Abstract
Developmental anomalies of the ocular anterior chamber angle may lead to an incomplete development of the structures that form the conventional aqueous outflow pathway. Thus, disorders that present with such dysfunction tend to be associated with glaucoma. Among them, Axenfeld-Rieger (ARS) malformation is a rare clinical entity with an estimated prevalence of one in every 200,000 individuals. The changes in eye morphogenesis in ARS are highly penetrant and are associated with 50% risk of development of glaucoma. Mutations in the cytochrome P4501B1 (CYP1B1) gene have been reported to be associated with primary congenital glaucoma and other forms of glaucoma and mutations in pituitary homeobox 2 (PITX2) gene have been identified in ARS in various studies. This case was negative for PITX2 mutations and compound heterozygote for CYP1B1 mutations. Clinical manifestations of this patient include bilateral elevated intraocular pressure (>40 mmHg) with increased corneal diameter (>14 mm) and corneal opacity. Patient also had iridocorneal adhesions, anteriorly displaced Schwalbe line, anterior insertion of iris, broad nasal bridge and protruding umbilicus. This is the first study from north India reporting CYP1B1 mutations in Axenfeld-Rieger syndrome with bilateral buphthalmos and early onset glaucoma. Result of this study supports the role of CYP1B1 as a causative gene in ASD disorders and its role in oculogenesis.
1. Introduction
Developmental anomalies of the anterior chamber angle may lead to an incomplete development of the structures that form the conventional aqueous outflow pathway. Thus, disorders that present with such dysfunction tend to be associated with glaucoma. Among them, Axenfeld-Rieger (ARS) malformation is a rare clinical entity with an estimated prevalence of one case in every 200,000 individuals [1]. The term ARS describes a group of congenital diseases that were historically summarized under the broader heading of anterior chamber cleavage syndromes (including Axenfeld anomaly, Rieger anomaly, and Rieger syndrome). ARS represents a spectrum of disorders involving ocular and, in some cases, extraocular structures caused by disruption of migration and differentiation of neural crest cells [2]. Several anterior segment anomalies known to involve tissues derived from neural crest cells have been described in literature and are thought to be due to inadequate regression of tissues and/or failure in differentiation, which results in distortions of normal iridocorneal angle anatomy. These disorders include Axenfeld-Rieger syndrome, Peters anomaly, familial iris hypoplasia associated with glaucoma, and iridogoniodysgenesis. The ocular structures involved in ARS include the cornea (posterior embryotoxon), the iridocorneal angle (peripheral iridocorneal adherences and ultrastructural abnormalities of the trabecular meshwork), and the iris (peripheral adhesions between the iris and the cornea, and stromal atrophy) [3].
Systemic manifestations more commonly associated with ARS include dental abnormalities (microdontia, hypodontia, and oligodontia) and facial malformations (hypoplasia of the maxillary bones) [3]. Redundant periumbilical skin, hypospadias, as well as other less frequent systemic alterations, may also be observed in ARS [4–6]. Pituitary homeobox 2 (PITX2) gene mutations have been identified in ARS in many studies [7, 8]. The changes in eye morphogenesis in ARS are highly penetrant and, anecdotally, have been associated with an approximately 50% risk of the development of glaucoma [9]. Glaucoma may appear during childhood, but it is more common during adolescence or at the beginning of adulthood. Glaucoma secondary to ARS is difficult to manage and may result in severe damage to the optic disc and visual field [3]. In 1997, Stoilov et al., described structural alterations in the CYP1B1 gene (MIM 601771), a member of the cytochrome P450 enzyme family, in patients with congenital glaucoma linked to the GLC3A locus [10]. Mutations in the CYP1B1 gene have been reported to be associated with other forms of glaucomas, including Peters' anomaly [11–13], and to act as a modifier gene in juvenile open-angle glaucoma [14]. Considering that Peters' anomaly is characterized by anterior ocular malformation because of defective migration of neural crest cells, it is possible to hypothesize that CYP1B1 gene could play a role in the pathogenesis of ARS.
