The subgingival periodontal pocket of humans harbors more than 500 bacterial species. Periodontitis is a chronic inflammation of the periodontium with multi-factorial etiology. It is initiated due to colonization as subgingival biofilms by a group of gram-negative anaerobes. The disease progresses as a result of the direct effects of bacterial virulence factors on host tissues as well as the self-damaging host responses to the colonizing bacteria (83). While no single species has been implicated as the primary pathogen and the available evidence is consistent with a polymicrobial disease etiology, the red-complex bacteria consisting of Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia has been strongly implicated in the onset of periodontitis (83). On the other hand, investigations utilizing the 16S ribotyping have also implicated novel phylotypes in the absence of detectable red-complex bacteria associated with periodontal lesions (44). In support of T. forsythia as being a suspected periodontal pathogen, the organism satisfies the necessary criteria postulated by Socransky and co-workers (82, 83). For instance: (i) its association and increased levels in periodontitis (83), (ii) evidence of host responses to its antigens (2, 81, 94) (iii) its ability to cause disease in animal models (2, 40, 79, 84), and (iv) expression of virulence factors which can potentially contribute to the disease process (described in detail in this review).
T. forsythia is an anaerobic gram-negative member of the Cytophaga-Bacteroides family which was initially described as Bacteroides forsythus by Tanner et al. (87) and later reclassified as Tannerella forsythia by Sakamoto et al. (74) based on 16S rRNA phylogenetic analysis. T. forsythia is associated more frequently and/or in higher levels with various forms of the disease, including gingivitis, chronic and aggressive periodontitis, than with health (for review see Ref. (86)). Several studies have also implicated T. forsythia in the progression of clinical attachment loss associated with periodontitis (11, 18, 19, 50, 85). Moreover, a recent study has suggested that T. forsythia infection is more likely to cause periodontitis in overweight women than in normal weight women (7). According to another recent study, overweight or obese individuals have an overgrowth of T. forsythia compared to normal weight individuals, thus subjecting overweight and obese individuals to a higher risk of developing periodontal disease (20). In spite of the overwhelming evidence implicating T. forsythia in pathogenesis, this bacterium remains an understudied organism. This is partly due to the fastidious growth requirement for culturing this bacterium, as well as the fact that genetic manipulations of this organism are difficult to perform (30, 73). Moreover, there are no gene complementation systems currently available for the organism. While T. forsythia is the sole member of the new genus Tannerella, uncultivated oral phylotypes BU045, BU063, 97 and 997 are its closest relatives (95). These phylotypes have long rod-like segmented structure, and though they are frequently found in various periodontal disease-associated plaques, they are present only in low numbers, do not proliferate to high densities and therefore, are considered not relevant to disease pathogenesis (95).
Studies in animal models have demonstrated the virulence potential of T. forsythia. For example, T. forsythia caused skin abscesses in rabbits (84) and mice (2, 93) as well as alveolar bone loss in mice (79) and rats (40). These in vivo studies also showed that the pathogenic potential of T. forsythia was enhanced in the presence of other bacteria. For instance, abscess formation in rabbits and in mice was enhanced synergistically when Fusobacterium nucleatum or P. gingivalis were the coinfecting partners of T. forsythia. Similarly, a synergy was observed relative to alveolar bone loss in rats following oral infection with the red-complex consortium, P. gingivalis, T. denticola and T. forsythia (40). These results demonstrate that T. forsythia is a pathogenic organism which might play synergistic roles in inflammation along with other periodontal pathogens. Therefore, in order to fully understand the mechanisms underlying the pathogenesis associated with T. forsythia, it would be important to identify the virulence functions of the organism and determine how these factors are regulated in response to coexisting bacteria and to host-derived factors. It is likely that T. forsythia might influence the physiology and virulence of coexisting periodontal pathogens. For this purpose, physical, chemical, and metabolic interactions are expected to occur, which might further involve bacterial two component sensor-regulator systems.
So far, only a few putative virulence factors have been identified in T. forsythia: trypsin-like (17) and PrtH proteases (72), sialidases SiaH (6, 35) and NanH (88), a leucine-rich repeat cell-surface-associated and secreted protein BspA (81), an apoptosis-inducing activity (61), alpha-D-glucosidase and N-acetyl-beta-glucosaminidase (32), a hemagglutinin (59), components of the bacterial S-layer (71, 73), and methylglyoxal production (53).
Protease and apoptosis inducing activity
T. forsythia with asachrolytic physiology would require peptides and free amino acids for growth. At least two proteolytic enzymes have been identified which might play roles in degradation of host proteins, providing essential amino acids, peptides and heme for growth of T. forsythia. (17)(72). In addition, these proteases might contribute to bacterial virulence in multiple ways; such as by degrading host periodontal tissues, activating host degradative enzymes, modifying host cell proteins to expose cryptotopes for bacterial colonization, cleaving components involved in innate (cytokines/chemokines, complement factors) and adaptive immunity (immunoglobulins) thus paralyzing host immunity and activating components involved in clotting/fibrinolyis. These and other putative functions of bacterial proteases are discussed in details elsewhere (28, 45, 66, 90).
A trypsin-like protease of T. forsythia was first characterized by Grenier (17). The trypsin activity was shown to be associated with a cell surface associated protein of 81-kDa molecular size and it required reducing agents for optimal activity. Moreover, this enzyme was shown to be a serine protease since its activity was sensitive to diisopropylfluorophosphate and other serine protease inhibitors treatments. The enzyme cleaved arginine or lysine bonds only in synthetic peptides but not in native proteins such as casein and gelatin. Therefore, it is believed that this enzyme is mainly involved in the degradation of smaller peptides released from proteolysis of larger proteins by other proteolytic enzymes and by itself may not play a major role in virulence (17). A protease with the ability to cleave larger protein substrates was later identified by screening of a T. forsythia genomic expression library in Escherichia coli (72). An expression clone displaying skim-milk hydrolytic activity was identified and the nucleotide sequence of the insert revealed that the protease activity corresponded to an open reading frame with an expected molecular weight of 47.8-kDa. The putative gene was termed prtH and the encoded protease PrtH. Further characterization of protease activity associated with the positive clone revealed that PrtH hydrolyzes, in addition to milk, the synthetic peptide N-benzoyl-Val-Gly-Arg-p-nitroanilide and causes hemolysis of horse blood (72). The cysteine proteinase nature of PrtH is based on inhibition studies with protease inhibitors p-toluenesulfonyl-L-lysine chloromethyl ketone hydrochloride and leupeptin (72). Later, PrtH was identified from the spent-medium initially as a detachment factor, called forsythia detachment factor (FDF), with an ability to cause detachment of adherent cells from the substratum (61). During biochemical characterization, two different activities were initially identified from the spent medium, one having the detachment activity, which was later confirmed to be the PrtH protein, and the other having cytopathic activity arresting cells in the G2 phase (61). The identity of the cytopathic factor is currently unknown. The cloning of the detachment factor gene fdf revealed that the gene encodes a 536-residue protein. It was discovered that the fdf gene contained the original prtH gene and since the translational start site for PrtH was incorrectly predicted in the previous study (72), fdh encodes for PrtH plus a region N-terminal of PrtH (61). Consistent with this finding, recombinant protein encoded by fdf showed cysteine protease activity. Thus, FDF and PrtH designate the same protein. Moreover, PrtH has been shown to increase the mitochondrial oxidative membrane potential in cells, resulting in IL-8 production from detached cells (89). In silico analyses have indicated the presence of a caspase-like fold (Pfam accession number PF00656) in the N-terminal region of PrtH (65). Also, the catalytic residues histidine and cysteine present in the C14 peptidase family are conserved in PrtH (65). Taken together, the above studies suggest that PrtH might be involved in the disintegration of the gingival epithelium and induction of chemokine IL-8 from detached cells. These activities might play roles in periodontal pathogenesis.
