Abstract

Escherichia coli NikR regulates cellular nickel uptake by binding to the nik operon in the presence of nickel and blocking transcription of genes encoding the nickel uptake transporter. NikR has two binding affinities for the nik operon: a nanomolar dissociation constant with stoichiometric nickel and a picomolar dissociation constant with excess nickel [Bloom, S. L., and Zamble, D. B. (2004) Biochemistry 43, 10029−10038; Chivers, P. T., and Sauer, R. T. (2002) Chem. Biol. 9, 1141−1148]. While it is known that the stoichiometric nickel ions bind at the NikR tetrameric interface [Schreiter, E. R., et al. (2003) Nat. Struct. Biol. 10, 794−799; Schreiter, E. R., et al. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13676−13681], the binding sites for excess nickel ions have not been fully described. Here we have determined the crystal structure of NikR in the presence of excess nickel to 2.6 Å resolution and have obtained nickel anomalous data (1.4845 Å) in the presence of excess nickel for both NikR alone and NikR cocrystallized with a 30-nucleotide piece of double-stranded DNA containing the nik operon. These anomalous data show that excess nickel ions do not bind to a single location on NikR but instead reveal a total of 22 possible low-affinity nickel sites on the NikR tetramer. These sites, for which there are six different types, are all on the surface of NikR, and most are found in both the NikR alone and NikR−DNA structures. Using a combination of crystallographic data and molecular dynamics simulations, the nickel sites can be described as preferring octahedral geometry, utilizing one to three protein ligands (typically histidine) and at least two water molecules.
Regulatory proteins play key roles within the cell, maintaining a crucial balance of nutrients and preventing the overload of any element or compound that could prove lethal to the cell at higher concentrations. One class of regulatory proteins is the ligand-binding transcription factor, which is often described as controlling gene expression in a strictly on−off manner. However, if a protein is able to fine-tune the degree of gene repression or induction, rather than inducing a strictly on−off response, this would make for a highly efficient transcription factor. Escherichia coli NikR,1 the nickel regulatory transcription factor, has been shown to have two distinct DNA binding affinities for its operon in vitro: nanomolar when stoichiometric nickel is bound and picomolar when excess nickel is present (1,2). Although in vivo numbers are not established, the in vitro data raise the possibility that the cellular response to nickel could be incrementally adjusted. Thus, understanding the molecular basis for these different observed NikR−operon binding affinities has been a point of interest in this system. One key unanswered question is the exact location and description of these low-affinity sites on the NikR protein.
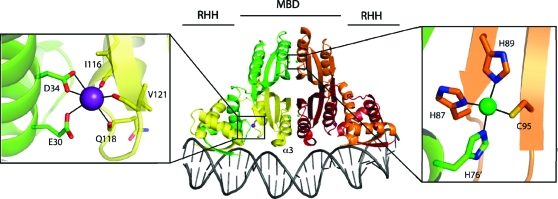
NikR is a homotetrameric protein composed of two types of domains: a central tetrameric metal binding domain (MBD) and two flanking dimeric ribbon−helix−helix (RHH) or DNA-binding domains (Figure 1) (2,3,5). NikR has multiple types of metal binding sites: high-affinity nickel binding sites for stoichiometric nickel (low picomolar dissociation constant) (2,6), potassium binding sites (4,7,8), and low-affinity nickel binding sites (dissociation constants from 30 μM to 30 nM) (1,2). The four identical square-planar high-affinity nickel binding sites are located at the central tetrameric interface in the MBD (Figure 1) (3,4,9). When nickel ions bind to the high-affinity sites, they order the helix α3 and the preceding loop, which then makes contacts with DNA, thereby initiating NikR−DNA binding (4,10).
Figure 1.
Overall topology of NikR with the metal binding domains (MBD) and ribbon−helix−helix (RHH) domains indicated. Representative structures of the potassium (left panel) and high-affinity nickel sites (right panel) with coordinating amino acids labeled.
When DNA binds, two identical octahedral metal binding sites are formed between the MBD and RHH domains (Figure 1) (4). A metal bound to this site could serve to stabilize the RHH domains in a DNA-binding conformation, increasing the affinity of NikR for DNA. Although potassium from the crystallization buffer was modeled in these sites in the NikR−DNA complex crystal structure (4), it was thought that this location could be the experimentally observed low-affinity nickel binding sites responsible for enhancing NikR’s affinity for DNA when excess nickel is present (11,12). However, recent biochemical studies show that NikR’s affinity for DNA is also dependent on potassium (8), and calculations examining metal stability, charge, and size preference suggest that these second metal binding sites are optimized for potassium binding (7). Thus, there is an emerging consensus that potassium is the relevant metal in the second metal binding site and, as such, is the metal responsible for stabilizing the RHH domains in a DNA-binding conformation.
Although it has not been possible to measure the levels of nikABCDE transcription as a function of cellular nickel concentration, in vitro studies suggest that NikR has a third type of metal binding site to which excess nickel ions bind, changing the dissociation constant of NikR for DNA from the nanomolar to picomolar range (1,2). In this work, we locate and describe these low-affinity nickel binding sites. Using X-ray crystallography and nickel anomalous maps, potential low-affinity nickel sites on NikR are identified. To visualize these sites on the NikR−DNA complex, we utilized a combination of the low-resolution X-ray anomalous data and molecular modeling. Through this work, we can describe the molecular basis for the in vitro low-affinity nickel effect and begin to consider possible physiological functions for low-affinity nickel sites, such as whether they might be involved in tuning NikR’s DNA affinity more precisely than a simple on−off mode for transcriptional regulation or, alternatively, whether they could act as a “sink” for excess nickel under certain cellular conditions.
