidase fusion proteins obtained from our yeast-two hybrid experiments as well as a trans-sialidase, TCTS 6 11/2 (Fig. 2A and 2B) and suggests that this antibody would be a useful reagent to investigate T. cruzi trans-sialidases.
Immunoprecipitation experiments demonstrated that the immune-complex precipitated by TcPKAc mAb from Triton X-100 extracts of trypomastigotes contained proteins recognized by the commercial sialidase antibody. There were four major bands recognized in this IP ranging from 85 kDa to 200 kDa (Fig.2 C). The immune-complex pulled down by this sialidase antibody contained a 40-kDa band recognized by immunoblot with a mAb to TcPKAc, indicating that this protein was TcPKAc (Fig. 2D). Similarly, TcPKAc mAb pulled down a complex containing an 85-kDa protein, which reacted with the SA85-1.1 antibody on immunoblot. Furthermore, TcPKAc was in the complex precipitated by the SA85-1.1 antibody (Fig.2 E and Fig. 2 F). Since the SA85-1.1 antibody may react with other gp85 candidates, the TcPKAc mAb pull down complex could have more than one gp85 proteins. The polyclonal antibody against active transsialidase, TCTS 611/2, recognized four bands in TcPKAc mAb immune-complex while a mAb to TcPKAc detected a 40-kDa band in anti-TCTS 6 11/2 pulled down complex (Fig.2 G and Fig.2 H). These data suggest that TcPKAc is able to interact with several trans-sialidase family members.
3.2. PKAc phosphorylates trans-sialidase in vitro
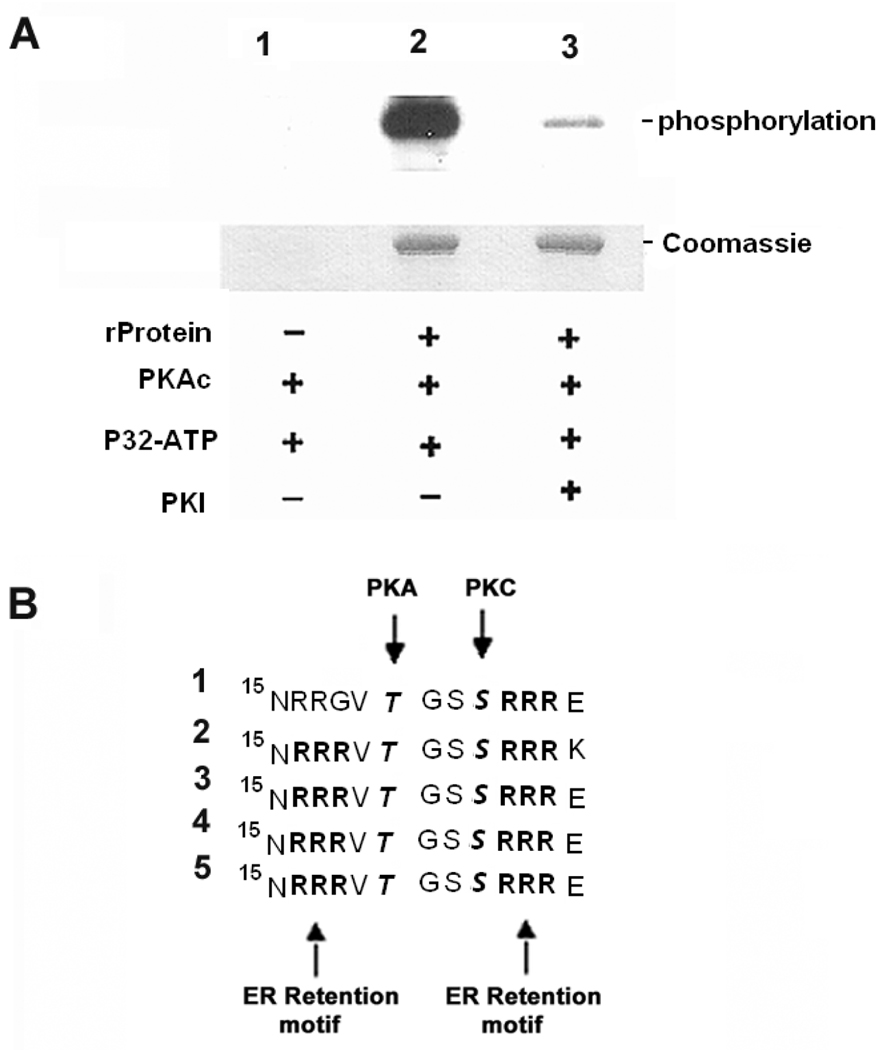
In silico sequence analysis of these trans-sialidase proteins confirmed the presence of consensus phosphorylation sites for PKA. All of the candidate genes identified by the yeast two hybrid assays interacting with TcPKAc possess typical PKA phosphorylation sites. There are two to four typical sites in the five full-length protein sequences of trans-sialidase and TCTS 6 11/2 contains one site (See table 1B). To confirm that trans-sialidase can be phosphorylated by PKA in vitro, we performed an in vitro phosphorylation assay using TCTS 6 11/2 recombinant protein (an enzymatically active trans-sialidase) and bovine PKAc. The results demonstrated that this protein could be phosphorylated in vitro by PKA and that PKI, a specific PKA inhibitor, inhibited this phosphorylation (Fig. 3A).
