Abstract
Glutamate transporters are integral membrane proteins that catalyze a thermodynamically uphill uptake of the neurotransmitter glutamate from the synaptic cleft into the cytoplasm of glial and neuronal cells by harnessing the energy of pre-existing electrochemical gradients of ions. The linchpin of the reaction is the conformational transition of the transporters between outward and inward facing states, in which the substrate binding sites are accessible from the extracellular space and the cytoplasm respectively. Here we describe a crystal structure of a double cysteine mutant of a bacterial homologue of glutamate transporters, GltPh, which is trapped in the inward facing state by cysteine cross-linking. Together with the previously determined crystal structure of GltPh in the outward facing state, the structure of the cross-linked mutant allows us to propose a molecular mechanism, by which GltPh and, by analogy, mammalian glutamate transporters, mediate sodium-coupled substrate uptake.
Glutamate is the predominant excitatory neurotransmitter in the brain responsible for learning, memory formation and higher cognitive function. Specialized membrane proteins, glutamate transporters, also termed excitatory amino acid transporters (EAATs), mediate the transmitter uptake from the extracellular space into the cytoplasm of astrocytes and neurons against steep concentration gradients to allow for rounds of neurotransmission and to prevent glutamate-mediated excitotoxicity1. EAATs are members of a ubiquitous SoLute Carrier 1 (SLC1) family of secondary solute transporters2, which catalyze concentrative uptake of the acidic and neutral amino acids and dicarboxylic acids in a reaction coupled to symport of sodium and/or protons and, in the case of EAATs, antiport of potassium. The conceptual mechanisms of protein-mediated solute transport were developed over 40 years ago3–5 and focus on the ability of transporters to undergo isomerization between two states: an outward facing state, in which the substrate-binding site is accessible from the external solution, and an inward facing state, in which it is reached from the cytoplasm. Although our understanding of the molecular mechanisms of transport have advanced significantly in the last decade6,7, the structural transitions between the outward and inward facing states remain poorly characterized.
The sodium/aspartate symporter from Pyrococcus horikoshii (GltPh), an archaeal homologue of the EAATs, was one of the first sodium-coupled transporters whose structure had been determined at a near atomic resolution8,9. The structural analysis of GltPh revealed that individual protomers assemble into a homotrimer, forming a deep solvent-filled bowl open to the extracellular solution and reaching approximately half way across the lipid bilayer (Fig. 1a). The first six transmembrane segments (TMs) of each GltPh protomer mediate all inter-subunit contacts and form a distorted cylinder within which the highly conserved carboxyl terminus of the protein is folded into a compact core. The core consists of an intracellular re-entrant helical hairpin (HP) 1, TM7 with an unwound segment, an extracellular hairpin, HP2, and an amphipathic TM8. Despite the trimeric assembly, shared by all characterized glutamate transporter homologues10–12, protomers function independently13–17. Consistently, in GltPh crystal structure an L-asp molecule and two sodium ions are bound within the core of each individual protomer. Positioned between the tips of HP1 and HP2 at the bottom of the extracellular bowl, the substrate is occluded from the aqueous milieu only by HP2. Strikingly, in a crystal structure of GltPh in complex with a competitive blocker, L-threo-β-benzyloxyaspartate (L-TBOA)9,18, HP2 assumes an open conformation, revealing its dynamic nature and suggesting that it may serve as an extracellular gate. Bound L-asp and L-TBOA are separated from the cytoplasm by over 15 Å of a compact protein core, suggesting that these structures correspond to the outward facing state of the transporter. However, the structure of the inward facing state and the manner in which the substrate and sodium ions are released into the intracellular solution, remain unknown.
Figure 1. Cross-linking of GltPh-K55C/A364C.
a, Cartoon representation of two substrate-bound GltPh protomers (PDB code 2NWX) viewed in the membrane plain. The third protomer and TM4 of the protomer on the right are removed for clarity. TMs 1 through 6 are coloured salmon and wheat; the carboxyl terminal cores are light blue; and the hairpins HP1 and HP2 are dark blue. Bound L-asp and Na+ are shown as sticks and purple spheres, respectively. Orange spheres correspond to the Cα atoms of residues 55 and 364. All molecular representations have been generated using Pymol46. b, SDS PAGE analysis of GltPh-K55C/A364C before and after incubation with 100 μM CuPhen for indicated periods of time. Detergent-solubilized purified GltPh-K55C/A364C (left) and unpurified transporter in crude E. coli membranes (right) were visualized by Coomassie staining and western blotting, respectively. c, Cross-linking of GltPh-K55C/A364C in the presence of 50 μM HgCl2. Samples were analyzed as in b.