Experimental Procedures
Refinement of the Crystal Structure of NikR Soaked with 8 mM NiCl2 and Identification of Nickel Sites
Full-length E. coli NikR with stoichiometric Ni2+ bound was purified and crystallized as previously described (3,4). NikR crystals were soaked in 2.5 μL of precipitation solution [200 mM MgCl2, 100 mM Hepes (pH 8.5), 30% (v/v) polyethylene glycol (PEG 400), and 70 mM detergent molecule cyclohexyl propyl β-d-maltoside (CYMAL-3)] supplemented with 8 mM NiCl2 at room temperature for 70 min and then cryocooled to 100 K in a gaseous N2 stream. Cocrystallization was not a possibility because of protein precipitation at high nickel concentrations. A data set was collected on this crystal at the nickel peak wavelength (1.4845 Å) at beamline 5.0.2 of the Advanced Light Source and processed keeping Friedel mates separate using DENZO/SCALEPACK (13).
The unit cell did not change significantly between the structure of stoichiometric Ni2+-bound NikR [Protein Data Bank (PDB) entry 2HZA] and the structure of NikR soaked with excess nickel (original NikR unit cell, space group P3121, a = b = 49.8 Å, c = 181.7 Å; 8 mM NiCl2-soaked NikR unit cell, space group P3121, a = b = 50.7 Å, c = 183.1 Å). Thus, the stoichiometric Ni2+-bound NikR structure with waters removed was refined against this data set using CNS (4,14). Rigid body refinement of the whole structure and individual domains was followed by simulated annealing, positional, and individual B-factor refinement for final Rwork and Rfree values of 23.1 and 28.7%, respectively, with no σ cutoff. Data collection and refinement statistics are listed in Table 1. The final model contains two protein chains, both missing the last C-terminal residue and one chain missing residues 46−48, which lie on a loop region connecting the RHH domain and MBD and are also disordered in the stoichiometric Ni2+-bound NikR structure (4). The model also contains a total of 11 nickel ions (two bound at the high-affinity sites and nine bound at the low-affinity sites), one copy of the detergent molecule CYMAL-3, and 63 water molecules. Phases from the refined protein model were used to calculate an anomalous difference Fourier map in CNS to locate the positions of all nickel ions in the structure. A 3.5σ contour of this map with the two crystallographically unique NikR subunits is shown in Figure 2a. Following positional and B-factor refinement of the structure, each nickel ion was assigned a B-factor corresponding to the average B-factor of the residues ligating that ion and the occupancies of the nickel ions were sequentially refined (Table 2). Composite omit, 2Fo − Fc, and Fo − Fc difference maps were used throughout refinement to verify the structure (an example of the composite omit map density is shown in Figure S2 of the Supporting Information). Figure 3 shows the low-affinity sites in the NikR alone structure with 2Fo − Fc electron density and anomalous nickel density, illustrating the quality of the density that was used to model the nickel ligands.
Table 1. Data Processing Statistics for NikR and the NikR−DNA Complex with Low-Affinity Nickel Sites Filled and Refinement Statistics for NikR with Low-Affinity Sites Filleda.
| NikR soaked with 8 mM NiCl2 | NikR−DNA complex soaked with 5 mM NiCl2 | |
|---|---|---|
| space group | P3121 | C2 |
| cell dimensions | ||
| a (Å) | 50.7 | 199.6 |
| b (Å) | 50.7 | 82.3 |
| c (Å) | 183.1 | 125.7 |
| α (deg) | 90 | 90 |
| β (deg) | 90 | 104.7 |
| γ (deg) | 120 | 90 |
| wavelength (Å) | 1.4845 | 1.4862 |
| temperature (K) | 100 | 100 |
| resolution range (Å)b | 50−2.6 (2.69−2.60) | 20−3.6 (3.73−3.60) |
| no. of unique reflectionsb | 8682 (649) | 43937 (3843) |
| average redundancy | 9.3 | 3.3 |
| completeness (%)b | 98.3 (89.3) | 97.5 (85.6) |
| I/σ(I)b | 16.3 (4.5) | 14.3 (1.6) |
| Rsym (%)b,c | 8.9 (28.8) | 10.6 (47.1) |
| no. of NikR monomers per asymmetric unit | 2 | 8 |
| Rcryst (Rfree) (%)d | 23.1 (28.7) | |
| average B-factor (Å2)e | ||
| overall | 71.7 (2105) | |
| protein | 70.0 (199) | |
| high-affinity nickels | 70.5 (2) | |
| excess nickels | 138.0 (9) | |
| waters | 59.7 (63) | |
| detergent molecule | 154.6 (32) | |
| root-mean-square deviation | ||
| bond lengths (Å) | 0.011 | |
| bond angles (deg) | 1.2 | |
| Ramachandran plot (%) | ||
| most favored | 84.2 | |
| additionally allowed | 14.6 | |
| generously allowed | 0.8 | |
| disallowed | 0.4 |
Both data sets were scaled anomalously.
The number in parentheses is for the highest-resolution shell.
Rsym = ∑ihkl|Ii(hkl) − ⟨I(hkl)⟩|/∑hkl⟨I(hkl)⟩, where Ii(hkl) is the ith measured diffraction intensity and ⟨I(hkl)⟩ is the mean of the intensity for the Miller index (hkl).
Rcryst = ∑hkl||Fo(hkl)| − |Fc(hkl)||/∑hkl|Fo(hkl)|. Rfree = Rcryst for a test set of reflections (5%) not included in the refinement.