A cross-sectional clinical study has suggested that the prtH genotype is associated with high levels of T. forsythia, subsequently subjecting individuals to an increased risk for periodontitis (23). This study showed that the prtH genotype was associated with almost every individual infected with T. forsythia and that the baseline levels of prtH were significantly lower in individuals with no attachment loss compared to those those who subsequently lost attachment. A separate longitudinal clinical study confirmed the association between prtH genotype levels and future periodontal attachment loss (22). This study demonstrated that baseline levels of the prtH genotype were significantly lower in subjects without loss of attachment compared to those who lost attachment over 1, 2, 4, or 5 years.
Surface components
Surface-layer associated glycoproteins
T. forsythia possesses a surface-layer (S-layer) consisting of serrated structural subunits (about 10 nm wide and 10 nm high) in either oblique or tetragonal lattices (39), and it lacks surface appendages such as fimbriae. The S-layer has been shown to be composed of at least two high molecular weight glycoproteins of 220 and 210 kDa size encoded by the tfsA and tfsB genes, respectively (26, 46). Experimental evidence has also indicated that the wecC gene associated with an exopolysaccharide operon (TF23052-TF2055) contributes to glycosylation of the S-later glycoproteins (29). S-layers in bacteria are thought to provide a protective shielding as well as ion-traps and molecular sieves for metabolites in the environment. S-layers also help bacteria in promoting adhesion. In this regard, the T. forsythia S-layer has been shown to promote epithelial cell adherence and invasion (71, 73). It is tempting to speculate that the glycan residues on the S-layer proteins might be important in recognition by lectin-like receptors on host cells for adhesion and invasion. The S-layer glycans may also serve as ligands for lectin-like receptors present on coaggregating bacteria F. nucleatum (60). This in turn might promote mixed species biofilm formation observed between the two species (80) and could lead to increased disease severity. However, the role of S-layer associated sugar residues in host cell and bacterial interactions has yet to established.
Leucine-rich repeat BspA protein
A surface as well as secreted protein BspA (Bacteroides surface protein A) belonging to the leucine-rich repeat family was indentified in T. forsythia (81). The encoded sequence of BspA contains two regions D1 and D2 in the N-terminal portion with 14- and 6-tandem repeats of a 23-amino acid long leucine-rich-repeat (LRR) motif, respectively. The C-terminal portion contains four immunoglobulin-like (Ig-like) domains found in bacteria (Big_2)(Fig. 1). Furthermore, analysis of the C-terminal region of BspA revealed the presence of a conserved C-terminal domain (CTD) (62, 63) predicted to be involved in trafficking of bacterial proteins to the outer membrane and secretion, as well as recognition by the protein glycosylation machinery. CTD is a 50-residue domain found in a family of proteins associated with P. gingivalis and Bacteroides sp. and it functions as a targeting domain for secretion, outer-membrane anchoring and recognition by the glycosylation machinery (63, 77). Recently, in agreement with in-silico predictions for BspA, a proteomic study of T. forsythia has shown that BspA is indeed glycosylated and is associated with the outer-membrane fraction (92). With regard to the LRR repeats, they are found in many eukaryotic and prokaryotic proteins with diverse functions and cellular localizations (41). The LRR-containing proteins represent an important superfamily involved in protein-protein interactions and in signal transduction. The domain structure of LRRs (Pfam ID: PF00560) reveals that LRRs correspond to β-α structural units which are arranged such that all of the β-strands and the helices are parallel to a common axis, resulting in a non-globular, horse-shoe shaped molecule with a curved parallel β-sheet lining the inner circumference of the horse-shoe and the helices flanking its outer circumference (Fig. 2). The Ig-like Big_2 domains are beta barrel like structures that have been identified in cell-surface intimins, the bacterial adhesin proteins of enteropathogenic and enterohaemorrhagic strains of E. coli (1, 10, 13-15, 24, 38). The bacterial LRR proteins whose functions have been defined include Listeria monocytogenes internalins (InlA and InlB) (55, 56), Shigella flexneri IpaH (12), host specific Salmonella sp. proteins (91), Group B streptococci LLR protein lrrG (76), L. monocytogenes InlJ protein (70), P. gingivalis (8) InlJ protein, and T. denticola LrrA protein (33). These studies indicate that the functional roles of different LRR sequences differ depending upon the amino acid sequence. For example, L. monocytogenes cell surface associated internalin A (InlA) is necessary and sufficient to promote adherence and entry into host cells expressing its receptor E-cadherin (57, 58). InlB binds to a different cell-surface receptor (75), and the LRR region of InlB, and not of InlA, triggers the release of proinflammatory cytokines from macrophages (54). The S. flexneri IpaH protein has been suggested to facilitate bacterial escape from the phagocytic vacuole of monocytes and macrophages (12) and Samonella enterica LRR proteins have been shown to be specific for host adaptation (91). Immunization with the Group B streptococci lrrG protein was shown to elicit protective immunity against the group B streptococci (76). While BspA is the first well-characterized LRR protein identified in oral bacteria, genes encoding LRR proteins have been identified in several recently sequenced oral bacterial genomes, including P. gingivalis, T. denticola, Prevotella intermedia, and F. nucleatum. In this regard, the LrrA protein from T. denticola (33) and the InlJ protein from P. gingivalis (8) have been shown to be involved in coaggregation and biofilm development in the respective bacterium. Homologues of BspA in P. intermedia have also been shown to be involved in bacterial adherence and invasion of epithelial cells (47).
Fig. 1.
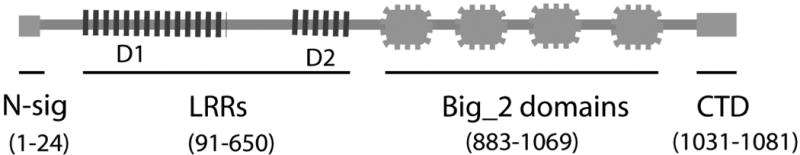
Schematic representation of the domain structure of BspA. Numbers in parenthesis indicate amino acid residues comprising each domain. Abbreviations: Big_2, bacterial Ig-like domains; CTD, C-terminal domain; LRR, leucine-rich repeats; N-sig; N-terminal secretion signal.