Cross-linking cysteines in TM2 and HP2
In a study predating the crystal structure determination of GltPh, Vandenberg and colleagues reported that two cysteines placed in TM2 and HP2 of EAAT1 formed a spontaneous disulfide bond suggesting close proximity of these residues19. Strikingly, in the crystal structures of L-asp and L-TBOA-bound GltPh the corresponding residues K55 in TM2 and A364 in HP2 are over 25 Å apart and cysteines at these positions are unlikely to become cross-linked. Moreover, the inter-subunit residue-to-residue distances are also 25–30 Å (Fig. 1a). Intrigued by these results, we reasoned that TM2 and HP2 become juxtaposed in an as yet structurally uncharacterized functional state of the transporter and set out to reproduce the double cysteine mutation in GltPh for crystallographic studies. The K55C/A364C mutant was generated within a cysteine-less GltPh, expressed in E. coli and purified in detergent solution. Oxidative cross-linking of GltPh-K55C/A364C in the presence of copper 1,10-phenantroline (CuPhen) yielded distinct protein species with a higher electrophoretic mobility on SDS PAGE (Fig. 1b), which we identified as intramolecularly cross-linked GltPh protomers (Supplementary Fig. 1a–c). Interestingly, CuPhen-catalyzed disulfide bond formation is greatly facilitated when the transporter is purified in the absence of sodium ions (Na+) and L-asp, suggesting that the state in which residues 55 and 364 are proximal, is more populated under these conditions (Fig. 1b and Supplementary Fig. 2a). Qualitatively similar results were also obtained using unpurified K55C/A364C mutant within E. coli crude membranes, demonstrating that GltPh, like EAAT1, adopts the new conformation in the context of a lipid bilayer. Moreover, cysteines at structurally adjacent positions did not form a disulfide bond, suggesting that the distance shortening between positions 55 and 364 occurs in a highly specific manner (Ref. 19 and Supplementary Fig. 2b). Strikingly, incubation of GltPh-K55C/A364C with divalent mercury (Hg2+), which serves as a bi-functional thiol-specific cross-linker, yields an essentially complete cross-linking both in the presence and in the absence of Na+ and L-asp in detergent and in crude membranes (Fig 1c). Furthermore, cross-linking is completed within seconds, suggesting that both substrate-free and bound GltPh rapidly sample the crosslinking-competent state.
Crystal structure determination
Hg2+-cross-linked K55C/A364C mutant (GltPh-55C/364CHg) in the presence of Na+ and L-asp yielded crystals diffracting to 3.5–3.9 Å (See Supplementary Table 1). A previous extensive study, conducted by Slotboom and colleagues, has suggested that the functionally relevant conformational transitions of a related bacterial glutamate transporter do not involve regions engaged in protein trimerization, because cross-linking of multiple interfacial residues had no effect on substrate transport17. Hence, we used a trimeric GltPh model comprised of TM-s 2, 4 and 5, which are responsible for all inter-subunit contacts (Fig. 1a), to search for a molecular replacement solution. The initial phases yielded distinct peaks within the anomalous difference Fourier map corresponding to Hg atoms at positions adjacent to cysteines 55 in TM2. Rounds of manual building and refinement resulted in a protein model lacking 5 and 8 residues on the amino and carboxyl termini, respectively, and including 98 % of the side chains. To verify the model, we crystallized the selenomethionine-substituted GltPh-55C/364CHg. The Se peaks within the anomalous difference Fourier maps are in perfect agreement with the location of 17 methionines in the protein model (Supplementary Fig. 3a). The distance between the Cα atoms of modelled cysteines 55 and 364 is 7.4 Å, and the Hg peak within the anomalous difference Fourier map is positioned between these residues with the sulphur to mercury distances estimated at 2 – 2.3 Å (Supplementary Fig. 3b), similar to those observed in small molecular weight compounds20 and peptides21, 22.