Numbers in parentheses are the numbers of atoms in each category.
Figure 2.
Nickel anomalous density maps of excess nickel-soaked NikR and the NikR−DNA complex. (a) Dimer found in the asymmetric unit of the NikR structure with excess nickel ions. (b) NikR−DNA complex structure with excess nickel ions. Nickel anomalous density is shown in blue mesh at 3.5σ. Excess nickel ions are shown as cyan spheres, high-affinity nickel ions as green spheres, and potassium ions as purple spheres. Each monomer chain is colored uniquely. Nickel sites are numbered corresponding to the site types in Table 2.
Table 2. Descriptions of the Types of Low-Affinity Nickel Sites Seen in the Structures of NikR and the NikR−DNA Complex.
| site typea | description | no. of sites per NikR dimerb | no. of sites per NikR−DNA tetramerb | protein ligandsc | average occupancy | average anomalous signald | |
|---|---|---|---|---|---|---|---|
| 0 | high-affinity site | 2 (2) | 4 (4) | H87, H89, C95, H76′ | 0.94 | 22 | 7 |
| 1 | low-affinity site 1 | 1 (1) | 2 (2) | H125, H125′ | 0.64 | 20 | 7 |
| 2 | low-affinity site 2 | 2 (2) | 4 (4) | H79, H92′ | 0.71 | 15 | 7 |
| 3 | low-affinity site 3 | 2 (2) | 4 (4) | H110, D114, H78 | 0.88 | 22 | 7 |
| 4 | NikR−DNA only site | 0 (2) | 2 (4) | H48 | not available | 0 | 3 |
| 5 | NikR only site | 2 (2) | 0 (4) | H123 | 0.45 | 7 | 0 |
| 6 | N-terminal site | 2 (2) | 2 (4) | M1 | 0.61 | 19 | 2 |
Site type numbers correspond to sites in Figure 2. Site types 0−3 and site type 6 are in both structures; site type 4 is only in the NikR−DNA complex structure, and site type 5 is only in the structure of NikR alone.
The number in parentheses is the total possible number of each type of site on the NikR dimer or NikR−DNA tetrameric complex.
The prime indicates that the residues are located on a different chain.
Average anomalous signal refers to the average maximum σ level of the nickel anomalous map at which density for a given ion disappears. σ levels determined from the nickel anomalous map for the NikR structure with excess nickel ions (maximum signal of 25σ) are listed in the left column, and σ levels from the nickel anomalous map for the NikR−DNA complex structure with excess nickel ions (maximum signal of 10σ) are listed in the right column. For site types 4−6, which have at least one potential site empty in either the NikR alone or NikR−DNA structure, 0 was averaged in for each empty site.
Figure 3.
Low-affinity nickel sites 1−3, 5, and 6 from the crystal structure of NikR without DNA. The 2Fo − Fc electron density maps at 1.0σ are colored gray around the coordinating protein ligands, water molecules, and metal ions. Nickel anomalous density maps at 3.5σ are colored blue. The coloring is the same as in Figure 2. The low-affinity nickel sites are numbered and correspond to the sites numbered in the NikR model (middle right) as well as the sites in Figure 2 and Table 2.
Identifying Nickel Sites on a NikR−DNA Complex Using Anomalous Scattering from Low-Resolution X-ray Data
While it was not possible to obtain a high-resolution structure of NikR bound to DNA with excess nickel ions, the low-resolution structural information and nickel anomalous maps were used to locate nickel sites on the NikR−DNA complex. Crystals of the NikR−DNA complex with stoichiometric Ni2+ bound were grown as described previously (4). One such crystal was soaked in precipitant solution [200 mM KCl, 50 mM MgCl2, 50 mM Tris (pH 7.5), and 10 (w/v) PEG 4000] supplemented with 5 mM NiCl2 at room temperature for 60 min before cryoprotection with 25% ethylene glycol and cryocooling in a gaseous N2 stream at 100 K. A data set was collected on this crystal at the nickel peak wavelength (1.4862 Å) at beamline X29A of the NSLS and processed anomalously using DENZO/SCALEPACK (13). Data processing statistics are listed in Table 1.
There was a significant change in the unit cell upon soaking with excess nickel (original NikR−DNA unit cell, a = 195 Å, b = 76 Å, c = 132 Å, and β = 110°; unit cell after soaking with 5 mM NiCl2, a = 120 Å, b = 82 Å, c = 126 Å, and β = 105°), so simple rigid body refinement with the previous NikR−DNA structure (PDB entry 2HZV) was not successful. Thus, we used molecular replacement in Phaser with a single copy of the NikR−DNA complex structure (PDB entry 2HZV) with waters removed as the search model (4,15). Two copies of the NikR−DNA complex were found in the asymmetric unit (asu), resulting in a Z score of 27. Further refinement of the structure was attempted in CNS, but poor data quality, likely due to the change in cell dimensions resulting from the nickel soaking, prevented full refinement of the structure. However, when phase information from molecular replacement is combined with the anomalous data, the resulting nickel anomalous map illustrates the approximate location of the nickel ions (Figure 2b). Many of these sites are also observed in the NikR alone structure (Table 2 and Figure 2a). Only one site (site 4) was present in the NikR−DNA complex but not in the NikR alone structure, and this site has a comparatively low anomalous signal and occupancy and appears to be stabilized by crystal contacts (Table 2 and Figure S1 of the Supporting Information).