2. Case Details
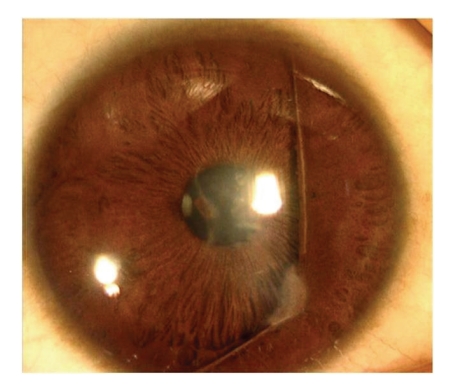
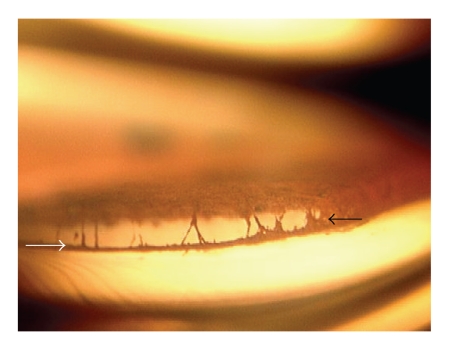
The proband was a 9 months old male offspring who presented with the bilateral buphthalmos with corneal edema. He had intraocular pressure of 48 mmHg and 40 mmHg in the right and left eye, respectively, with corneal diameter of 14.0 × 14.5 mm and 15.0 × 15.0 mm. He had iridocorneal adhesions (Figures 1 and 2), anteriorly displaced Schwalbe line (Figures 1 and 2), and anterior insertion of iris.
Figure 1.
Slit-lamp photograph of eye showing anterior segment dysgenesis.
Figure 2.
Gonioscopic photograph of eye showing anteriorly displaced Schwalbe line (white arrow) and irido-corneal adhesions (black arrow).
On general examination, he had broad nasal bridge and protruding umbilicus. On the basis of ocular and general examination clinical diagnosis of congenital glaucoma with Axenfeld-Rieger syndrome was made. The proband underwent bilateral trabeculotomy + trabeculectomy + mitomycin C (MMC) treatment. Patient is under regular follow up since then. After two years of follow-up he had IOP of 34 mmHg and 20 mmHg in the right and left eye, respectively, with 0.7 : 1 optic nerve cupping in both eyes. Recent corneal diameter was 15.0 × 14.0 mm in both eyes. He also had malaligned teeth.
3. Methods
The entire coding region including exon-intron boundaries of CYP1B1 and PITX2 gene from patient and his parents were amplified. PCR for CYP1B1 gene was done by using three sets of overlapping primers described elsewhere [15]. The primers used were set I (1F-1R, 786 bp), set II (2F-2R, 787 bp), and set III (3F-3R, 885 bp). PCR amplifications for primer sets I and II were performed in a 40 μL volume containing 1.0 μL of 20 μM stock solution for each primer, 100 ng of genomic DNA, 1 unit of Taq polymerase (Banglore Genei), 0.1 mM of each dNTP, 4 μL of 10X PCR buffer (with 15 mM MgCl2), and 4 μL of dimethyl sulphoxide (Sigma), by means of 35 cycles of amplification, each consisting of 30 seconds denaturation at 94°C, 30 seconds annealing at 56°C, and 1 minute extension at 72°C [14], while conditions for set III were initial denaturation at 94°C for 3 minutes followed by 30 cycles each at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute and final extension at 72°C for 5 minutes. PCR primers for PITX2 gene (Table 1) were designed using NCBI PRIMER3 program (available at http://www.ncbi.nlm.nih.gov/tools/primer-blast/). PCR amplifications for PITX2 all primer sets were performed in a 40 μL volume containing 1.0 μL of 20 μM stock solution for each primer, 100 ng of genomic DNA, 1 unit of Taq polymerase (Banglore Genei), 0.1 mM of each dNTP, and 4 μL of 10X PCR buffer (with 15 mM MgCl2), by means of 40 cycles of amplification, each consisting of 30 seconds denaturation at 94°C, 50 seconds annealing at 56–57°C, and 50 seconds extension at 72°C and final extension at 72°C for 5 minutes.
Table 1.