Fig. 3. PKA phosphorylates trans-sialidase recombinant protein in vitro PKAc phosphorylation sites were found to flank ER retention motifs in the members of trans-sialidase superfamily.
A. In vitro phosphorylation of trans-sialidase recombinant protein by PKA is confirmed.
Lane1, Absence of trans-sialidase recombinant protein as negative control;
Lane 2, Trans-sialidase recombinant protein was phosphorylated in absence of PKI;
Lane 3, PKI inhibited the phosphorylation; − absence of the reagent; + presence of the reagent (for details see MATERIALS AND METHODS).
B. Five trans-sialidase protein sequences interacting with TcPKAc possess RXR (Arg-X-Arg) ER retention motifs in N-terminal. A PKAc phosphorylation site and a PKC phosphorylation site flank with these sites. It has been reported that phosphorylation by PKA and PKC in ER and /or Golgi at these sites suppresses ER retention and regulates surface delivery to membrane. The sequences were lined up at N-terminal position 15, RXR is a typical ER retention signal; RRXT or RXXT (position17–20) is a PKA phosphorylation site and SXR (position 23–25) is a PKC phosphorylation site.
(1). Trans-sialidase Tc00.1047053508563.20;
(2). Trans-sialidase Tc00.1047053509187.10;
(3). Trans-sialidase Tc00.1047053507839.40;
(4). Trans-sialidase Tc00.104753510111.20;
(5). Trans-sialidase Tc00.1047053508285.60.
3.3. TcPKAc phosphorylates candidates of the trans-sialidase family in vivo
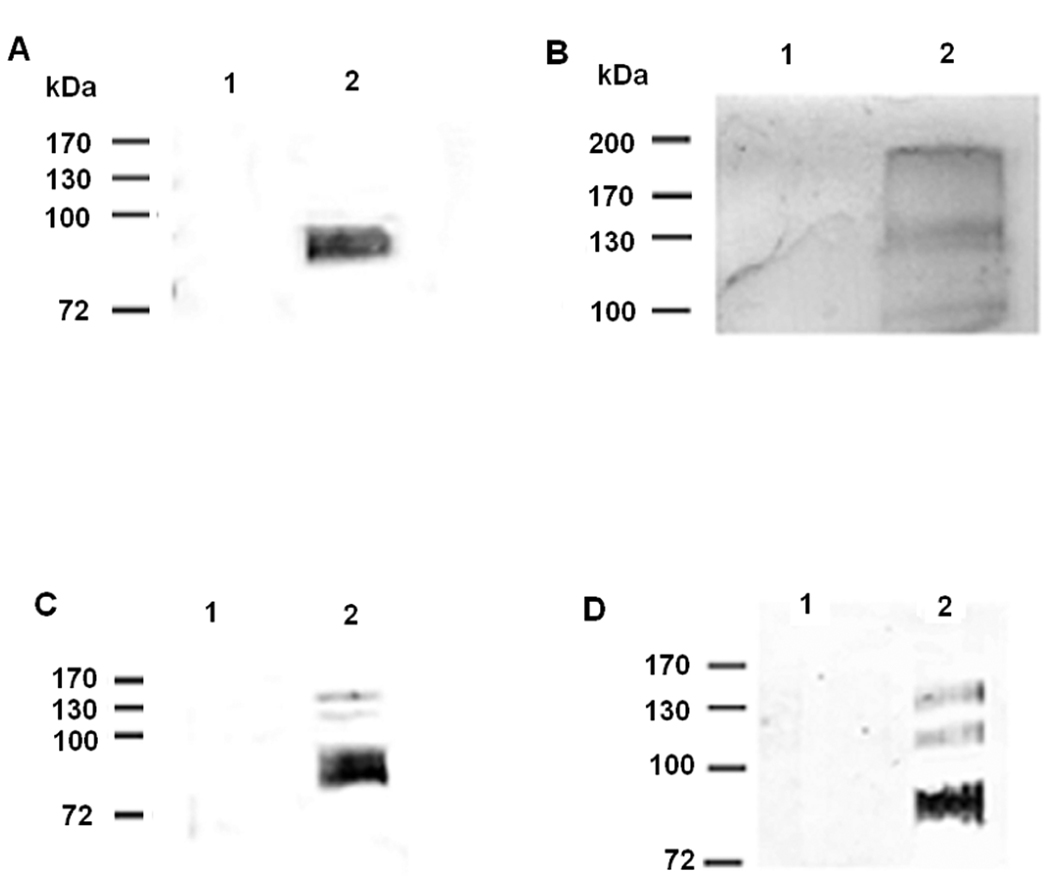
To detect whether TcPKAc phosphorylates trans-sialidase super family proteins, we examined trypomastigote lysate as well as culture media from L6E9 myoblasts heavily infected with trypomastigotes. Triton X-100 was used to extract the membrane bound proteins of the trans-sialidase family and enrich the contents of trans-sialidase and gp85 proteins in the soluble fraction for co-IP experiments. Complexes immunoprecipitated by the SA85-1.1 antibody demonstrated a broad around 85 kDa which reacted with the phospho-(Ser/Thr) PKA substrate antibody (Fig. 4A) while the complex immunoprecipitated by anti- TCTS 6 11/2 revealed three major bands detected by a phospho-(Ser/Thr) PKA substrate antibody (Fig. 4B). The commercial sialidase antibody precipitated several trans-sialidase proteins from the media (Fig. 4C) and these proteins were recognized by a phospho-(Ser/Thr) PKA substrate antibody (Fig. 4D). These data demonstrate that in this cell culture system trans-sialidases were phosphorylated at PKA consensus sites. These data also suggest that phosphorylation by PKA of trans-sialidase super family proteins occurs in vivo, consistent with the in silico analysis prediction. Recently, a paper on the T. cruzi phosphoproteome of epimastigotes demonstrated that four trans-sialidases were phosphorylated [22].