Fig. 2.
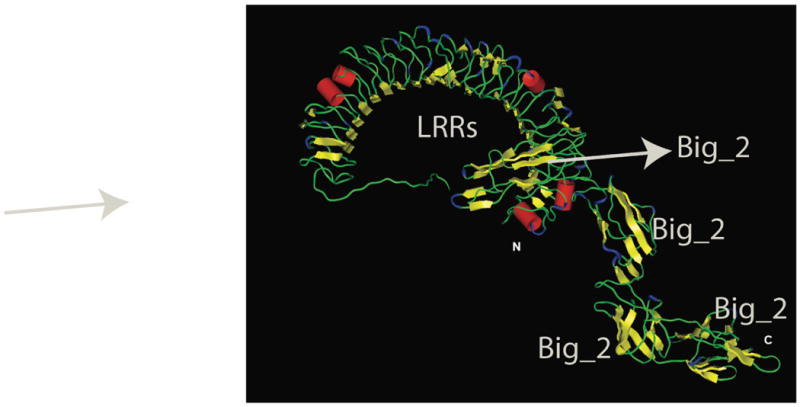
Three-dimensional computer model of the BspA protein showing horse-shoe shaped LRR domain in the N-terminal and four Ig-like domains in the C-terminal region. As mentioned in the text, at least one of the adjacent Ig-like domains is predicted to stabilize the LRR domain (courtesy of Dr. J. P. Malone, University at Buffalo).
A computer generated three-dimensional structure model generated by Dr. J. P. Malone (University at Buffalo, personnel communication) is shown in Fig. 2. This model predicts that the LRR region is uninterrupted (i. e. the intervening region between D1 and D2 also assumes the LRR conformation) and that the LRR region is likely to be stabilized by one of the adjacent Ig-like domains fused C-terminally. The function of the Ig-like domains (Pfam ID PF08191) adjacent to the LRR region is predicted to be mainly structural in nature, i.e. to stabilize the LRR region, and these domains play no role in protein-protein interactions themselves.
The presence of LRR domains suggested that the BspA protein could be involved in protein-protein interactions important in mediating T. forsythia interactions with the host factors and/or components of other bacteria. Consequently, BspA was shown to bind to the extracellular matrix component fibronectin and the clotting factor fibrinogen (81). Based on the computer modeling of an orthologue of BspA found in Trichomonas vaginalis, it is predicted that the Fn-3 domain of fibronectin binds to amino acid residues on the concave side of the LRR domain (27). In relation to bacterial-bacterial interactions, homotypic binding between the LRR region of BspA and that of the T. denticola LrrA protein was shown to mediate coaggregation between T. forsythia and T. denticola (33). In addition, BspA has been shown to be involved in interactions with the oral bacteria F. nucleatum (80). In cellular activation, BspA has been shown to trigger the release of bone-resorbing pro-inflammatory cytokines from monocytes (21) and chemokine IL-8 from gingival epithelial cells (64) by activating the TLR-2-dependent pathway. TLR1 serves as a coreceptor for TLR2 in this activation, and the BspA LRR domain-1 is involved in activation of TLR2/1 heterodimer (64). Thus, host may recognize BspA as a microbial-associated-molecular-pattern through TLR2/1 for secretion of inflammatory cytokines and chemokines. In addition, BspA has been shown to mediate bacterial adherence and invasion into epithelial cells (34). Interestingly, P. gingivalis or its outer membrane vesicles promoted BspA-mediated invasion of epithelial cells by T. forsythia (34). A direct evidence for an in vivo role of BspA in pathogenesis came from a study which showed that a BspA-defective mutant was significantly less potent than the wild-type strain in inducing alveolar bone loss in mice (79).
Recent studies have shown that conserved LRR motifs associated with bacterial proteins, including BspA, are recognized as novel patterns by the human pattern recognition receptor gp340 of the innate immune system (52). This recognition involves protein-protein interaction between LRRs and conserved peptide domains of gp340. Gp340 appears to be an important mediator of host immune responses to various microbes. It is expressed by epithelial cells and cells of the immune system and has been shown to inhibit bacterial invasion into epithelial cells and blocks secretion of proinflammatory cytokines (69). Gp340 is also found saliva, and the salivary form has been shown to function as a salivary agglutinin important in oral biofilm formation and thus suggested to have a role in dental caries development (36, 48, 67). However, a direct role for gp340-BspA interactions in T. forsythia-mediated pathogenesis has yet to be determined. In summary, the interactions of BspA with the pattern recognition receptors TLR2 and gp340 are expected to play important roles in the pathogenesis of periodontal disease.
Interestingly, several BspA homologues have been identified in the T. forsythia genome. The genome sequencing of T. forsythia has now predicted six other putative BspA-like homologues in T. forsythia. In this regard, a homologue TF1843 requires special mention. The PSORT program predicts that this homologue is surface localized. Both BspA (annotated as TF2998 with 97% identity) and TF1843 have LRR domains in their N-terminal regions and Big_2 domains at the C-terminal ends of the molecules. While significant primary amino acid sequence differences are observed in LRRs, both proteins show 99% sequence identity in their Big_2 domains. TF1843 also possesses a CTD domain as in BspA. The differences in the primary amino acid sequences within LRRs suggest that these homologues might have disparate biological functions. While the functional role of Big_2 domains in BspA is currently unknown, it is likely that this domain having a rigid beta-barrel structure serves mainly a structural role and facilitates the presentation of the adjacent LRRs for protein-protein interactions. Genes encoding LRR domain proteins have also been identified in several recently sequenced oral bacterial genomes, including P. intermedia and F. nucleatum. Although the functions of LRR proteins may differ, LRR repeats appear to be conserved and relatively invariant despite negative selective pressure by the host immune system. Therefore, LLRs may be critical for bacterial survival.
Surface lipoproteins
Previous studies have shown that T. forsythia surface-lipoproteins activate host cells to release proinflammatory cytokines and induce cellular apoptosis (25). It was further shown that the lipoprotein fractions from T. forsythia contain ester-bound fatty acids and stimulate human gingival fibroblasts and monocytic cells to release interleukin-6 and tumor necrosis factor alpha (25). Activation of the transcription factor nuclear factor-{kappa} B due to TLR2 (but not CD14 or TLR4)-mediated signaling was demonstrated to be responsible for lipoprotein-mediated cytokine production by host cells. Furthermore, the lipoprotein fraction from T. forsythia has been shown to cause apoptotic cell death of human gingival fibroblasts, KB cells (an epithelial cell line), HL-60 cells (a human myeloid leukemia cell line) and THP-1 cells (monocytic cell line) but not MOLT4 cells (a T-cell leukemia cell line) (25). The lipoprotein induced cell death involved activation of caspase-8, an initiator of the caspase cascade in apoptosis. Thus, it has been suggested that T. forsythia lipoproteins might play roles in the pathogenesis of periodontal disease by induction of cell activation and apoptosis.