Structure of the inward facing state
The most striking structural feature of GltPh-K55C/364CHg compared to the WT GltPh, is a ca 18 Å movement of the substrate binding sites from near the extracellular solution at the bottom of the extracellular bowl, to near the cytoplasm (Figs. 2a, b). Structural superposition using trimerization TM-s 2, 4 and 5, along with the peripheral TM1 reveals that these segments remain largely unchanged, with the root mean square deviation (RMSD) for the atomic positions of 1.2 Å. The remainder of the protein, including TMs 3 and 6, HP1, TM7, HP2 and TM8, does not align and has undergone a substantial movement (Fig. 2a). Remarkably, when these regions alone are superposed, their structures are essentially identical, yielding RMSD of 0.6 Å. These results suggest that GltPh can be partitioned into two structural domains, which we termed the trimerization and transport domains (Fig. 2c). The rigid body movements of the transport domains relative to the trimerization domain dominate the structural changes between the substrate-bound WT GltPh and GltPh-55C/364CHg (Fig. 2a,b and Supplementary Movie). The structural environment of the ion- and substrate-binding sites, which are positioned entirely within the transport domains, is preserved. The Fo-Fc difference Fourier map reveals excess electron density at these sites (Supplementary Fig. 4), suggesting that the transporter remains bound to L-asp and to two Na+ ions. Consistently, substrate-free GltPh-55C/364CHg in solution binds L-asp with high affinity and in a sodium-dependent manner (Fig 2d). In the structure of GltPh-55C/364CHg the substrate binding sites lie near the bottom of deep crevices formed within each protomer between HP1 and the cytoplasmic portion of TM7 of the transport domain and TMs 2 and 5 of the trimerization domain (Fig. 2b). Bound L-asp and Na1 are completely occluded from the solution by the tips of HP1 and HP2, while Na2 is partially exposed to the solvent (Fig. 2a, b). Because substrate and ion binding sites are near the intracellular solution, we propose that the conformation of GltPh-55C/364CHg corresponds to an inward facing state of the transporter.
Figure 2. GltPh-55C/364CHg in the inward facing substrate-bound state.
Cartoon representation of the single protomers (a) and surface representation of the trimers sliced through the binding sites (b). WT GltPh and GltPh-55C/364CHg are shown on the left and right, respectively. In (a), TMs 1, 2, 4 and 5 are coloured wheat with the remainder of the protomer light blue. Na+ are shown as purple spheres. c, Extracellular view of GltPh-55C/364CHg with straight and curved lines delineating individual protomers and transport domains, respectively. The trimerization domains (wheat) and transport domains (blue) are connected by short cytoplasmic (solid arrows) and extracellular (open arrows) loops, highlighted in red. The long TM3–4 loops (simple arrows) cross over the transport domains. d, Isothermal titration calorimetry analysis of L-asp binding to GltPh-55C/364CHg. Shown are the binding heats in 10 mM NaCl. The linear dependence (slope = −2.6 ± 0.7) of the log of the apparent dissociation constant, Kd, on the log of Na+ concentration is shown in the inset. GltPh-55C/364CHg was exchanged into Na+/L-asp-free buffer, diluted to 15–20 μM in the reaction cell of the Microcal ITC200, supplemented with indicated concentrations of Na+, and titrated with L-asp at 25 °C. The binding enthalpy and the apparent number of binding sites were 23.6 ± 0.8 kcal/mol and 0.4 ± 0.03 (n=6), respectively.
Because the trimerization domain undergoes little conformational transitions, it is unlikely to change its position relative to the membrane plane. Hence, we estimate that a ~25 Å thick apolar region of the lipid bilayer, which in the WT GltPh aligns approximately with the hydrophobic portion of TM1, is similarly positioned in GltPh-55C/364CHg. Such bilayer placing suggests that several polar residues at the extracellular ends of the lipid-facing TMs 3 and 6 move into the hydrophobic region of the membrane. It is possible that the lack of the lipid bilayer in the crystals removes important constrains, resulting in a non-native positioning of the transport domain. However, cross-linking of residues 55C and 364C in GltPh and corresponding residues in EAAT1 occurs efficiently in lipid membranes, suggesting that a sufficient range of motions of the transport domain is allowed. Movement of the transport domain toward the cytoplasm also significantly increases its exposure to the intracellular solvent. Strikingly, although the total exposed cytoplasmic surface area increases from ~1,300 in the WT to ~4,200 Å2 in the GltPh-55C/364CHg, the fraction of the apolar area changes only modestly from 57 to 63 %. Both values are well within the range reported for the solvent exposed area of soluble proteins23.