Modeling Ligands to Potential Low-Affinity Nickel Sites on the NikR−DNA Complex
The higher-resolution electron density maps of the NikR alone structure allowed us to model the protein ligands and potential water ligands to each of the excess nickel sites. While anomalous scattering indicates the approximate positions, low-resolution electron density prevented modeling of ligands in the NikR−DNA structure. Therefore, we modeled low-affinity nickel ion site types 1−3 on the NikR−DNA complex. These low-affinity nickel sites are filled at least two-thirds of the time or have occupancies of >66% (Table 2). In addition, site types 1−3 were found in both the NikR alone and NikR−DNA complex nickel anomalous maps at all potential sites, suggesting that these sites are the most preferred low-affinity nickel sites.
To model these sites, the NikR−DNA complex structure with stoichiometric nickel was aligned with the NikR structure with excess nickel ions in COOT (16). The low-affinity nickel ions (site types 1−3) were modeled onto the NikR−DNA complex structure using the NikR structure with low-affinity nickel ions as a guide. To ensure correct metal coordination, the amino acid side chains in the NikR−DNA complex responsible for metal ligation were reoriented to match the geometries observed in the unbound NikR structure, and water molecules coordinating metal ions in the NikR structure were added to the NikR−DNA complex structure.
Water molecules refined in the crystal structure (Figure 2) were included in the minimization studies and molecular dynamics simulations. The structure was then minimized using 200 steps of steepest descent in CHARMM to remove bad contacts (17). After the structure had been solvated in a 57 Å radius water sphere, a total of 21276 TIP3P water molecules (including the 32 crystallographic waters) were explicitly modeled in our system and the water boundary was maintained using a spherical stochastic boundary potential (18). Finally, to allow the low-affinity sites to rearrange before dynamics, all water molecules and protein residues within 5 Å of each low-affinity nickel site were minimized using 500 steps of conjugate gradient minimization.
Molecular dynamics simulations were conducted using CHARMM to obtain a sampling of different conformations of each low-affinity nickel site (17). To maintain the histidine ligands seen in the crystal structure, nickel−histidine distances were restrained with a strong force constant of 100 kcal mol−1 Å−2, using the distances measured in the crystal structure as a guide (17). All protein atoms were held fixed except atoms in residues within 5 Å of each low-affinity site. In addition, water molecules and low-affinity nickel ions underwent full molecular dynamics. The protein was first gently heated from 0 to 300 K at a rate of 5 K/ps and the temperature maintained using a Nosé-Hoover heat bath (19). Dynamical simulations were then run at 300 K until the system was fully equilibrated. The total run time was 1.1 ns, and structures were saved every 10 ps. All calculations in CHARMM were performed with an atom-based cutoff where the electrostatic interactions were brought to zero between 8 and 12 Å, van der Waals interactions were turned off at 12 Å, and the nonbonded list cutoff was at 13 Å. In addition, the SHAKE command was used to restrain bonds involving hydrogen atoms near their equilibrium values (20).
Results
Crystal Structure of NikR Soaked with 8 mM NiCl2 in the Absence of DNA
To visualize the low-affinity nickel binding sites, we soaked stoichiometric Ni2+-bound NikR crystals with excess nickel chloride and collected data at the nickel peak wavelength (1.4845 Å). A NiCl2 concentration of 8 mM was used in the soaking experiments because this was the maximum concentration that could be used without destroying the crystal. We were not able to cocrystallize NikR or the NikR−DNA complex with excess nickel due to precipitation of the protein under these conditions. Similar to data for the stoichiometric Ni2+-bound full-length NikR, the excess nickel-soaked NikR data indicated a dimer, or half of the full NikR tetramer, in the same asymmetric unit (asu) and in space group P3121. Data processing and refinement statistics are listed in Table 1. The overall fold of NikR did not change when it was soaked with excess Ni2+, with an all-atom root-mean-square deviation (rmsd) of 0.50 Å between the stoichiometric and excess Ni2+-soaked structures.
The nickel anomalous maps generated from the structure following the soaking experiment indicate nine potential low-affinity nickel binding sites on the surface of a NikR dimer (Figure 2a). Because of the symmetry of the dimer, the nine sites can be categorized into five types on NikR. The site types are numbered in Figure 2a and correspond to the descriptions in Table 2. After refining the NikR structure, we were able to estimate the occupancies of each of the low-affinity sites as described in Experimental Procedures. Nickel ions in the stoichiometric or high-affinity sites (site type 0) refine to the highest occupancy of 0.94, while nickels in the potential low-affinity nickel sites have varying occupancies (45−88%) (Table 2). These sites are described in detail below.
Nickel Anomalous Density in the NikR−DNA Structure Soaked with 5 mM NiCl2
To visualize the low-affinity nickel binding sites when NikR is complexed with DNA, we soaked stoichiometric Ni2+−NikR−DNA crystals with excess nickel chloride and collected data at the nickel peak wavelength (1.4845 Å). A NiCl2 concentration of 5 mM was used in these soaking experiments rather than 8 mM, because these crystals were not stable at 8 mM NiCl2. The excess nickel-soaked NikR−DNA crystals were indexed in the same space group, C2, as those of the published NikR−DNA complex (4), yet with sufficiently different cell parameters such that simple rigid body refinement could not be used to solve the phase problem. Thus, the phases were determined with molecular replacement using a single copy of the published NikR−DNA complex as the search model (4). Data processing statistics are listed in Table 1. The low resolution of this crystal structure (3.6 Å) and poor data quality did not warrant refinement past the initial molecular replacement solution. However, phases found using molecular replacement were used to calculate a nickel anomalous map, which indicated the location of the excess nickel ions (Figure 2b). Comparing the low-affinity sites from the NikR−DNA complex structure with the structure of NikR alone, we find that most sites are conserved (site types 1−3 and site type 6 described in Figure 2 and Table 2).