PITX2 primers used in this study.
| Primer Name | Primer Sequence | Product Size |
|---|---|---|
| PITX1F | 5′ CAC TCC CGC TGC CAT TGC GT 3′ | 618 bp |
| PITX1R | 5′ GGG GGC TTC GGT ACA CAG CG 3′ | |
| PITX2F | 5′ ACC GGG GAG GCG CAG AAA GA 3′ | 635 bp |
| PITX2R | 5′ GCC GAG GTT TGC TGG AGC GT 3′ | |
| PITX3F | 5′ GCA GCC CAG CTC TTC CAC GG 3′ | 559 bp |
| PITX3R | 5′ GTG AGA TCG CGG GAT GGC GG 3′ | |
| PITX4F | 5′ CTG CGC TTG GTG GAG ACC CG 3′ | 812 bp |
| PITX4R | 5′ GTT GCC CCA TCC GGC AAG GT 3′ |
Amplified PCR products were purified using a gel/PCR DNA fragments extraction kit (number DF100; Geneaid Biotech Ltd., Sijhih City, Taiwan). Purified PCR products were sent for sequencing to MCLAB (Molecular Cloning Laboratories, South San Francisco, CA). CYP1B1 DNA sequences were analyzed against the CYP1B1 gene reference sequence ENSG00000138061 (available at http://www.ensembl.org/Homosapiens/Gene/Sequence?g=ENSG00000138061) and PITX2 DNA sequences against PITX2 gene reference sequence ENSG00000164093 (available at http://www.ensembl.org/Homo_sapiens/Gene/Sequence?g=ENSG00000164093) using ClustalW2 (multiple sequence alignment program for DNA; European Molecular Biology Laboratory (EMBL)-European Bioinformatics Institute (EBI).
4. Computational Assessment of Missense Mutations
Two homology based programs PolyPhen (Polymorphism Phenotyping), available at http://genetics.bwh.harvard.edu/pph/, and SIFT (Sorting Intolerant From Tolerant) analysis tool, available at http://sift.jcvi.org/ were used to predict the functional impact of missense changes identified in this study. PolyPhen structurally analyzes an amino acid polymorphism and predicts whether that amino acid change is likely to be deleterious to protein function [16–18]. The prediction is based on the position-specific independent counts (PSIC) score derived from multiple sequence alignments of observations. PolyPhen scores of >2.0 indicate the polymorphism is probably damaging to protein function. Scores of 1.5–2.0 are possibly damaging, and scores of <1.5 are likely benign. SIFT is a sequence homology-based tool that sorts intolerant from tolerant amino acid substitutions and predicts whether an amino acid substitution in a protein will have a phenotypic effect [19–22]. SIFT is based on the premise that protein evolution is correlated with protein function. Positions important for function should be conserved in an alignment of the protein family, whereas unimportant positions should appear diverse in an alignment. Positions with normalized probabilities less than 0.05 are predicted to be deleterious and those greater than or equal to 0.05 are predicted to be tolerated.
5. Results
PITX2 gene analysis showed heterozygous T > A change at g.11553261 (in Intron 5). No pathogenic PITX2 mutation was identified in this patient. CYP1B1 analysis showed presence of two heterozygous pathogenic mutations (p.Arg355Stop + p.Arg368His). Both of these mutations (CYP1B1) have been reported in cases with primary congenital glaucoma [15]. Father of the proband was negative for these mutations but mother was heterozygous for p.R368H mutation.
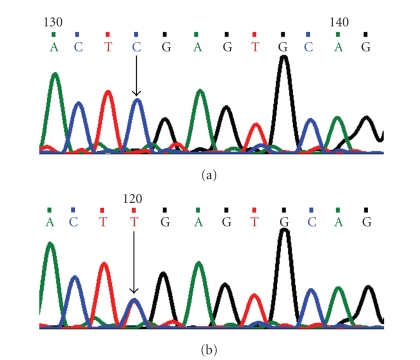
5.1. Arginine355Stop (p.R355X) Mutation
In this mutation, a single-base cytosine (C) was replaced by thymine (T) (Figure 3) at genomic position g.38151938; coding nucleotide number c.1063. This resulted in a codon change from CGA to TGA and p.R355X a non-sense mutation in CYP1B1 protein. This resulted in a truncated CYP1B1 protein of 354 amino acids.
Figure 3.