Domain interaction interfaces
Both WT GltPh and GltPh-55C/364CHg bury approximately equal surface areas of ~2,500 Å2 on the interface between the trimerization and transport domains. Furthermore, in both structures essentially the same residues of the trimerization domain are involved, forming a smooth relatively featureless interaction surface, which is over 80 % hydrophobic (Fig. 3a, b). Although TM1 and TM4 contribute to the contact area, TM2 and TM5, lying parallel to each other and crossing the membrane at an oblique angle, provide the bulk of the interactions, contributing 77 and 67 % to the interaction surface in the WT and GltPh-K55C/A364CHg, respectively. In contrast, the transport domains in the two structures present interaction interfaces with little overlap, but similar in area (1250 Å2) and hydrophobicity (70%). They are dominated by HP1 and the cytoplasmic portion of TM7 in the WT GltPh (Fig. 3a), and by HP2 and the extracellular portion of TM8 in GltPh-55C/364CHg (Fig 3b), which provide 92 and 88 % of the total buried area, respectively. The analysis of the evolutionary conservation reveals that the interacting residues within the trimerization domain as well as within the two alternative interfaces of the transport domain are all highly conserved (Fig. 3c, d). The high conservation of HP2 surface residues and the reported transport inhibition of the mammalian EAATs upon chemical modification of cysteines in HP224–28 are consistent with this region being involved in specific protein contacts as observed in GltPh-55C/364CHg. The disengagement of HP1 and the amino-terminal part of TM7 and their transition to the cytoplasmic surface of the transporter is also consistent with the previously reported intracellular solvent accessibility of these regions in related prokaryotic and mammalian transporters29–32.
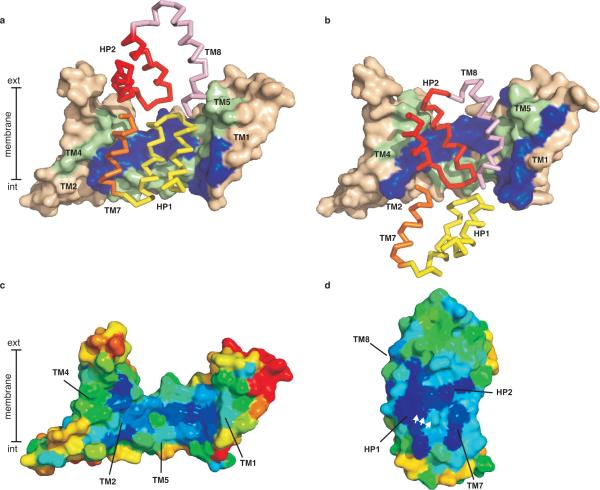
Figure 3. Domain interaction surfaces.
Trimerization domains of WT GltPh (a) and GltPh-55C/364CHg (b) are shown in surface representation and coloured wheat. Residues involved in domain contacts, identified by ProFace server47, are coloured blue (TM1 and 2) and green (TM4 and 5). Interacting structural elements of the transport domain are shown in ribbon representation: HP1 (yellow) and TM7 (orange) in WT GltPh, and HP2 (red) and TM8 (pink) in GltPh-55C/364CHg. Surface representation of the trimerization (c) and transport (d) domains coloured according to evolutionary conservation. Dark blue and red correspond to the highly conserved and variable residues, respectively. The interacting surfaces are facing the viewer and the white arrows mark the highly conserved serine-rich signature motif in HP1. Conservation scores were calculated using Consurf server48 and 212 SLC1 sequences with less than 60 % identity harvested from Pfam database49 and aligned in ClustalW250.
It has been noted previously that the carboxyl terminal core of GltPh contains structurally symmetrical elements8. Specifically, HP1 and the amino terminal half of TM7 can be superimposed on HP2 and the amino terminal portion of TM8 with RMSD of 2.5 Å. It is precisely these triplets of helices that constitute the two alternative interaction interfaces of the transport domain (Fig. 3a, b). Strikingly, also the amino terminal portion of the transporter, comprised of the first 6 TMs, exhibits a pseudo two-fold structural symmetry despite the lack of detectable sequence conservation. TMs 4 through 6 can be superimposed upon TMs 1 through 3 with RMSDs of ~6 and 4.3 Å for the WT and mutant, respectively (Fig. 4a, b). Hence, unlike other characterized secondary transporters33–35, glutamate transporters contain not one, but two inverted structural repeats (Fig. 4d). Because of the amino-terminal symmetry, the interaction interface of the trimerization domain can be partitioned into structurally related extracellular and cytoplasmic portions (Fig. 4c), which contribute approximately equally to the total buried surface area.