Fig. 4. PKA phosphorylates the members of trans-sialidase super-family in vivo.
Triton X −100 protein extracts from trypomastigotes and L6E9 myoblast culture media heavily infected with trypomastigotes were used for immunoprecipitation by antibodies to trans-sialidase family and then immunoblot by a phospho-(Ser/Thr) PKA substrate antibody (rabbit poly-clonal antibody) (dilutions: 1:100 for immunoprecipitation; 1:1000 for immunoblot).
A. Co-IP in Triton X −100 protein extracts from trypomastigotes with a SA85-1.1 antibody. Precipitated complex by a SA85-1.1 antibody contained a broad band recognized by a phospho-(Ser/Thr) PKA substrate antibody.
Lane 1, Negative control using pre-immune rabbit serum;
Lane 2, Immunoprecipitation with SA85-1.1 antibody.
B. Co-IP in Triton X −100 protein extracts from trypomastigotes with a TCTS 6 11/2 antibody. Precipitated complex by a TCTS 6 11/2 antibody contained three bands recognized by a phospho-(Ser/Thr) PKA substrate antibody
Lane 1, Negative control using pre-immune mouse serum;
Lane 2, Immunoprecipitation with TCTS 6 11/2 antibody
C. Immunoprecipitation using L6E9 myoblast culture media heavily infected with trypomastigotes. A commercial sialidase antibody precipitated several proteins of trans-sialidases in the media with T. cruzi infected L6E9 myoblast.
Lane 1, Uninfected L6E9 myoblast culture medium as negative control.
Lane 2, Media from L6E9 myoblast heavily infected with trypomastigotes.
Note that there were several bands precipitated by the sialidase antibody and these bands reacted with the same antibody on immunoblot, indicating that they were shed trans-sialidases.
D. Co-IP using L6E9 myoblast culture media heavily infected with trypomastigotes with a commercial sialidase antibody followed by immunoblot with a phospho-(Ser/Thr) PKA substrate antibody. The shed trans-sialidases reacted with the phospho-(Ser/Thr) PKA substrate antibody, indicating that they were phosphorylated by TcPKAc before being shed into the media.
Lane 1, Uninfected L6E9 myoblast culture medium as negative control.
Lane 2, Medium from L6E9 myoblast heavily infected with trypomastigotes.
3.4. A PKAc phosphorylation site flanks endoplasmic reticulum (ER) retention motifs
Examination of the five trans-sialidase protein sequences interacting with TcPKAc demonstrated that all of them possessed an RXR (Arg-X-Arg) ER retention motif near their N-terminus. A PKAc phosphorylation site and a PKC phosphorylation site flanked these sites. It has been reported, in other eukaryotes, that phosphorylation of Serine and/or Threonine near ER retention sites by PKA and PKC suppresses ER retention and regulates surface delivery to membranes [23–24] (Fig.3B).