Glycosidic acivity
Although an asaccharolytic bacterium, T. forsythia has been shown to express a variety of glycosidases. It expresses exo-alpha-sialidases or neuraminidases SiaHI (35) and NanH (88), α-D-glucosidase (SusB) upstream of the Sus cluster involved in starch uptake and metabolism, and N-acetyl β-D glucosaminidase (hexA) (32). In addition to these glycosidases which were identified experimentally, the genome of T. forsythia has been predicted to encode several other putative glycosidases. In principle, these glycosidases can hydrolyze terminal glycosidic linkages in complex oligosaccharides and proteoglycans abundantly found in saliva, gingival crevicular fluid and periodontal tissue. The degradation of oligosaccharides and proteoglycans would affect the functional integrity of periodontium and may promote disease progression. The degradation of host oligosaccharides and proteoglycans by these glycosidases can also provide nutrients for other community bacteria. Moreover, glycosidase treatment can expose protein epitopes for adherence and colonization by bacteria. An N-acetylneuraminyllactose senstivive hemagglutinin has been identified in T. forsythia (59), which might promote bacterial binding to host cell surface sugars exposed by bacterial glycosidases. While T. forsythia glycosidases in theory can contribute to pathogenicity in different ways, it remains to be determined whether they truly play important functions in the pathobiology of periodontal diseases.
T. forsythia in the presence of glucose accumulates high levels of toxic methylglyoxal product in vitro (53). Methylglyoxal product accumulates in cultures of a variety of microorganisms but this activity is pronounced in T. forsythia during its growth in the presence of glucose because of the possible imbalance between the rate of methylglyoxal product synthesis and detoxification by the bacterium (5). Methylglyoxal product is a highly reactive electrophile with high reactivity with cysteine residues and lesser reactivity with arginine and lysine residues within proteins (51). This modification can result in protein cross-linking and loss of protein function. Thus, methylglyoxal product production by T. forsythia could be toxic to the host and this may contribute to tissue damage observed in periodontal disease. In support of this, methylglyoxal product has been detected at higher concentrations in the crevicular fluid of individuals with periodontitis compared to healthy individuals (37).
Biofilm activity
T. forsythia has been shown to form poor monospecies biofilms in vitro (29). However, when an operon involved in exopolysaccharide synthesis in T. forsythia was disrupted a significant increase in the biofilm formation was observed (29). Moreover, in comparison to the wild-type strain an exopolysaccharide-deficient mutant displayed enhanced microcolony formation and showed increased surface-hydrohobicity (29). This study suggested that surface-exopolysaccharides in T. forsythia play inhibitory roles in biofilm development by T. forsythia. Interestingly, it was found that T. forsythia also forms mixed synergistic biofilms with F. nucleatum (80). F. nucleatum is considered to be a bridge-bacterium due its ability to coaggregate with both early and late colonizing bacteria, thereby facilitating dental plaque formation (3). It was demonstrated that cell to cell contact is important for mixed species biofilm formation between T. forsythia and F. nucleatum, although the role of diffusible molecules cannot be ruled out completely (80). Interestingly, the T. forsythia genome (www.oralgen.org) neither possesses an enzyme homologue of LuxS for the synthesis of universal quorum sensing AI-2 molecule or a detection system for sensing environmental AI-2. Together, this suggests that AI-2 mediated signaling may not be responsible for enhanced T. forsythia-F. nucleatum mixed biofilm formation. While the underlying mechanisms of interspecies communication leading to enhanced biofilm formation are not yet well understood, our recent study has suggested that the oxidative response regulator OxyR might be involved (31). It was observed that a T. forsythia OxyR-defective mutant as compared to the wild-type strain had significantly reduced ability to form mixed biofilms with F. nucleatum (31). The mutant also showed reduced ability to autoaggregate compared to the wild-type strain, suggesting OxyR-mediated regulation of surface adhesins in T. forsythia. It is plausible that the oxidative-stress regulator OxyR in T. forsythia also regulates expression of membrane porins or transporters which are responsible for uptake of interspecies signaling molecules or metabolites released by F. nucleatum. While these predictions are purely speculative in explaining the underlying mechanisms of interspecies biofilm formation by T. forsythia, OxyR has been shown to control biofilm formation in several bacteria. For example, in E. coli OxyR regulates the expression of a surface adhesin Ag43 (9), in Serratia marcescens, OxyR regulates biofilm formation through induction of fimbriae production (78); and In P. gingivalis OxyR acts as a repressor for surface adhesin FimA involved in biofilm formation and therefore an oxyR deletion mutant forms increased biofilms compared to its parental strain (49).
In-vivo induced virulence factors
By utilizing In-vivo Induced Antigen Technology (IVIAT), T. forsythia antigens specifically expressed during an infection in patients with periodontal disease were identified (94). IVIAT is an attractive antibody-based screening technique which can be utilized to identify antigens of a pathogen specifically expressed during an infection (68). Briefly, pooled patient sera is first adsorbed with the antigens isolated from in-vitro grown pathogen and then preadsorbed sera is used as antibody probe for screening a genomic library of pathogen in plasmid or phage vector. With IVIAT technology, twelve in- vivo induced proteins from T. forsythia were identified (94). These included; BspA, a well-defined virulence factor of T. forsythia; enzymes involved in housekeeping functions; enzymes implicated in tissue destruction (DppIV, dipeptidyl peptidase IV); a DNA mismatch repair protein; and a putative TonB-dependent outer membrane receptor (Gene ID: TF1439). While in vivo roles for many of these genes identified by IVIAT have yet to be determined, at least two of the factors, namely BspA and DppIV, have been well characterized and shown to be important in virulence. The role for BspA in virulence has been described above. With regard to DppIV, it is a serine protease that cleaves X-Pro or X-Ala dipeptides at the N-terminal end of the polypeptide chain. Although the exact functions of DppIV in T. forsythia virulence have yet to be determined, DppIV has been shown to be involved in the virulence of P. gingivalis (42). Mice challenged with a DppIV-deficient mutant of P. gingivalis developed reduced abscesses than those challenged with the wild-type strain. Moreover, mice injected with the mutant exhibited faster recovery from the infection, as assessed by weight gain and the rate of lesion healing. Mechanistically, DppIV has been suggested to activate host-derived matrix metalloproteinases, thereby promoting degradation of tissue collagen (43). Also, DppIV can bind to fibronectin and mediate bacterial binding to fibronectin (43). Therefore, DppIV-mediated tissue destruction and bacterial adherence may play important roles in the pathogenesis. It would be interesting to determine if DppIV expression induced in vivo plays roles in periodontitis associated with T. forsythia infections.