Figure 4. Amino-terminal inverted structural repeat.
Cartoon representation of TMs 1–3 (blue) and TMs 4–6 (green) in the GltPh-55C/364CHg viewed in the membrane plain (a), and their structural superposition (b). c, Symmetrical helices, TMs 1–2 and TMs 4–5, form the interaction surface within the transport domain, which is partitioned into intracellular and extracellular halves delineated by the dotted line. (d) Schematic representation of GltPh trimerization (orange) and the transport (light blue) domains. Two inverted structural repeats are emphasized by blue and green, and yellow and red trapezoids. Structure of TM2–3 and TM5–6 loops in WT GltPh (e) and GltPh-55C/364CHg (f). TMs 2 through 6 are shown in cartoon representation with 4 omitted for clarity. The transporter core is shown in transparent surface representation. Bound L-asp and the highly conserved glycines are shown as spheres.
Hinge movements
Within the transport domain, structurally symmetrical TM3 and TM6 serve as two arms holding the transporter core and extending from the trimerization domain, the former from the cytoplasm and the latter from the extracellular solution (Fig. 4e, f). The loops, which connect these transmembrane segments to the trimerization domain, enable the inward movement. The long flexible loop between TMs 3 and 4 is poorly structured in the L-asp-bound WT GltPh crystals, and in GltPh-55C/364CHg its structure is largely determined by crystal contacts. In contrast, the structurally symmetrical loops between TMs 2 and 3 and between TMs 5 and 6 are short and undergo well-defined transitions to accommodate the movements of the transport domain (Fig. 4e, f and Supplementary Movie). In GltPh-55C/364CHg, unwinding of the TM3 α-helix by approximately one turn extends TM2–3 loop by 4 residues and allows for the descent of TM3 toward the cytoplasm. In contrast, the TM5–6 loop is shortened by 2 residues as a result of simultaneous unwinding of TM5 by a half of a turn and extension of TM6 by a turn of a helix. Both loops contain conserved glycines, G69, G79 and G82 in TM2–3 loop, and G221 in TM5–6 loop, which may facilitate the folding/unfolding of the helices and serve as hinges. Indeed, the transport domain movement can be decomposed into a 16 Å descent toward the cytoplasm followed by a 37 ° rotation around an axis passing simultaneously through the transport domain centre of mass and TM2–3 and TM5–6 loops. Consistent with the importance of the structural rearrangements in the loop regions, chemical modifications of cysteines in TM5–6 loop of EAAT3 inhibited uptake36.
Transport mechanism
In the structure of the WT GltPh, L-asp is bound ~5 Å beneath the surface of the extracellular bowl occluded from the solution by HP2. In contrast, in the structure of GltPh-55C/364CHg, L-asp is bound ~5 Å beneath the intracellular surface occluded from the solution by HP1. Hence, we propose that L-asp-bound GltPh structure corresponds to the state right after binding of the substrate from the extracellular side, outward-facing occluded, and GltPh-55C/364CHg structure - to the state right before the release of the substrate into the cytoplasm, inward-facing occluded (Fig 5). The isomerization between these states involves a combination of a cross-membrane movement and rotation of the transport domain, comprised of the substrate-binding transporter core and peripheral TMs 3 and 6. During this movement, the lipid-facing hydrophobic TMs 3 and 6 traverse the bilayer directly, moving toward the cytoplasm by ~12 Å. In contrast, passage of the relatively polar HP1 and HP2, with their tips crossing as much as 20 Å of the lipid bilayer, is facilitated by the intra-protein track provided by the trimerization domain. The transport domains are expected to move stochastically and independently within each protomer, so that the crystal structure represents a statistically rare case, when all three transport domains are in the inward orientation. The rigid trimerization domain is anchored in the membrane and provides a counterbalance to the movements of the bulky transport domains, suggesting an explanation for the obligatory oligomeric assembly of SLC1 family of transporters.
Figure 5. Schematic transport mechanism.
Shown is a single transporter protomer. Substrate and sodium binding to the outward and inward facing states is coupled to the closure of the extracellular and intracellular gates, HP2 (red) and HP1 (yellow), respectively. Isomerization between the outward and inward facing occluded states occurs upon movement of the transport domain (blue), relative to the trimerization domain (grey). The inward facing open state has not been structurally characterized and is hypothetical.