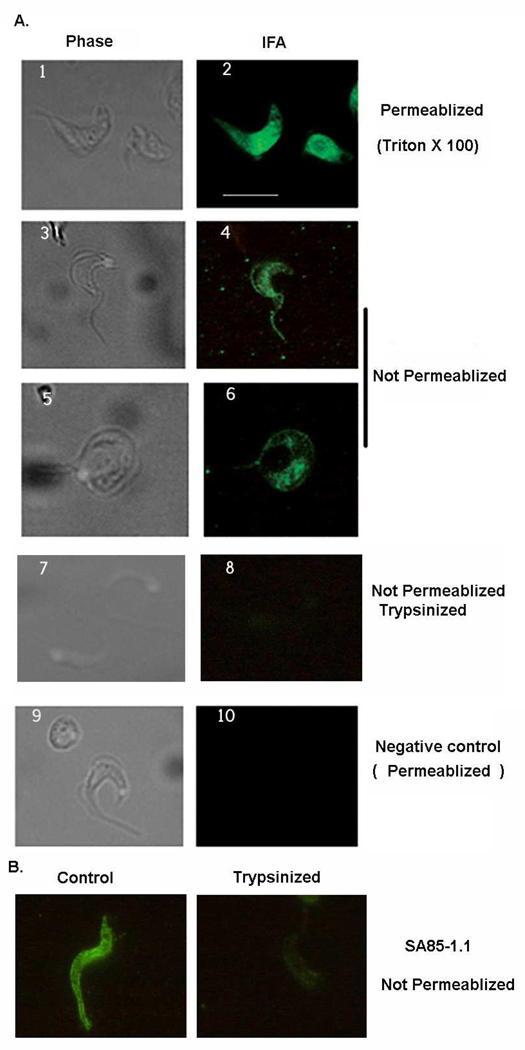
3.5. TcPKAc localizes both to the cytoplasm and membrane
IFA was performed in trypomastigotes (Fig. 5). With permeabilization using Triton X-100 cytoplasmic TcPKAc staining is evident. Without permeabilization the antibody also recognized TcPKAc on the surface membrane and flagellum, indicating that some TcPKAc may be located in surface membrane and flagellum. Thus, it is possible that TcPKAc interacts and phosphorylates surface proteins in this parasite. These observations were further confirmed by trypsin treatment of trypomastigotes. Treatment of trypsin wiped out the surface staining of TcPKAc on trypomastigotes in non-permeabilized parasites (Fig. 5A). In addition, a SA85-1.1 antibody which recognizes some trans-sialidase superfamily surface antigens was used to examine the effect of the treatment of trypsin on trypomastigotes by IFA. We found that treatment of trypsin dramatically reduced the staining with this antibody, confirming that the trypsin treatment method was able to remove surface proteins (Fig. 5B).
Fig. 5. TcPKAc localizes both in cytoplasm and on membrane.
A. IFA was performed in trypomastigotes. With treatment of Triton X-100 for permeabilization, TcPKAc staining is evident in the cytoplasm (1–2 panel). Without permeabilization (no Triton X-100), the antibody also recognized TcPKAc on the surface membrane and flagellum (3–6 panels). Trypsin treatment dramatically reduced surface staining with TcPKAc mAb (7–8 panels). Unrelated mAb (anti-bag5 of Toxoplasma gondii) was used as negative control (9–10 panels).
Scale bars = 10 µm.
B. SA85-1.1 rabbit polyclonal antibody was used to confirm the effect of trypsin treatment of trypomastigotes. Control IFA was performed on parasites without trypsin treatment. Trypsinized parasites were obtained by the process described in Material and Method. Note that Trypsin treatment dramatically reduced surface staining with the SA85-1.1 antibody.
4. DISCUSSION
PKA is a highly conserved serine/threonine protein kinase, which is able to phosphorylate a variety of proteins and thereby affect their activity; however, PKA is held in an inactive form by association of two catalytic subunits (PKAc) with their regulatory subunit (PKAr) dimer. At higher cAMP concentrations, the regulatory subunit binds cAMP at two sites and dissociates from the catalytic subunit [25]. Only when the internal concentration of cAMP increases in cells is PKAc active. T. cruzi has a complex life cycle involving four morphogenetic stages [26]. The epimastigote and metacyclic trypomastigote are insect-specific stages, whereas the blood form trypomastigote and amastigotes are mammalian host-specific, extracellular and intracellular stages, respectively. In T. cruzi, the level of cAMP is elevated in metacyclic trypomastigotes in contrast to log phage epimastigotes [27]. This increase in cAMP level occurs after epimastigotes enter stationary phase, but precedes the onset of metacyclic morphological changes. It has also been observed that the addition of membranepermeable cAMP analogs or adenylate cyclase activators can stimulate metacyclogenesis [11–12]. Although PKA function has been found to be important in T. cruzi, the pathways and proteins with which PKAc interacts have not been elucidated.