Miscellaneous functions
T. forsythia growth requires an exogenous source of N-acetyl muramic acid, an important amino sugar in almost all eubacteria, which together with N-acetylglucosamine forms the peptidoglycan (murein) cell wall. Unlike other bacteria which synthesize their own N-acetyl muramic acid, T. forsythia lacks a metabolic pathway to synthesize its own MurNac. This is because homologues of key enzymes UDP-N-acetylglucosamine-enolpuruvate transferase and UDP-enolpyruvate reductase, involved in a two-step synthesis of MurNac from GlucNA in bacteria, are lacking in the T. forsythia based genome sequence. This implies that T. forsythia might possess unique systems to scavenge peptidoglycan degradation products released during cell-wall recycling of oral biofilm bacteria.
Interestingly, while scavenging of exogenous N-acetyl muramic acid/peptidoglycan by T. forsythia may be critical for the growth of the organism, additionally this might have downstream repercussions on periodontal inflammation and disease pathogenesis. Since bacterial peptidoglycan and its degradation products such as muramylpeptides can act as inflammatory mediators by activating host innate pattern-recognition receptors (surface toll-like receptors and intracellular (NOD1) (4), scavenging of peptidoglycan within the gingival crevice by T. forsythia might help to dampen inflammation and assist in evading the host immune response. It is tempting to speculate that the growth of T. forsythia might contribute to the maintenance of immunological homeostasis and the modulation of periodontal disease progression.
It is likely that T. forsythia releases metabolites beneficial for the growth of the other red-complex species. In this regard, it is possible that succinate, a precursor for the synthesis of membrane lipids and phospholipids which promotes the growth P. gingivalis (16) could likely be produced by T. forsythia through reduction of fumarate by a putative fumarate dehydrogenase enzyme encoded by TF2650. Fumarate could be generated in T. forsythia during the arginine utilization pathway involving the activity of an N-(L-Argininosuccinate) arginine-lyase (EC 4.3.2.1) homologue encoded by TF1489. Therefore, the growth of P. gingivalis, a highly proteolytic organism, would further lead to the destruction of host proteins releasing peptides and amino acids which provide nutrients for T. forsythia. Together, the scavenging of peptidoglycan and production of succinate by T. forsythia might explain the increased risk for periodontitis in the presence of the red-complex bacteria.
A recent study has identified several surface proteins associated with the bacterial outer membrane by proteomic analysis (92). The functions of only few of the proteins such as TfsA, TfsB, and BspA have been characterized. The functions of the rest of the identified proteins will need to be determined in the future to understand their potential roles in virulence.
Animal models demonstrating the virulence of T. forsythia
An important criterion to be satisfied by T. forsythia to be considered as a periodontal pathogen is confirmation of its virulence potential in animal models. In this regard, one of the earliest studies performed by Takemoto et al. in rabbits (84) showed that when T. forsythia was coinoculated with F. nucleatum or P. gingivalis, a significant abscess formation was observed in animals. Monoinfections with T. forsythia, F. nucleatum or P. gingivalis strains used did not induce significant abscess in animals. This study successfully demonstrated the pathogenic potential of T. forsythia for the first time in an animal system and also provided evidence indicating that pathogenicity of individual bacterial species can be modulated following interactions with other bacteria. Later, utilizing a skin abscess model in mice the virulence potential of T. forsythia was demonstrated (2, 93). The study by Yoneda et al. (93) showed pathological synergy between T. forsythia and P. gingivalis (93). This study also suggested a role for P. gingivalis gingipains in synergism with T. forsythia.
The pathogenic potential of T. forsythia in inducing alveolar bone loss was demonstrated in a mouse model (79). Mice orally infected with T. forsythia showed increased alveolar bone loss as compared to sham-infected mice. Furthermore, a T. forsythia mutant defective in the expression of the BspA protein showed significantly less bone loss as compared to mice infected with the wild-type strain, indicating that BspA is an important virulence determinant of the bacterium. This study also reported that T. forsythia did not induce any alveolar bone loss in gnotobiotic germ free animals (79). This suggests that interactions of T. forsythia with other bacteria might be important for bacterial growth and expression of virulence. In this respect, T. forsythia's dependence on environmental MurNac released by co-inhabiting bacteria could be critical for its growth. In addition, interactions involving other metabolic pathways, physical coaggregations for biofilm development and/or chemical communication might also be critical. This notion is further supported by a study which showed that polymicrobial infections with the red-complex consortium resulted in synergism with respect to immunoinflammatory responses and alveolar bone loss in rats (40).
Conclusions
It is now well established that periodontitis involves a polymicrobial etiology, mainly due to association with the red complex bacterial consortium. The availability of the genome sequence containing 3034 ORFs (http://www.oralgen.lanl.gov/under Tannerella forsythensis) and the development of specific gene deletion system for T. forsythia (29-31, 73) should pave the way for identifying additional virulence factors of this bacterium in the near future. Thus, comparative genomic approaches could now be utilized to identify homologous pathogenic genes in T. forsythia. Moreover, it is likely that many of the virulence genes in T. forsythia might express only when the organism comes in direct contact with partner community bacteria or with the host. In this respect, indentifying two-component regulatory networks regulating expression of such virulence genes would be important for future development of novel therapeutic strategies. For identification of in vivo induced genes, approaches involving gene expression profiling might be necessary. Currently, microarrays for the organism are not available; however, alternative gene expression profiling strategies such as RNA subtractive hybridization or quantitative reverse-transcription PCR might be exploited. In addition, antibody based IVIAT technique mentioned above may help to identify genes preferentially expressed under different stages of the disease. In addition to identifying the T. forsythia factors involved in host interactions, it is equally important to identify factors responsible for the integrity of the community structure. In this respect, factors that mediate physical, metabolic and chemical communication between T. forsythia and its community partners need to be identified. It is tempting to speculate that identification of such molecules would help to design novel strategies for eliminating or reducing the levels of T. forsythia from the bacterial consortium. Theoretically, this in turn might disrupt the community structure and homeostasis as well as reduce disease pathogenesis.
In conclusion, complementary strategies involving computational and wet-lab experimental approaches will be necessary to identify such factors that govern interactions of T. forsythia with the host as well as other community bacteria. Finally, better insight into the mechanisms of these factors in pathogenesis will allow development of future strategies to control periodontitis.
Acknowledgments
I sincerely thank Professor Howard Kuramitsu for critical reading of this manuscript, and Dr. James P. Malone for computer modeling of the BspA protein. The work in author's laboratory was supported by a grant (DE014749) from the National Institute of Dental and Craniofacial Research.