Substrate binding/unbinding on either side of the membrane is associated with additional conformational changes, or gate openings. The crystallographic data and molecular dynamic simulations9, 37, 38 suggest that substrate and ion dissociation on the extracellular side is associated with the opening of HP2, defining it as an extracellular gate. We hypothesize that in a functionally symmetrical manner, movements of HP1 allow for dissociation of the substrate and ions from the inward facing state, defining it as a bona fide intracellular gate. The alternate opening of the extracellular and intracellular gates is strictly maintained: in the outward facing state, when HP2 opens to expose the substrate and ion binding sites to the extracellular solution, HP1 is secured in the closed state, packed against TMs 2 and 5. Conversely, in the inward facing state HP2, the extracellular gate, is locked closed upon displacing HP1.
As has been suggested previously39, coupling of the substrate transport to the energy of the ionic gradients is established via synergistic binding of substrate and sodium ions on the extracellular and intracellular sides of the membrane coupled to the closure of the corresponding gates. The steep sodium dependence of the substrate binding, observed both in WT GltPh9 and in GltPh-55C/364CHg (Fig. 2d), is responsible for the differential affinity for the substrate on the extracellular side of the membrane, where sodium concentration is high, and the intracellular side, where it is low. We further propose that the isomerization between the outward and inward facing states is sodium independent and may simply be driven by the thermal energy alone. The observation that Hg2+-mediated cross-linking of K55C/A364C mutant is completed within seconds both in membranes and in detergent suggests that this transition is rapid and does not limit the rate of transport in GltPh, which has a turnover time of ~3 minutes at ambient temperatures40. Finally, we hypothesize, that a similar isomerization reaction may also occur in the apo-transporter to complete the transport cycle. The structural re-arrangements within the transport domain, which would allow for the closure of the extracellular and the intracellular gates in the absence of bound substrate and ions, remain to be elucidated.
Methods
Cysteine cross-linking
GltPh-K55C/C321A/A364C was expressed as His8 fusion and purified as described previously8. Transporter samples were exchanged by size exclusion chromatography (SEC) into buffer, containing (in mM) 10 HEPES/NaOH or KOH, pH 7.4, 1 n-dodecyl-β-D-maltopyranoside and either 100 NaCl and 0.1 L-asp or 100 KCl. Cross-linking was initiated by addition of 1:2 molar ratio of Cu2+ and 1,10 phenantroline or HgCl2. Reactions were quenched with 100 mM N-ethyl maleimide prior to SDS PAGE analysis. Crude E. coli membranes were isolated by centrifugation, washed either in a Na+/L-asp-containing or free buffer and cross-linked as in detergent. Protein bands were visualized by western blotting using antibodies against histidine tag.
Crystallography
K55C/C321A/A364C mutation was introduced within a heptahistidine mutant of GltPh, used in earlier crystallographic studies8,9, to which we refer as “wild type” for brevity. Purified protein was cross-linked in the presence of 10 fold molar excess of HgCl2, dialyzed against buffer containing 10 mM HEPES/NaOH, 7 mM n-decyl-β-D-maltopyranoside, 100 mM NaCl and 100 μM L-asp, diluted to the final concentration of 2–4 mg/ml and supplemented with 0.5 mM E. coli total polar lipid extract and 100 mM NaBr. Protein solution was mixed at 1:1 (v:v) ratio with the reservoir solution, containing 100 mM MES, pH 5.0, 18–20 % PEG 350 MME and 200 mM CaCl2, and crystallized at 4 °C by hanging drop vapour diffusion. Crystals were cryoprotected by allowing the drop to dry until its volume was reduced by 50 %. Selenomethionine-substituted protein was expressed as described previously8 and crystallized as above. Diffraction data were indexed, integrated and scaled using HKL-2000 package41. Further analyses were performed using CCP4 programs42. Initial phases were obtained using Phaser43, and the protein model built manually in Coot44 and refined using REFMAC42 with TLS45 and three fold NCS restrains.
Supplementary Material
Acknowledgements
We thank Drs. D. Patel and H. Li for the help with ITC and Dr. H. Weinstein for constructive criticism. X-ray diffraction data were measured at X25 beamline at the National Synchrotron Light Source. This work was supported by the National Institute of Health (O.B.) and by a Jane Coffin Childs Memorial Fund postdoctoral fellowship (N.R.).