The yeast two-hybrid system has been employed by other investigators to identify PKAc downstream interacting proteins. This approach successfully identified downstream targets of PKAc in the yeast Saccharomyces cerevisiae [28]. In addition, these methods were successfully applied in T. cruzi to find the Cdc2p-related protein kinase 1 interacting molecules, resulting in the identification of three novel associating cyclins [29]. Recently, we reported identification of thirty-eight proteins that interact with TcPKAc. Some of these proteins are important in metabolism, environmental adaptation, growth and differentiation [10]. Using this approach, we have now found several genes encoding members of trans-sialidase super-family interacted with TcPKAc. TcPKAc and these trans-sialidase proteins interacted in yeast two-hybrid system under the highest selection stringency. Immunoprecipitation demonstrated that trans-sialidase proteins interacted with TcPKAc in vivo. PKA phosphorylated trans-sialidase proteins both in vitro and in vivo. In addition, by in silico analysis, each of these proteins contained PKA phosphorylation sites. This work demonstrates that these groups of important proteins are likely substrates for PKA. In support of this concept, a recently published paper on the T. cruzi phosphoproteome demonstrated that in epimastigotes four trans-sialidases were phosphorylated [22], one of which has a consensus GSK3 site. The phosphopeptides of the other trans-sialidases were examined by our group and we found that one has a KXXS# (X=any amino acid and S#=phospho-serine) and another has a KXS# motif. While typical PKA consensuses are R-R/K-X-S/T, R-X-X-S/T and R-X-S/T, in many cases R can be replaced by K [17,30]. The trans-sialidase gene family consists of hundreds of genes, including active and inactive trans-sialidases which are anchored by glycosylphosphatidylinositol (GPI) [8] to the surface membrane and are shed by the parasites into the external milieu to transfer sialic acid from host sialoglycoconjugates to β-galactose in the glycoconjugates of the parasites. This action mediates its evasion of the early complement-mediated immune response and its host cell adhesion/invasion [31–32] as well as its host responses by activating phosphoinositide-3 kinase-Akt signaling [33–35]. A parasite-derived neurotrophic factor (PDNF), a trans-sialidase that is located on the surface of T. cruzi, is both a substrate and an activator of the serine-threonine kinase Akt, an anti- apoptotic molecule, in the cytosol [36].
The correlation that exists between the increased cAMP level, elevated PKA activity and trans-sialidase expression in late stage epimastigotes before they differentiate into invasive trypomastigotes is interesting, for it is essential for the survival of this parasite. As trans-sialidases are GPI anchored proteins, which eventually anchor the proteins to the cell membrane before shedding, it is possible that PKA interacts and modifies trans-sialidases intracellularly during protein synthesis and delivery. GPI anchors were found to play an important role in directing the secretory pathway of Trypanosoma. The deletion of a C-terminal GPI addition peptide signal in the variant surface glycoprotein of Trypanosoma brucei significantly reduced its transport kinetics [37]. Other mechanisms regulating the trafficking of GPI anchored proteins probably exist in Trypanosoma. For example, it has been reported that phosphorylation of a major GPI-anchored surface protein, GPEET, by a protein kinase in procyclic forms of Trypanosoma brucei mediates the transport to the plasma membrane. Unphosphorylated GPEET was predominantly in the flagella pocket whereas the phosphorylated form was distributed over the cell surface [38].
PKA has been reported to regulate secretory transport, delivering receptors and ion channels to the plasma membrane in eukaryotic cells [23–24, 39]. Coordinated PKA and PKC phosphorylation suppresses RXR-mediated ER retention and regulate the surface delivery of NMDA receptors [24]. Interestingly, ER retention motifs (RXR) exist at the beginning of N-terminal in the five candidate proteins as interacting with TcPKAc. In addition, PKA and PKC phosphorylation sites are located in these proteins near or between these ER retention motifs (Fig. 3B). It is possible that similar protein trafficking mechanisms that described for NMDA receptors for surface protein delivery exist in Trypanosoma. TCTS 6 11/12 has a homolog in the genome (Tc00.1047053509495.30) and phosphorylation sites also exist at the beginning of the N-terminal (PKA 44–47 and PKC37–39) and an RXR retention signal exists at position (346–348). PKA and PKC phosphorylation may regulate the cellular trafficking of these proteins. As PKA and PKC phosphorylation sites are found in the beginning of the N-terminus of these proteins, phosphorylation may occur in cytoplasm before these proteins target to ER. Examination of T. cruzi genome demonstrates that the ER retention motif with associated PKA and PKC phosphorylation sites is highly conserved and exists in many trans-sialidase protein sequences (data not shown). It is possible that coordinated PKA and PKC phosphorylations may promote trans-sialidase surface delivery. These mechanism(s) clearly require further investigation in this parasite system.