Low-Affinity Ni2+ Sites from the Crystal Structures
From an analysis of both the NikR alone and NikR−DNA complex structures with excess nickel ions, there are a total of six types of low-affinity nickel sites, five of which have clear 2Fo − Fc electron density for coordinating ligands because these sites are present in the 2.6 Å resolution NikR structure in the absence of DNA (Figure 2a, site types 1−3, 5, and 6). Site type 1 is present in both structures and corresponds to a nickel ion coordinated by H125, one from each of the monomers composing the NikR dimer (Figures 2 and 3). There is 2Fo − Fc electron density for two water molecules acting as additional ligands to the nickel (Figure 3). This site most closely resembles a distorted seesaw geometry with a H125−Ni−H124 angle of 71°. While these sites are solvent accessible, they are the most buried of all the observed excess nickel binding sites. Site type 1 is well-coordinated with four ligands to the nickel ion and has an average occupancy of 0.64 in the NikR structure with strong nickel anomalous electron density peaks in both the NikR (20σ) and NikR−DNA complex structures (7σ) (Figure 2 and Table 2).
Site type 2 is located in a central region of the structure near the tetrameric interface. Each site is coordinated by H79 of one monomer and H92 of the monomer across the tetrameric interface (Figure 3). Four water molecules visible in the electron density maps complete an octahedral geometry around the nickel ion (Figure 3). The occupancies of the type 2 sites refine to an average of 0.71 (Table 2). In addition, there is nickel anomalous density at every potential site type 2 location in both NikR and the NikR−DNA complex (Figure 2).
Coordinated by H110, H78, and D114 from the same monomer, site type 3 also has nickel anomalous density to at least 20σ in both monomers of the NikR dimer (maximum signal at 25σ) and at least 6σ in all four monomers of the NikR−DNA complex (maximum signal at 10σ) (Figure 2 and Table 2). This site utilizes three amino acid ligands and three water ligands, creating a common octahedral geometry for the nickel ion (Figure 3). This excess nickel binding site appears to be filled in all of the NikR monomers within the crystal, with an average occupancy of 0.88.
Because site type 4 is only visualized in the nickel anomalous maps of the NikR−DNA complex (Figure 2), the exact coordination of this site is undetermined. However, nickel is likely to be coordinated by nearby H48. This site is stabilized by crystal contacts with a symmetry-related DNA molecule (Figure S1 of the Supporting Information), potentially explaining why this site is occupied only in the NikR−DNA complex crystal structure and suggesting that this site may not be a true low-affinity nickel binding site. In addition, only two of the four H48 residues in the molecule have associated nickel anomalous density (Figure 2), and intensities of the nickel anomalous signal, when present, are weak (Table 2). The average nickel anomalous level of site type 4 is 3σ, in comparison to site types 1−3 that have peaks visible to 7σ. Because this site is present only in the structure of the NikR−DNA complex, the low resolution of the data prohibited refinement of the occupancy of a nickel ion in this site.
The low-affinity nickel site present only in the NikR structure without DNA, site type 5, is also coordinated by a histidine, specifically H123. An octahedral geometry is completed at this site by five water molecules, which are stabilized by R122 and other nearby residues (Figure 3). This site is occupied in less than half of the NikR monomers in the crystal with a refined average occupancy of 0.45 (Table 2).
Site type 6 is located at the N-terminus of the protein and is the only excess nickel binding site not coordinated by a histidine residue. Instead, the N-terminal amine and carbonyl from M1 and water molecules coordinate site type 6 in an octahedral geometry (Figure 3). While this site is present in both structures, the anomalous density for it is very weak in the NikR−DNA structure, with an average nickel anomalous density of 2σ (Table 2), and is empty in two of the four possible locations (Figure 2b).
Molecular Dynamics Simulations of Site Types 1−3 on the NikR−DNA Complex
A 1 ns molecular dynamics trajectory of the solvated NikR−DNA complex yielded representative structures of the complex when nickel ions were bound at the three most highly occupied low-affinity nickel sites (site types 1−3). Throughout the trajectory, all low-affinity sites (10 sites were modeled on the basis of the three site types) either attained (site type 1) or maintained (site types 2 and 3) an octahedral geometry (Figure 4). Type 1 sites maintained the two H125 ligands and the two water molecules seen in the crystal structure and gained two additional water ligands, converting it to an octahedral site (Figure 4). Type 2 sites maintained the original protein ligands, H92 and H79, with either four water molecules or three water molecules and a third coordinating protein ligand to complete the octahedral geometry (Figure 4). The third coordinating protein ligand (D80) was not present in the crystal structure (Figure 3). Finally, type 3 sites maintained an octahedral geometry nearly identical to what was seen in the crystal structure with the same protein ligands and same number of water molecules (Figures 3 and 4).
Figure 4.
Representative structures following 1 ns molecular dynamics simulation of three types of low-affinity nickel binding sites. Simulations resulted in two types of ligand arrangements for site 2. Site types correspond to numbers in Figures 2 and 3 and Table 2.