References
- 1.Agin TS, Wolf MK. Identification of a family of intimins common to Escherichia coli causing attaching-effacing lesions in rabbits, humans, and swine. Infect Immun. 1997;65:320–326. doi: 10.1128/iai.65.1.320-326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird PS, Shakibaie F, Gemmell E, Polak B, Seymour GJ. Immune response to Bacteroides forsythus in a murine model. Oral Microbiol Immunol. 2001;16:311–315. doi: 10.1034/j.1399-302x.2001.016005311.x. [DOI] [PubMed] [Google Scholar]
- 3.Bolstad AI, Jensen HB, Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55–71. doi: 10.1128/cmr.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boneca IG. The role of peptidoglycan in pathogenesis. Curr Opin Microbiol. 2005;8:46–53. doi: 10.1016/j.mib.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Booth IR, Ferguson GP, Miller S, Li C, Gunasekera B, Kinghorn S. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem Soc Trans. 2003;31:1406–1408. doi: 10.1042/bst0311406. [DOI] [PubMed] [Google Scholar]
- 6.Braham PH, Moncla BJ. Rapid presumptive identification and further characterization of Bacteroides forsythus. J Clin Microbiol. 1992;30:649–654. doi: 10.1128/jcm.30.3.649-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan RM, Genco RJ, Wilding GE, Hovey KM, Trevisan M, Wactawski-Wende J. Bacterial species in subgingival plaque and oral bone loss in postmenopausal women. J Periodontol. 2007;78:1051–1061. doi: 10.1902/jop.2007.060436. [DOI] [PubMed] [Google Scholar]
- 8.Capestany CA, Kuboniwa M, Jung IY, Park Y, Tribble GD, Lamont RJ. Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect Immun. 2006;74:3002–3005. doi: 10.1128/IAI.74.5.3002-3005.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danese PN, Pratt LA, Dove SL, Kolter R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol. 2000;37:424–432. doi: 10.1046/j.1365-2958.2000.02008.x. [DOI] [PubMed] [Google Scholar]
- 10.DeVinney R, Stein M, Reinscheid D, Abe A, Ruschkowski S, Finlay BB. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect Immun. 1999;67:2389–2398. doi: 10.1128/iai.67.5.2389-2398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dzink JL, Socransky SS, Haffajee AD. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–323. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Prada CM, Hoover DL, Tall BD, Hartman AB, Kopelowitz J, Venkatesan MM. Shigella flexneri IpaH(7.8) facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect Immun. 2000;68:3608–3619. doi: 10.1128/iai.68.6.3608-3619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel G, Candy DC, Fabiani E, Adu-Bobie J, Gil S, Novakova M, Phillips AD, Dougan G. Molecular characterization of a carboxy-terminal eukaryotic-cell- binding domain of intimin from enteropathogenic Escherichia coli. Infect Immun. 1995;63:4323–4328. doi: 10.1128/iai.63.11.4323-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frankel G, Lider O, Hershkoviz R, Mould AP, Kachalsky SG, Candy DC, Cahalon L, Humphries MJ, Dougan G. The cell-binding domain of intimin from enteropathogenic Escherichia coli binds to beta1 integrins. J Biol Chem. 1996;271:20359–20364. doi: 10.1074/jbc.271.34.20359. [DOI] [PubMed] [Google Scholar]
- 15.Frankel G, Philips AD, Novakova M, Batchelor M, Hicks S, Dougan G. Generation of Escherichia coli intimin derivatives with differing biological activities using site-directed mutagenesis of the intimin C- terminus domain. Mol Microbiol. 1998;29:559–570. doi: 10.1046/j.1365-2958.1998.00950.x. [DOI] [PubMed] [Google Scholar]
- 16.Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun. 1992;60:5298–5301. doi: 10.1128/iai.60.12.5298-5301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grenier D. Characterization of the trypsin-like activity of Bacteroides forsythus. Microbiology. 1995;141:921–926. [Google Scholar]
- 18.Grossi SG, Genco RJ, Machtei EE, Ho AW, Koch G, Dunford RG, Zambon J, Hausmann E. Assessment of risk for periodontal disease. II. Risk indicators for alveolar bone loss. J Periodontol. 1995;66:23–29. doi: 10.1902/jop.1995.66.1.23. [DOI] [PubMed] [Google Scholar]
- 19.Grossi SG, Zambon JJ, Ho AW, Koch G, Dunford RG, Machtei EE, Norderyd OM, Genco RJ. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- 20.Haffajee AD, Socransky SS. Relation of body mass index, periodontitis and Tannerella forsythia. Journal of Clinical Periodontology. 2009;36:89–99. doi: 10.1111/j.1600-051X.2008.01356.x. [DOI] [PubMed] [Google Scholar]
- 21.Hajishengallis G, Martin M, Sojar HT, Sharma A, Schifferle RE, DeNardin E, Russell MW, Genco RJ. Dependence of bacterial protein adhesins on toll-like receptors for proinflammatory cytokine induction. Clin Diagn Lab Immunol. 2002;9:403–401. doi: 10.1128/CDLI.9.2.403-411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamlet SM, Ganashan N, Cullinan MP, Westerman B, Palmer JE, Seymour GJ. A 5-year longitudinal study of Tannerella forsythia prtH genotype: association with loss of attachment. J Periodontol. 2008;79:144–149. doi: 10.1902/jop.2008.070228. [DOI] [PubMed] [Google Scholar]
- 23.Hamlet SM, Taiyeb-Ali TB, Cullinan MP, Westerman B, Palmer JE, Seymour GJ. Tannerella forsythensis prtH genotype and association with periodontal status. J Periodontol. 2007;78:344–350. doi: 10.1902/jop.2007.060161. [DOI] [PubMed] [Google Scholar]
- 24.Hartland EL, Huter V, Higgins LM, Goncalves NS, Dougan G, Phillips AD, MacDonald TT, Frankel G. Expression of intimin gamma from enterohemorrhagic Escherichia coli in Citrobacter rodentium. Infect Immun. 2000;68:4637–4646. doi: 10.1128/iai.68.8.4637-4646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasebe A, Yoshimura A, Into T, Kataoka H, Tanaka S, Arakawa S, Ishikura H, Golenbock DT, Sugaya T, Tsuchida N, et al. Biological activities of Bacteroides forsythus lipoproteins and their possible pathological roles in periodontal disease. Infect Immun. 2004;72:1318–1325. doi: 10.1128/IAI.72.3.1318-1325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higuchi N, Murakami Y, Moriguchi K, Ohno N, Nakamura H, Yoshimura F. Localization of major, high molecular weight proteins in Bacteroides forsythus. Microbiol Immunol. 2000;44:777–780. doi: 10.1111/j.1348-0421.2000.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 27.Hirt RP, Harriman N, Kajava AV, Embley TM. A novel potential surface protein in Trichomonas vaginalis contains a leucine-rich repeat shared by micro-organisms from all three domains of life. Mol Biochem Parasitol. 2002;125:195–199. doi: 10.1016/s0166-6851(02)00211-6. [DOI] [PubMed] [Google Scholar]
- 28.Holt SC, Bramanti TE. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit Rev Oral Biol Med. 1991;2:177–281. doi: 10.1177/10454411910020020301. [DOI] [PubMed] [Google Scholar]