Immunofluorescence indicated that in addition to its characteristic cytoplasmic localization some of the TcPKAc resided on the plasma membrane of trypomastigotes (Fig. 5). The observation was confirmed by trypsin treatment of the parasites. This suggests that TcPKAc could interact or phosphorylate surface proteins such as the anchored trans-sialidases. T. cruzi PKA also localizes in flagellum and there is an adenylyl cyclase that also localizes in flagellum [40]. Adenylyl cyclase generates cAMP, which in turn activates PKA. Although the majority of biochemical mechanisms and effects of cAMP and PKA presumably occur inside cells; and PKA is known predominantly to be an intracellular enzyme [41], extracellular cell surface PKA activity has been reported in many cell types [41]. N-myristoylation is required for PKAc export [20] and is reported to mediate PKAc to membrane localization [41]. To this end, in silico sequence analysis identified several potential myristoylation sequences in each of the TcPKAc isoforms (Table 1C). Although these myristoylation sites are not at the N-terminus, TcPKAc Isoform 2 has a predicted myristoylation at position 25. If TcPKAc isoform 2 is post-translationally proteolytically processed it could be modified by N-myristoylation resulting in the membrane localization of a subset of the expressed TcPKAc. Therefore, a similar mechanism of PKA targeting to the plasma membrane may exist in this organism as has been reported in other organisms [41]. Finally, TcPKAc may also be associated with the plasma membrane via its association with its regulatory subunit, which also localizes in membrane and flagellum in trypomastigotes [14]. The significance and mechanism(s) of TcPKAc targeting to the membrane is a topic of active investigation by our laboratory group.
In summary, this is the first demonstration that several members of trans-sialidase super family are probable PKA substrates. These proteins are unique molecules in T. cruzi mediating invasion and immuno-evasion. Cyclic AMP-PKA signaling is a fundamental pathway regulating many essential functions such as metabolism, adaptation, growth and differentiation in eukaryotic cells. It is quite possible that phosphorylation of the members of trans-sialidase super family by PKA regulates their intracellular trafficking, protein turnover or enzymatic activities. Our laboratory group will continue to investigate these mechanisms, as an increased understanding of the regulation of PKA on these molecules in T. cruzi could lead to a novel approach to potential therapeutic targets.
ACKNOWLEDGMENTS
We thank Drs. Laura Ratier and Alberto C. C. Frasch for providing reagents. National Institutes of Health Grants AI 058893, AI076248 and AI05739 supported this work.
The abbreviations used in this manuscript are
- PKA
Protein Kinase A
- PKAc
Protein Kinase A catalytic subunit
- T. cruzi
Trypanosoma cruzi
- mAb
monoclonal antibody
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Weiss LM, Wittner M, Coyle C, Shah S, Tanowitz HB. Parasitic infections of the nervous system and the eye in AIDS. Neurological Infections and Epidemiology. 1997;2:113–127. [Google Scholar]
- 2.Ferreira MS. Infections by protozoa in immunocompromised hosts. Mem Inst Oswaldo Cruz. 2000;95 Suppl 1:159–162. doi: 10.1590/s0074-02762000000700026. [DOI] [PubMed] [Google Scholar]
- 3.Lages-Silva E, Ramirez LE, Silva-Vergara M, Chiari E. Chagasic meningoencephalitis in a patient with acquired immunodeficiency syndrome, diagnosis, follow-up, and genetic characterization of Trypanosoma cruzi. Clin Inf Dis. 2002;34:118–123. doi: 10.1086/324355. [DOI] [PubMed] [Google Scholar]
- 4.Sartori AM, Lopes MH, Benvenuti LA, Caramelli B, di Pietro A, Nunes EV, Ramirez LP, Shikanai-Yasuda MA. Reactivation of Chagas’ disease in a human immunodeficiency virus-infected patient leading to severe heart disease with a late positive direct microscopic examination of the blood. Am J Trop Med Hyg. 1998;59:784–786. doi: 10.4269/ajtmh.1998.59.784. [DOI] [PubMed] [Google Scholar]