Discussion
Playing a crucial role in metal homeostasis in the cell, metalloregulatory transcription factors have proven to be much more complex than one may originally have imagined. E. coli NikR, for example, has three types of metal binding sites: high-affinity nickel binding sites, potassium binding sites, and low-affinity nickel binding sites. While prior work has outlined the location and role of the stoichiometric or high-affinity nickel sites (2−4,6,10,12) and the second, potassium-containing metal binding sites (4,7,8), until now, the exact locations and coordination states of the third type of metal binding site on NikR, the low-affinity sites where excess nickel ions bind, have not been thoroughly described. This work serves to complete the description of NikR’s metal binding sites, allows us to consider the molecular basis by which excess nickel increases NikR’s affinity for DNA in vitro, and provides an opportunity to consider possible roles for these sites in vivo.
The crystallographic data described here suggest that there is not a single low-affinity nickel site on NikR, but rather a number of low-affinity nickel binding sites. Nickel anomalous maps indicate six types of potential low-affinity nickel sites on NikR (Figure 2). These sites are all on the surface of the protein, and five of the six site types have at least one histidine ligand (Table 2); all nickels are coordinated by oxygen-containing ligands, typically water molecules. Most sites in the crystal structures are six-coordinate, consistent with XAS data on the low-affinity nickel sites that are best fit with a model in which the low-affinity sites are six-coordinate with N and O donors as ligands (12). With these sites located across the surface of NikR, one can envision a variety of roles for them in explaining the in vitro binding data. Because site type 1 is located near the RHH domain−MBD interface, it is important to consider whether this site could play a role in stabilizing these domains in a DNA-bound conformation. Comparison of site type 1 in the NikR alone (with RHH domains in an “out” orientation) and NikR−DNA complex structure (with RHH domains in a “down” orientation), however, shows that this site does not change when the RHH domains are in the two different orientations (Figure 2), suggesting that this site does not play a role in stabilizing a DNA-bound form of NikR. Previous work has indicated that ordering of helix α3 (residues 65−78) is important for NikR binding to DNA (4,10), and both low-affinity site types 2 and 3 utilize residues from helix α3 or the following loop. A possible mechanism explaining how the binding of excess nickel ions stabilizes the NikR−DNA complex could be the ability of excess nickel to further stabilize helix α3. However, stoichiometric nickel alone does order the α3 helices as indicated by circular dichroism spectroscopy and B-factor comparison (3,6,10). In the apo crystal form of NikR, the B-factors of the α3 helical residues are significantly greater (88 Å2) than those for the rest of the protein (62 Å2), whereas in the stoichiometric Ni2+-bound forms, the B-factors of the α3 helical residues are very similar (33 Å2) to the average B-factor of the protein (28 Å2) (21), indicating that stoichiometric nickel alone imparts stability to these helices. In general, the binding of excess nickel ions does not induce significant conformational changes in NikR, although the change in cell dimension for the NikR−DNA complex indicates that the crystal lattice can accommodate rearrangements. Thus, it seems that the mechanism of action of excess nickel is not structural, suggesting instead an electrostatic mechanism to explain the increased affinity of NikR for DNA with the addition of excess nickel that is observed in vitro.
Our structural data are consistent with these surface nickel sites being low-affinity nickel sites. The refined occupancies for these nickel binding sites, and in some cases the weak nickel anomalous density (peaks disappear at relatively low σ levels), suggest that these sites are not filled 100% of the time (Figure 2 and Table 2), despite the high concentrations of NiCl2 used in these soaking experiments. The incomplete occupancies are consistent with all sites being solvent-exposed and not maintaining a coordination sphere of exclusively protein ligands. The greatest number of protein ligands used by any given site is three as is seen in site type 3, which also has the highest occupancy of nickel of all the low-affinity sites (0.88), consistent with the notion that a larger number of coordinating protein ligands correlates with increased site stability (Figure 3 and Table 2). However, no low-affinity site has an occupancy for nickel as high as that of the high-affinity sites, which utilize only protein ligands (Figure 1, Table 2, and Figure S2 of the Supporting Information). There are data to suggest that the low-affinity sites on NikR are nickel-specific (1), and although our work here does not directly address the question of specificity, histidine is a common nickel ligand (22). Regardless of whether the high number of histidine ligands imparts a preference for nickel over other metals, these sites are not designed to bind any metal tightly. In fact, because the physiological relevance of these sites is controversial (1,2,23), one could argue that the in vitro observation of increased NikR−DNA affinity could be an artifact of nickel binding to surface histidine residues. On the other hand, it is possible to imagine physiological roles for these weaker metal sites in either fine-tuning NikR’s affinity for DNA or as a sink for extra nickel ions in the cell, allowing Ni2+ to easily dissociate in case more nickel is required for use by other nickel proteins. While it is unlikely that the nickel concentration in an E. coli cell would ever be high enough that the NikR structure depicted in Figure 2b, with 14 surface nickels, would ever be realized, all sites identified here are easily accessible to excess nickel ions, even if NikR is already bound to DNA. In this sense, the binding of low-affinity nickel to the surface of NikR could help tune the affinity of NikR for the nik operon in response to the nickel concentration in the cell, either by facilitating the NikR−DNA binding interaction or by stabilizing the bound NikR−DNA complex.