- 29.Honma K, Inagaki S, Okuda K, Kuramitsu HK, Sharma A. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog. 2007;42:156–166. doi: 10.1016/j.micpath.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Honma K, Kuramitsu HK, Genco RJ, Sharma A. Development of a gene inactivation system for Bacteroides forsythus: construction and characterization of a BspA mutant. Infect Immun. 2001;69:4686–4690. doi: 10.1128/IAI.69.7.4686-4690.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honma K, Mishima E, Inagaki S, Sharma A. The OxyR homologue in Tannerella forsythia regulates expression of oxidative stress responses and biofilm formation. Microbiology. 2009;155:1912–1922. doi: 10.1099/mic.0.027920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes CV, Malki G, Loo CY, Tanner AC, Ganeshkumar N. Cloning and expression of alpha-D-glucosidase and N-acetyl-beta-glucosaminidase from the periodontal pathogen, Tannerella forsythensis (Bacteroides forsythus) Oral Microbiol Immunol. 2003;18:309–312. doi: 10.1034/j.1399-302x.2003.00091.x. [DOI] [PubMed] [Google Scholar]
- 33.Ikegami A, Honma K, Sharma A, Kuramitsu HK. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect Immun. 2004;72:4619–4627. doi: 10.1128/IAI.72.8.4619-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis Vesicles Enhance Attachment, and the Leucine-Rich Repeat BspA Protein is Required for Invasion of Epithelial Cells by “Tannerella forsythia”. Infect Immun. 2006;74:5023–5028. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikura H, Arakawa S, Nakajima T, Tsuchida N, Ishikawa I. Cloning of the Tannerella forsythensis (Bacteroides forsythus) siaHI gene and purification of the sialidase enzyme. J Med Microbiol. 2003;52:1101–1107. doi: 10.1099/jmm.0.05349-0. [DOI] [PubMed] [Google Scholar]
- 36.Jonasson A, Eriksson C, Jenkinson HF, Kallestal C, Johansson I, Stromberg N. Innate immunity glycoprotein gp-340 variants may modulate human susceptibility to dental caries. BMC Infect Dis. 2007;7:57. doi: 10.1186/1471-2334-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kashket S, Maiden MF, Haffajee AD, Kashket ER. Accumulation of methylglyoxal in the gingival crevicular fluid of chronic periodontitis patients. J Clin Periodontol. 2003;30:364–367. doi: 10.1034/j.1600-051x.2003.00322.x. [DOI] [PubMed] [Google Scholar]
- 38.Kelly G, Prasannan S, Daniell S, Fleming K, Frankel G, Dougan G, Connerton I, Matthews S. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat Struct Biol. 1999;6:313–318. doi: 10.1038/7545. [DOI] [PubMed] [Google Scholar]
- 39.Kerosuo E, Haapasalo M, Alli K, Lounatmaa K. Ingestion of Bacteroides buccae, Bacteroides oris, Porphyromonas gingivalis, and Fusobacterium nucleatum by human polymorphonuclear leukocytes in vitro. Oral Microbiology & Immunology. 1990;5:202–207. doi: 10.1111/j.1399-302x.1990.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 40.Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M, Ebersole JL. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun. 2007;75:1704–1712. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 42.Kumagai Y, Konishi K, Gomi T, Yagishita H, Yajima A, Yoshikawa M. Enzymatic properties of dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis and its participation in virulence. Infect Immun. 2000;68:716–724. doi: 10.1128/iai.68.2.716-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumagai Y, Yagishita H, Yajima A, Okamoto T, Konishi K. Molecular mechanism for connective tissue destruction by dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2005;73:2655–2664. doi: 10.1128/IAI.73.5.2655-2664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuramitsu HK, Yoneda M, Madden T. Proteases and collagenases of Porphyromonas gingivalis. Adv Dent Res. 1995;9:37–40. doi: 10.1177/08959374950090010701. [DOI] [PubMed] [Google Scholar]
- 46.Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene. 2006;371:102–111. doi: 10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 47.Lewis JP, Iyer D, He H, Miyazaki H, Yeudall A, Anaya C. Identification and Characterization of adhesins from Prevotella intermedia 17. 37th American Asociation for Dental Research; Dallas, Texas, USA. 2008. [Google Scholar]
- 48.Ligtenberg TJ, Bikker FJ, Groenink J, Tornoe I, Leth-Larsen R, Veerman EC, Nieuw Amerongen AV, Holmskov U. Human salivary agglutinin binds to lung surfactant protein-D and is identical with scavenger receptor protein gp-340. Biochem J. 2001;359:243–248. doi: 10.1042/0264-6021:3590243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin X, Lamont RJ, Wu J, Xie H. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J Bacteriol. 2008;190:4367–4371. doi: 10.1128/JB.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Listgarten MA, Lai CH, Young V. Microbial composition and pattern of antibiotic resistance in subgingival microbial samples from patients with refractory periodontitis. J Periodontol. 1993;64:155–161. doi: 10.1902/jop.1993.64.3.155. [DOI] [PubMed] [Google Scholar]
- 51.Lo T, Westwood M, McLellan A, Selwood T, Thornalley P. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with N alpha- acetylarginine, N alpha-acetylcysteine, and N alpha-acetyllysine, and bovine serum albumin. J Biol Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- 52.Loimaranta V, Hytonen J, Pulliainen AT, Sharma A, Tenovuo J, Stromberg N, Finne J. Leucine-rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J Biol Chem. 2009;284:18614–18623. doi: 10.1074/jbc.M900581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maiden MFJ, Pham C, Kashket S. Glucose toxicity effect and accumulation of methylgloxal by the periodontal pathogen Bacteroides forsythus. Anaerobe. 2004;10:27–32. doi: 10.1016/j.anaerobe.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Mansell A, Braun L, Cossart P, O'Neill LA. A novel function of InIB from Listeria monocytogenes: activation of NF-kappaB in J774 macrophages. Cell Microbiol. 2000;2:127–136. doi: 10.1046/j.1462-5822.2000.00038.x. [DOI] [PubMed] [Google Scholar]
- 55.Marino M, Braun L, Cossart P, Ghosh P. Structure of the lnlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes. Mol Cell. 1999;4:1063–1072. doi: 10.1016/s1097-2765(00)80234-8. [DOI] [PubMed] [Google Scholar]
- 56.Marino M, Braun L, Cossart P, Ghosh P. A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc Natl Acad Sci U S A. 2000;97:8784–8788. doi: 10.1073/pnas.97.16.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mengaud J, Lecuit M, Lebrun M, Nato F, Mazie JC, Cossart P. Antibodies to the leucine-rich repeat region of internalin block entry of Listeria monocytogenes into cells expressing E-cadherin. Infect Immun. 1996;64:5430–5433. doi: 10.1128/iai.64.12.5430-5433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mengaud J, Ohayon H, Gounon P, Mege RM, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 59.Murakami Y, Higuchi N, Nakamura H, Yoshimura F, Oppenheim FG. Bacteroides forsythus hemagglutinin is inhibited by N-acetylneuraminyllactose. Oral Microbiol Immunol. 2002;17:125–128. doi: 10.1046/j.0902-0055.2001.00093.x. [DOI] [PubMed] [Google Scholar]