Footnotes
Supplementary Information Description. Supplementary Information, including 4 Supplementary Figures, Supplementary Table and Supplementary Movie, accompanies the paper.
Coordinate Deposition. The coordinates for the structure and the structure factors are deposited in the Protein Data Bank under accession code 3KBC.
REFERENCES
- 1.Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 2.Saier MH, Jr., Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–6. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180:134–6. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- 4.Patlak CS. Contributions to the theory of active transport: II. The gate type non-carrier mechanism and generalizations concerning tracer flow, efficiency, and measurement of energy expenditure. Bulletin of Mathematical Biology. 1957;19:209–235. [Google Scholar]
- 5.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–70. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 6.Sobczak I, Lolkema JS. Structural and mechanistic diversity of secondary transporters. Curr Opin Microbiol. 2005;8:161–7. doi: 10.1016/j.mib.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–55. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–8. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 9.Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–93. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 10.Yernool D, Boudker O, Folta-Stogniew E, Gouaux E. Trimeric subunit stoichiometry of the glutamate transporters from Bacillus caldotenax and Bacillus stearothermophilus. Biochemistry. 2003;42:12981–8. doi: 10.1021/bi030161q. [DOI] [PubMed] [Google Scholar]
- 11.Gendreau S, et al. A trimeric quaternary structure is conserved in bacterial and human glutamate transporters. J Biol Chem. 2004;279:39505–12. doi: 10.1074/jbc.M408038200. [DOI] [PubMed] [Google Scholar]
- 12.Raunser S, et al. Structure and function of prokaryotic glutamate transporters from Escherichia coli and Pyrococcus horikoshii. Biochemistry. 2006;45:12796–805. doi: 10.1021/bi061008+. [DOI] [PubMed] [Google Scholar]
- 13.Koch HP, Larsson HP. Small-scale molecular motions accomplish glutamate uptake in human glutamate transporters. J Neurosci. 2005;25:1730–6. doi: 10.1523/JNEUROSCI.4138-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewer C, et al. Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry. 2005;44:11913–23. doi: 10.1021/bi050987n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leary GP, Stone EF, Holley DC, Kavanaugh MP. The glutamate and chloride permeation pathways are colocalized in individual neuronal glutamate transporter subunits. J Neurosci. 2007;27:2938–42. doi: 10.1523/JNEUROSCI.4851-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch HP, Brown RL, Larsson HP. The glutamate-activated anion conductance in excitatory amino acid transporters is gated independently by the individual subunits. J Neurosci. 2007;27:2943–7. doi: 10.1523/JNEUROSCI.0118-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groeneveld M, Slotboom DJ. Rigidity of the subunit interfaces of the trimeric glutamate transporter GltT during translocation. J Mol Biol. 2007;372:565–70. doi: 10.1016/j.jmb.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 18.Shimamoto K, et al. DL-threo-beta-benzyloxyaspartate, a potent blocker of excitatory amino acid transporters. Mol Pharmacol. 1998;53:195–201. doi: 10.1124/mol.53.2.195. [DOI] [PubMed] [Google Scholar]
- 19.Ryan RM, Mitrovic AD, Vandenberg RJ. The chloride permeation pathway of a glutamate transporter and its proximity to the glutamate translocation pathway. J Biol Chem. 2004;279:20742–51. doi: 10.1074/jbc.M304433200. [DOI] [PubMed] [Google Scholar]
- 20.Manceau A, Nagy KL. Relationships between Hg(II)-S bond distance and Hg(II) coordination in thiolates. Dalton Trans. 2008:1421–5. doi: 10.1039/b718372k. [DOI] [PubMed] [Google Scholar]
- 21.Dieckmann CR, et al. De Novo Design of Mercury-Binding Two- and Three-Helical Bundles. J Am Chem Soc. 1997;119:6195–6196. [Google Scholar]
- 22.Rosenzweig AC, et al. Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution. Structure. 1999;7:605–17. doi: 10.1016/s0969-2126(99)80082-3. [DOI] [PubMed] [Google Scholar]