Interestingly, a comparison of the results from gel shift assays studying the excess nickel binding effect on Helicobacter pylori NikR and E. coli NikR indicates that H. pylori NikR does not exhibit an enhanced DNA binding affinity in the presence of excess nickel ions, despite the fact that H. pylori NikR can bind up to 15 excess nickel ions (1,2,24). A sequence alignment of H. pylori and E. coli NikR illustrates that while the ligands to the high-affinity nickel site and the potassium binding site are conserved between these two species, the histidine residues that act as ligands to the low-affinity nickel sites in E. coli NikR are not present in H. pylori NikR (Figure 5). Instead, a crystal structure of H. pylori NikR from crystals soaked with 100 mM NiSO4 at pH 4.6 indicates that nickel ions can bind at a number of sites inside the tetramer NikR molecule (25), rather than on the surface as we observe. Therefore, it is likely that H. pylori NikR is not regulated in vitro by excess nickel because, unlike E. coli NikR, it does not possess the surface low-affinity nickel sites. While differences in nickel concentrations and pH (8 mM and pH 8.5 for E. coli and 100 mM and pH 4.6 for H. pylori) can easily explain the differences in nickel positions inside the NikR tetramer in the X-ray structures, if one function of low-affinity nickel sites is to act as a sink for excess nickel, the location of these sites would not have to be conserved. Along those same lines, the lack of sequence conservation of the surface histidine residues does not necessarily indicate the relative importance of “low-affinity nickel” to the regulation of DNA binding in E. coli, since the function of E. coli NikR differs from that of H. pylori NikR. In particular, E. coli NikR’s only known function is to repress the transcription of a nickel uptake transporter, while H. pylori NikR plays a variety of roles, downregulating a nickel uptake protein, NixA, while also being responsible for inducing the transcription of a number of nickel utilization genes (26−30). Given this variation in function, the lack of sequence conservation unfortunately does not provide insight into the importance of these sites in E. coli.
Figure 5.
Sequence alignment of NikR from E. coli (EcNikR) and H. pylori (HpNikR). High-affinity nickel ligands are highlighted in green, potassium ligands in purple, and low-affinity nickel site ligands from crystal structures in cyan, and the ligand added to site type 2 during molecular dynamics simulations (D80) is colored yellow (refer to Figure 4).
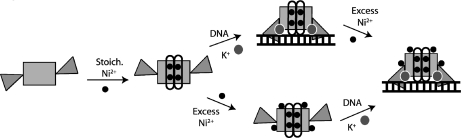
In conclusion, our data show that excess nickel ions bind to multiple, nonconserved sites on the surface of E. coli NikR. These sites, which utilize a small number of protein ligands in addition to water molecules, are consistent with low-affinity nickel binding. The observed lack of significant structural changes when excess nickel binds is consistent with the notion that nickel binding at low-affinity sites enhances NikR’s affinity for DNA in vitro primarily through an electrostatic effect. Hence, in vitro studies suggest that NikR can use three distinct metal binding sites to regulate its affinity for DNA (Figure 6). Stoichiometric or high-affinity nickel ions assist in stabilizing the folded state of the α3 helix and preceding loops that are disordered when no metal or the incorrect metal is bound (Figure 6) (3,10). The ordered helices and preceding loops can then make nonspecific hydrogen bonding contacts with the phosphate backbone, localizing NikR on DNA (4,10). Potassium binding to the octahedral sites at the MBD−RHH domain interface helps to stabilize the RHH domains in a “down” DNA-binding conformation (Figure 6) (4,7). Low-affinity nickel sites on the exterior of the protein could further enhance the affinity of the protein for the operator, potentially through an electrostatic mechanism (Figure 6). Alternatively, these sites could act as a nickel sink, protecting the DNA from nickel while at the same time storing the nickel for use by other proteins. By considering the final series of metal binding sites on NikR, we now present a detailed mechanism of action for this metal responsive transcription factor that is consistent with the in vitro biochemical data.
Figure 6.
Proposed scheme for NikR binding DNA considering all three types of metal binding sites. Nickel ions (black circles), potassium ions (gray circles), DNA (ladder), RHH domains (triangles), ordered α3 helices (white ovals), and the central MBD (rectangle) are illustrated. As structural data indicate that excess nickel ions can bind to the protein before and after DNA binding, two pathways are depicted.
Acknowledgments
Portions of this research were conducted at the Advanced Light Source (Contract DE-AC02-05CH11231), and the National Synchrotron Light Source at Brookhaven National Laboratory (Contract DE-AC02-98CH10886) was supported by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences.
Supporting Information Available
Crystallographic contacts that likely assist in stabilizing low-affinity nickel site type 4 (Figure S1) and a representative view of the composite omit electron density at the high-affinity nickel binding sites in the unbound NikR crystal structure with excess nickel (Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
This research was supported by National Institutes of Health Grant GM69857 to C.L.D.
Coordinates have been deposited in the Protein Data Bank for excess Ni2+-bound NikR (entry 3OD2).
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: NikR, nickel regulator protein; RHH, ribbon−helix−helix; MBD, metal binding domain.