- 60.Murray PA, Kern DG, Winkler JR. Identification of a galactose-binding lectin on Fusobacterium nucleatum FN-2. Infect Immun. 1988;56:1314–1319. doi: 10.1128/iai.56.5.1314-1319.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakajima T, Tomi N, Fukuyo Y, Ishikura H, Ohno Y, Arvind R, Arai T, Ishikawa I, Arakawa S. Isolation and identification of a cytopathic activity in Tannerella forsythia. Biochem Biophys Res Commun. 2006:133–139. doi: 10.1016/j.bbrc.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen KA, Travis J, Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of gram-negative bacteria? J Bacteriol. 2007;189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen KA, Zylicz J, Szczesny P, Sroka A, Hunter N, Potempa J. Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology. 2009;155:328–337. doi: 10.1099/mic.0.024323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, Sharma A. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pei J, Grishin NV. Prediction of a caspase-like fold in Tannerella forsythia virulence factor PrtH. Cell Cycle. 2009;8 doi: 10.4161/cc.8.9.8243. [DOI] [PubMed] [Google Scholar]
- 66.Potempa J, Pike R. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, Frängsmyr L, Holmskov U, Leffler H, Nilsson C, Borén T, Wright JR, Strömberg N, Fisher SJ. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem. 2000;275:39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- 68.Rollins SM, Peppercorn A, Hang L, Hillman JD, Calderwood SB, Handfield M, Ryan ET. In vivo induced antigen technology (IVIAT) Cell Microbiol. 2005;7:1–9. doi: 10.1111/j.1462-5822.2004.00477.x. [DOI] [PubMed] [Google Scholar]
- 69.Rosenstiel P, Sina C, End C, Renner M, Lyer S, Till A, Hellmig S, Nikolaus S, Fölsch UR, Helmke B, Autschbach F, Schirmacher P, Kioschis P, Hafner M, Poustka A, Mollenhauer J, Schreiber S. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178:8203–8211. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- 70.Sabet C, Lecuit M, Cabanes D, Cossart P, Bierne H. LPXTG Protein InlJ, a newly Identified internalin involved in Listeria monocytogenes virulence. Infect Immun. 2005;73:6912–6922. doi: 10.1128/IAI.73.10.6912-6922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sabet M, Lee SW, Nauman RK, Sims T, Um HS. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology. 2003;149:3617–3627. doi: 10.1099/mic.0.26535-0. [DOI] [PubMed] [Google Scholar]
- 72.Saito T, Ishihara K, Kato T, Okuda K. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect Immun. 1997;65:4888–4891. doi: 10.1128/iai.65.11.4888-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 2007;153:866–876. doi: 10.1099/mic.0.29275-0. [DOI] [PubMed] [Google Scholar]
- 74.Sakamoto M, Suzuki M, Umeda M, Ishikawa L, Benno Y. Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002;52:841–849. doi: 10.1099/00207713-52-3-841. [DOI] [PubMed] [Google Scholar]
- 75.Sasaki H, Hou L, Belani A, Wang CY, Uchiyama T, Muller R, Stashenko P. IL-10, but not IL-4, suppresses infection-stimulated bone resorption in vivo. J Immunol. 2000;165:3626–3630. doi: 10.4049/jimmunol.165.7.3626. [DOI] [PubMed] [Google Scholar]
- 76.Seepersaud R, Hanniffy SB, Mayne P, Sizer P, Le Page R, Wells JM. Characterization of a novel leucine-rich repeat protein antigen from group B streptococci that elicits protective immunity. Infect Immun. 2005;73:1671–1683. doi: 10.1128/IAI.73.3.1671-1683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, Reynolds EC. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol. 2006;188:6376–6686. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shanks RMQ, Stella NA, Kalivoda EJ, Doe MR, O'Dee DM, Lathrop KL, Guo FL, Nau GJ. A Serratia marcescens OxyR Homolog Mediates Surface Attachment and Biofilm Formation. J Bacteriol. 2007;189:7262–7272. doi: 10.1128/JB.00859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma A, Inagaki S, Honma K, Sfintescu C, Baker PJ, Evans RT. Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J Dent Res. 2005;84:462–467. doi: 10.1177/154405910508400512. [DOI] [PubMed] [Google Scholar]
- 80.Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol Immunol. 2005;20:39–42. doi: 10.1111/j.1399-302X.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 81.Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect Immun. 1998;66:5703–5710. doi: 10.1128/iai.66.12.5703-5710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Socransky SS. Criteria for the infectious agents in dental caries and periodontal disease. J Clin Periodontol. 1979;6:16–21. doi: 10.1111/j.1600-051x.1979.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 83.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 84.Takemoto T, Kurihara H, Dahlen G. Characterization of Bacteroides forsythus isolates. J Clin Microbiol. 1997;35:1378–1381. doi: 10.1128/jcm.35.6.1378-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanner A, Maiden MF, Macuch PJ, Murray LL, Kent RL., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 86.Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 87.Tanner ACR, Listgarten MA, Ebersole JL, Strzempko MN. Bacteroides forsythus sp. nov., a slow growing, fusiform Bacteroides sp. from the human oral cavity. Int J Syst Bacteriol. 1986;36:213–221. [Google Scholar]
- 88.Thompson H, Homer KA, Rao S, Booth V, Hosie AH. An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J Bacteriol. 2009;191:3623–3628. doi: 10.1128/JB.01618-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tomi N, Fukuyo Y, Arakawa S, Nakajima T. Pro-inflammatory cytokine production from normal human fibroblasts is induced by Tannerella forsythia detaching factor. J Periodontal Res. 2008;43:136–142. doi: 10.1111/j.1600-0765.2007.01003.x. [DOI] [PubMed] [Google Scholar]
- 90.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32:120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 91.Tsolis RM, Townsend SM, Miao EA, Miller SI, Ficht TA, Adams LG, Baumler AJ. Identification of a putative Salmonella enterica serotype typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Veith PD, O'Brien-Simpson NM, Tan Y, Djatmiko DC, Dashper SG, Reynolds EC. Outer membrane proteome and antigens of Tannerella forsythia. J Proteome Res. 2009 Aug 11; doi: 10.1021/pr900372c. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 93.Yoneda M, Hirofuji T, Anan H, Matsumoto A, Hamachi T, Nakayama K, Maeda K. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J Periodontal Res. 2001;36:237–243. doi: 10.1034/j.1600-0765.2001.036004237.x. [DOI] [PubMed] [Google Scholar]
- 94.Yoo JY, Kim HC, Zhu W, Kim SM, Sabet M, Handfield M, Hillman J, Progulske-Fox A, Lee SW. Identification of Tannerella forsythia antigens specifically expressed in patients with periodontal disease. FEMS Microbiol Lett. 2007;275:344–352. doi: 10.1111/j.1574-6968.2007.00906.x. [DOI] [PubMed] [Google Scholar]
- 95.Zuger J, Luthi-Schaller H, Gmur R. Uncultivated Tannerella BU045 and BU063 are slim segmented filamentous rods of high prevalence but low abundance in inflammatory disease-associated dental plaques. Microbiology. 2007;153:3809–3816. doi: 10.1099/mic.0.2007/010926-0. [DOI] [PubMed] [Google Scholar]