- 23.Sadeghi M, Naderi-Manesh H, Zarrabi M, Ranjbar B. Effective factors in thermostability of thermophilic proteins. Biophys Chem. 2006;119:256–70. doi: 10.1016/j.bpc.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Grunewald M, Menaker D, Kanner BI. Cysteine-scanning mutagenesis reveals a conformationally sensitive reentrant pore-loop in the glutamate transporter GLT-1. J Biol Chem. 2002;277:26074–80. doi: 10.1074/jbc.M202248200. [DOI] [PubMed] [Google Scholar]
- 25.Borre L, Kavanaugh MP, Kanner BI. Dynamic equilibrium between coupled and uncoupled modes of a neuronal glutamate transporter. J Biol Chem. 2002;277:13501–7. doi: 10.1074/jbc.M110861200. [DOI] [PubMed] [Google Scholar]
- 26.Seal RP, Shigeri Y, Eliasof S, Leighton BH, Amara SG. Sulfhydryl modification of V449C in the glutamate transporter EAAT1 abolishes substrate transport but not the substrate-gated anion conductance. Proc Natl Acad Sci U S A. 2001;98:15324–9. doi: 10.1073/pnas.011400198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan RM, Vandenberg RJ. Distinct conformational states mediate the transport and anion channel properties of the glutamate transporter EAAT-1. J Biol Chem. 2002;277:13494–500. doi: 10.1074/jbc.M109970200. [DOI] [PubMed] [Google Scholar]
- 28.Leighton BH, Seal RP, Shimamoto K, Amara SG. A hydrophobic domain in glutamate transporters forms an extracellular helix associated with the permeation pathway for substrates. J Biol Chem. 2002;277:29847–55. doi: 10.1074/jbc.M202508200. [DOI] [PubMed] [Google Scholar]
- 29.Slotboom DJ, Sobczak I, Konings WN, Lolkema JS. A conserved serine-rich stretch in the glutamate transporter family forms a substrate-sensitive reentrant loop. Proc Natl Acad Sci U S A. 1999;96:14282–7. doi: 10.1073/pnas.96.25.14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunewald M, Bendahan A, Kanner BI. Biotinylation of single cysteine mutants of the glutamate transporter GLT-1 from rat brain reveals its unusual topology. Neuron. 1998;21:623–32. doi: 10.1016/s0896-6273(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 31.Seal RP, Leighton BH, Amara SG. Transmembrane topology mapping using biotin-containing sulfhydryl reagents. Methods Enzymol. 1998;296:318–31. doi: 10.1016/s0076-6879(98)96024-4. [DOI] [PubMed] [Google Scholar]
- 32.Shlaifer I, Kanner BI. Conformationally sensitive reactivity to permeant sulfhydryl reagents of cysteine residues engineered into helical hairpin 1 of the glutamate transporter GLT-1. Mol Pharmacol. 2007;71:1341–8. doi: 10.1124/mol.106.032607. [DOI] [PubMed] [Google Scholar]
- 33.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–5. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–23. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 35.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 36.Shachnai L, Shimamoto K, Kanner BI. Sulfhydryl modification of cysteine mutants of a neuronal glutamate transporter reveals an inverse relationship between sodium dependent conformational changes and the glutamate-gated anion conductance. Neuropharmacology. 2005;49:862–71. doi: 10.1016/j.neuropharm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Huang Z, Tajkhorshid E. Dynamics of the Extracellular Gate and Ion-Substrate Coupling in the Glutamate Transporter. Biophys J. 2008 doi: 10.1529/biophysj.108.133421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shrivastava IH, Jiang J, Amara SG, Bahar I. Time-resolved mechanism of extracellular gate opening and substrate binding in glutamate transporter. J Biol Chem. 2008 doi: 10.1074/jbc.M800889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gouaux E. The molecular logic of sodium-coupled neurotransmitter transporters. Philos Trans R Soc Lond B Biol Sci. 2009;364:149–54. doi: 10.1098/rstb.2008.0181. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan RM, Compton EL, Mindell JA. Functional characterization of a Na+-dependent aspartate transporter from Pyrococcus horikoshii. J Biol Chem. 2009;284:17540–8. doi: 10.1074/jbc.M109.005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:308–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.No4, C. p. The CCP4 Suite: Programs for X-ray crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–33. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. [Google Scholar]
- 47.Saha RP, Bahadur RP, Pal A, Mandal S, Chakrabarti P. ProFace: a server for the analysis of the physicochemical features of protein-protein interfaces. BMC Struct Biol. 2006;6:11. doi: 10.1186/1472-6807-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.