Supplementary Material
References
- Bloom S. L.; Zamble D. B. (2004) Metal-Selective DNA-Binding Response of Escherichia coli NikR. Biochemistry 43, 10029–10038. [DOI] [PubMed] [Google Scholar]
- Chivers P. T.; Sauer R. T. (2002) NikR Repressor: High-Affinity Nickel Binding to the C-Terminal Domain Regulates Binding to Operator DNA. Chem. Biol. 9, 1141–1148. [DOI] [PubMed] [Google Scholar]
- Schreiter E. R.; Sintchak M. D.; Guo Y.; Chivers P. T.; Sauer R. T.; Drennan C. L. (2003) Crystal structure of nickel-responsive transcription factor NikR. Nat. Struct. Biol. 10, 794–799. [DOI] [PubMed] [Google Scholar]
- Schreiter E. R.; Wang S. C.; Zamble D. B.; Drennan C. L. (2006) NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc. Natl. Acad. Sci. U.S.A. 103, 13676–13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers P. T.; Sauer R. T. (1999) NikR is a ribbon-helix-helix DNA-binding protein. Protein Sci. 8, 2494–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. C.; Dias A. V.; Bloom S. L.; Zamble D. B. (2004) Selectivity of Metal Binding and Metal-Induced Stability of Escherichia coli NikR. Biochemistry 43, 10018–10028. [DOI] [PubMed] [Google Scholar]
- Phillips C. M.; Nerenberg P. S.; Drennan C. L.; Stultz C. M. (2009) Physical Basis of Metal-Binding Specificity in Escherichia coli NikR. J. Am. Chem. Soc. 131, 10220–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. C.; Li Y.; Robinson C. V.; Zamble D. B. (2010) Potassium is Critical for the Ni(II)-Responsive DNA-Binding Activity of Escherichia coli NikR. J. Am. Chem. Soc. 132, 1506–1507. [DOI] [PubMed] [Google Scholar]
- Carrington P. E.; Chivers P. T.; Al-Mjeni F.; Sauer R. T.; Maroney M. J. (2003) Nickel coordination is regulated by the DNA-bound state of NikR. Nat. Struct. Biol. 10, 126–130. [DOI] [PubMed] [Google Scholar]
- Phillips C. M.; Schreiter E. R.; Guo Y.; Wang S. C.; Zamble D. B.; Drennan C. L. (2008) Structural Basis of the Metal Specificity for Nickel Regulatory Protein NikR. Biochemistry 47, 1938–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers P. T.; Tahirov T. H. (2005) Structure of Pyrococcus horikoshii NikR: Nickel Sensing and Implications for the Regulation of DNA Recognition. J. Mol. Biol. 348, 597–607. [DOI] [PubMed] [Google Scholar]
- Leitch S.; Bradley M. J.; Rowe J. L.; Chivers P. T.; Maroney M. J. (2007) Nickel-Specific Response in the Transcriptional Regulator, Escherichia coli NikR. J. Am. Chem. Soc. 129, 5085–5095. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.; Minor W. (1997) Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Brünger A. T.; Adams P. D.; Clore G. M.; DeLano W. L.; Gros P.; Grosse-Kunstleve R. W.; Jiang J. S.; Kuszewski J.; Nilges M.; Pannu N. S.; Read R. J.; Rice L. M.; Simonson T.; Warren G. L. (1998) Crystallography & NMR system (CNS): A new software suite for macromolecular structure determination. Acta Crystallogr. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Storoni L. C.; McCoy A. J.; Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D60, 432–438. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Crowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. (1983) CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 4, 187–217. [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. (1983) Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 79, 926–935. [Google Scholar]
- Hoover W. G. (1985) Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 31, 1695–1697. [DOI] [PubMed] [Google Scholar]
- Ryckaert J. P.; Ciccotti G.; Berendsen H. J. C. (1977) Numerical-Integration of Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-Alkanes. J. Comput. Phys. 23, 327–341. [Google Scholar]
- Phillips C. M., and Drennan C. L. (2008) Nickel Regulatory Transcription Factor, NikR. In Handbook of Metalloproteins (Messerschmidt A., Ed.) John Wiley & Sons, Ltd., New York. [Google Scholar]
- Rulisek L.; Vondrasek J. (1998) Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J. Inorg. Biochem. 71, 115–127. [DOI] [PubMed] [Google Scholar]
- Helmann J. D. (2002) Sensing nickel. NikRs with two pockets. Chem. Biol. 9, 1055–1057. [DOI] [PubMed] [Google Scholar]
- Abraham L. O.; Li Y.; Zamble D. B. (2006) The metal- and DNA-binding activities of Helicobacter pylori NikR. J. Inorg. Biochem. 100, 1005–1014. [DOI] [PubMed] [Google Scholar]
- Dian C.; Schauer K.; Kapp U.; McSweeney S. M.; Labigne A.; Terradot L. (2006) Structural Basis of the Nickel Response in Helicobacter pylori: Crystal Structures of HpNikR in Apo and Nickel-bound States. J. Mol. Biol. 361, 715–730. [DOI] [PubMed] [Google Scholar]
- Davis G. S.; Flannery E. L.; Mobley H. L. T. (2006) Helicobacter pylori HP1512 is a nickel-responsive NikR-regulated outer membrane protein. Infect. Immun. 74, 6811–6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I.; Ieva R.; Soragni A.; Hilleringmann M.; Rappuoli R.; Scarlato V. (2005) In Vitro Analysis of Protein-Operator Interactions of the NikR and Fur Metal-Responsive Regulators of Coregulated Genes in Helicobacter pylori. J. Bacteriol. 187, 7703–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst F. D.; Kuipers E. J.; Heijens A.; Sarwari R.; Stoof J.; Penn C. W.; Kusters J. G.; van Vliet A. H. M. (2005) The Nickel-Responsive Regulator NikR controls Activation and Repression of Gene Transcription in Helicobacter pylori. Infect. Immun. 73, 7252–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst F. D.; Stoof J.; Horrevoets W. M.; Kuipers E. J.; Kusters J. G.; van Vliet A. H. M. (2006) NikR Mediates Nickel-Responsive Transcriptional Repression of the Helicobacter pylori Outer Membrane Proteins FecA3 (HP1400) and FrpB4 (HP1512). Infect. Immun. 74, 6821–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram L.; Haas E.; Bauerfeind P. (2006) Nickel Represses the Synthesis of the Nickel Permease NixA of Helicobacter pylori. J. Bacteriol. 188, 1245–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.