- 5.Sartori AM, Lopes MH, Caramelli B, Duarte MI, Pinto PL, Neto V, Amato Shikanai-Yasuda M. Simultaneous occurrence of acute myocarditis and reactivated Chagas’ disease in a patient with AIDS. Clin Inf Dis. 1995;21:1297–1299. doi: 10.1093/clinids/21.5.1297. [DOI] [PubMed] [Google Scholar]
- 6.Rocha A, de Meneses AC, da Silva AM. Pathology of patients with Chagas’ disease and acquired immunodeficiency syndrome. Am J Trop Med Hyg. 1994;50:261–268. doi: 10.4269/ajtmh.1994.50.261. [DOI] [PubMed] [Google Scholar]
- 7.Leiby DA, Herron RM, Read EJ, Lenes BA, Stumpf RJ. Trypanosoma cruzi in Los Angeles and Miami blood donors: impact of evolving donor demographics on seroprevalence and implications for transfusion transmission. Transfusion. 2002;42:549–555. doi: 10.1046/j.1537-2995.2002.00077.x. [DOI] [PubMed] [Google Scholar]
- 8.Frasch ACC. Functional Diversity in the Trans-sialidase and Mucin Families in Trypanosoma cruzi. Parasitol Today. 2000;16:282–286. doi: 10.1016/s0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- 9.Thevelein JM, Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 10.Bao Y, Weiss LM, Braunstein VL, Huang H. The role of Protein kinase A in Trypanosoma cruzi. Infect. Immun. 2008;76:4757–4763. doi: 10.1128/IAI.00527-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naula C, Seebeck T. Cyclic AMP Signaling in Trypanosomatides. Parasitol. Today. 2000;16:35–38. doi: 10.1016/s0169-4758(99)01582-3. [DOI] [PubMed] [Google Scholar]
- 12.Gonzales-Perdomo M, Romero P, Goldenberg S. Cyclic AMP and adenylate cyclase activators stimulate Trypanosoma cruzi differentiation. Exp. Parasitol. 1988;66:205–212. doi: 10.1016/0014-4894(88)90092-6. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Werner C, Weiss LM, Wittner M, Orr GA. Molecular cloning and expression of hte catalytic subunit of protein kinase A from Trypanosoma cruzi. Int J Parasitol. 2002;32:1107–1115. doi: 10.1016/s0020-7519(02)00085-1. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Weiss LM, Nagajyothi F, Tanowitz HB, Wittner M, Orr GA, Bao Y. Molecular cloning and characterization of the protein kinase A regulatory subunit of Trypanosoma cruzi. Mol. Biochem Parasitol. 2006;149:242–245. doi: 10.1016/j.molbiopara.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Kahn S, Colbert TG, Wallace JC, Hoagland NA, Eisen H. The major 85-kDa surface antigen of the mammalian-stage forms of Trypanosoma cruzi is a family of sialidases. Proc Natl Acad Sci U S A. 1991;88:4481–4485. doi: 10.1073/pnas.88.10.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremona ML, Sánchez DO, Frasch AC, Campetella O. single tyrosine differentiates active and inactive Trypanosoma cruzi trans-sialidases. Gene. 1995;160:123–128. doi: 10.1016/0378-1119(95)00175-6. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi H, Yamashita T, Yoshikawa H, Tohyama M. PKA phosphorylates the p75 receptor and regulates its localization to lipid rafts. EMBO J. 2003;22:1790–1800. doi: 10.1093/emboj/cdg177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews NW, Katzin AM, Colli W. Mapping of surface glycoproteins of Trypanosoma cruzi by two-dimensional electrophoresis. Eur. J. Biochem. 1984;140:599–6049. doi: 10.1111/j.1432-1033.1984.tb08144.x. [DOI] [PubMed] [Google Scholar]
- 20.Cho YS, Park YG, Lee YN, Kim MK, Bates S, Tan L, Cho-Chung YS. Extracellular protein kinase A as a cancer biomarker: its expression by tumor cells and reversal by a myristate-lacking Calpha and RIIbeta subunit overexpression. Proc Natl Acad Sci U S A. 2000;97:835–840. doi: 10.1073/pnas.97.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briones MR, Egima CM, Eichinger D, Schenkman S. Trans- genes expressed in mammalian forms of Trypanosoma cruzi evolved from ancestor genes expressed in insect forms of the parasite. J Mol Evol. 1995;41:120–131. doi: 10.1007/BF00170663. [DOI] [PubMed] [Google Scholar]
- 22.Nakayasu E.S. ES, Gaynor MR, Sobreira TJ, Ross JA, Almeida IC. Phosphoproteomic analysis of the human pathogen Trypanosoma cruzi at the epimastigote stage. Proteomics. 2009;9:3489–3506. doi: 10.1002/pmic.200800874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott DB, Blanpied TA, Ehlers MD. Coordinated PKA and PKC phosphorylation suppresses RXR-mediated ER retention and regulates the surface delivery of NMDA receptors. Neuropharmacology. 2003;45:755–767. doi: 10.1016/s0028-3908(03)00250-8. [DOI] [PubMed] [Google Scholar]
- 25.Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 1990;9:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 26.Tanowitz HB, Kirchhoff LV, Simon D, Morris SA, Weiss LM, Wittner M. Chagas’ disease. Clinical Microbiol Rev. 1992;5:409–419. doi: 10.1128/cmr.5.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flawia MM, Tellez-Inon MT, Torres HN. Signal transduction mechanisms in Trypanosoma cruzi. Parasitol. Today. 1997;13:30–33. doi: 10.1016/s0169-4758(96)10070-3. [DOI] [PubMed] [Google Scholar]
- 28.Laura SR, Gerald RF. The three yeast A linase have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci U S A. 1998;23:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gómez EB, Santori MI, Laría S, Engel JC, Swindle J, Eisen H, Szankasi P, Téllez-Iñón MT. Characterization of the Trypanosoma cruzi Cdc2p-related protein kinase 1 and identification of three novel associating cyclins. Mol Biochem Parasit. 2001;113:97–108. doi: 10.1016/s0166-6851(00)00382-0. [DOI] [PubMed] [Google Scholar]
- 30.Kang JH, Toita R, Jiang Y, Niidome T, Katayama Y. Simultaneous Analysis of Phosphorylated Peptides by MALDI-TOF-MS. Chromatographia. 2006;63:595–598. [Google Scholar]
- 31.Tomlinson S, Raper J. Natural human immunity to trypanosomes. Parasitol. Today. 1998;14:354–359. doi: 10.1016/s0169-4758(98)01295-2. [DOI] [PubMed] [Google Scholar]
- 32.Schenckman S, Eichinger D. Trypanosoma cruzi trans-sialidase and cell invasion. Parasitol. Today. 1993;9:218–222. doi: 10.1016/0169-4758(93)90017-a. [DOI] [PubMed] [Google Scholar]
- 33.Chuenkova MV, Pereira MA. A trypanosomal protein synergizes with the cytokines ciliary neurotrophic factor and leukemia inhibitory factor to prevent apoptosis of neuronal cells. Mol. Biol. Cell. 2000;11:1487–1498. doi: 10.1091/mbc.11.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chuenkova MV, Furnari FB, Cavenee WK, Pereira MA. Trypanosoma cruzi trans-sialidase: a potent and specific survival factor for human Schwann cells by means of phosphatidylinositol 3-kinase/Akt signaling. Proc. Natl. Acad. Sci. U S A. 2001;98:9936–9941. doi: 10.1073/pnas.161298398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chuenkova MV, PereiraPerrin MA. A synthetic peptide modeled on PDNF, Chagas' disease parasite neurotrophic factor, promotes survival and differentiation of neuronal cells through TrkA receptor. Biochemistry. 2005;44:15685–15694. doi: 10.1021/bi0512039. [DOI] [PubMed] [Google Scholar]
- 36.Chuenkova MV, PereiraPerrin M. Trypanosoma cruzi targets Akt in host cells as an intracellular antiapoptotic strategy. Sci Signal. 2009;2:1–9. doi: 10.1126/scisignal.2000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDowell MA, Ransom DM, Bangs JD. Glycosylphosphatidylinositoldependent secretory transport in Trypanosoma brucei. Biochem J. 1998;335:681–689. doi: 10.1042/bj3350681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bütikofer P, Vassella E, Ruepp S, Boschung M, Civenni G, Seebeck T, Hemphill A, Mookherjee N, Pearson TW, Roditi I. Phosphorylation of a major GPI-anchored surface protein of Trypanosoma brucei during transport to the plasma membrane. J Cell Sci. 1999;112:1785–1795. doi: 10.1242/jcs.112.11.1785. [DOI] [PubMed] [Google Scholar]
- 39.O'Connell AD, Leng Q, Dong K, MacGregor GG, Giebisch G, Hebert SC. Phosphorylation-regulated endoplasmic reticulum retention signal in the renal outermedullary K+ channel (ROMK) Proc Natl Acad Sci U S A. 2005;102:9954–9959. doi: 10.1073/pnas.0504332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Angelo MA, Montagna AE, Sanguineti S, Torres HN, Flawiá MM. A novel calcium-stimulated adenylyl cyclase from Trypanosoma cruzi, which interacts with the structural flagellar protein paraflagellar rod. J Biol Chem. 2002;277:35025–35034. doi: 10.1074/jbc.M204696200. [DOI] [PubMed] [Google Scholar]
- 41.Chin KV, Yang WL, Ravatn R, Kita T, Reitman E, Vettori D, Cvijic ME, Shin M, Iacono L. Reinventing the wheel of cyclic AMP: novel mechanisms of cAMP signaling. Ann N Y Acad Sci. 2002;968:49–64. doi: 10.1111/j.1749-6632.2002.tb04326.x. [DOI] [PubMed] [Google Scholar]