DNA sequence chromatogram of CYP1B1 exon 3 equivalent to codon 354–357. (a) The reference sequence derived from control is shown. (b) Sequence derived from ARS patient shows heterozygous c.1063C > T, which predicts a codon change CGA > TGA and heterozygous p.R355X, a non-sense mutation.
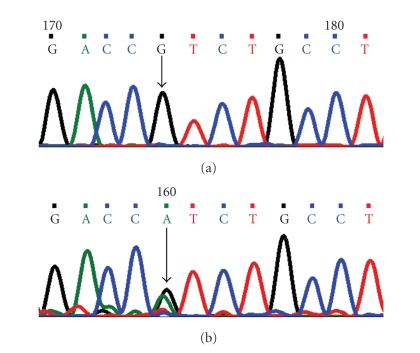
5.2. Arginine368Histidine (p.R368H) Mutation
A single-base guanine (G) was replaced by adenine (A) at genomic position g.38151898; coding nucleotide number c.1103. This resulted in a codon change from CGT to CAT and amino acid change from arginine to histidine (p.R368H) a nonsynonymous mutation (Figure 4). PSIC score of this mutation was 2.653 indicating that this change is probably damaging to protein function. SIFT score of p.R368H is 0.00 and is predicted to be deleterious for the protein function.
Figure 4.
DNA sequence chromatogram of CYP1B1 exon 3 equivalent to codon 367–370. (a) The reference sequence derived from control is shown. (b) Sequence derived from ARS patient shows heterozygous c.1103G > A, which predicts a codon change from CGT > CAT and heterozygous p.R368H mutation.
Since, missense mutation in PITX2 was absent no computational assessment was done for PITX2 protein.
6. Discussion
Over the last several years, the identification of genes and loci involved in the different forms of glaucoma has led to a better understanding of the pathogenetic mechanisms of primary open-angle glaucoma, congenital glaucoma, and developmental glaucoma, such as the one associated with ARS [2]. Although AR has an autosomal dominant inheritance there have been reports of sporadic cases in the literature [23].
Mutations of CYP1B1 are a major cause of primary congenital glaucoma in various studies from different populations [15, 24–29]. CYP1B1 is a member of the cytochrome P450 super family of drug metabolizing enzymes. It catalyzes several oxidative reactions, some of which are biosynthetic, producing necessary hormones or compounds of intermediary metabolism in most living organisms [30]. It also metabolizes vitamin A in two steps to all-trans-retinal and all-trans-retinoic acid. The latter is a potent morphogen and regulates in utero development of tissue growth and differentiation [31–33]. It is involved in the metabolism of the endogenous and exogenous substrates that take part in early ocular differentiation [31–33]. This case was compound heterozygote for p.R355X and p.R368H mutations. In p.R355X mutation, a truncated protein of 354 amino acids is produced. The arginine residue at position 355 lies in the carboxyl terminal of the J helix, carboxyl terminal of the J helix is involved in the functionally important heme-binding domain. This change (p.R355X) was first reported in PCG in Germany and after that from India [15, 34]. This truncating mutation results in a loss of the heme-binding domain and a functionally inactive protein [10, 34–36]. Mutations in CYP1B1 gene emerged as a largest cause of PCG and p.R368H has been reported as most predominant CYP1B1 mutation in PCG in Indian population [24].
Arginine residue at 368 position lies in between the helices J and K in an exposed loop [24, 29]. Consequences of this change are not immediately apparent except that the positively charged amino acid arginine is replaced by histidine whose charge state depends upon its protonation state. In the WT, arginine at 368 interacts with G-365, D-367, V-363, and D-374. Because of the p.R368H mutation, interactions between D-367 and D-374 are weakened [36]. PolyPhen and SIFT score of p.R368H showed that this mutation is deleterious for the protein function. An alteration of CYP1B1 expression due to a sequence change might alter the metabolic activity of CYP1B1, thus cause ocular development defects. Membrane-bound cytochromes, such as CYP1B1, have a molecular structure containing a transmembrane domain located at the N-terminal end of the molecule. This is followed by a proline-rich “hinge” region, which permits flexibility between the membrane-spanning domain and the cytoplasmic portion of the protein molecule. The COOH-terminal ends are highly conserved among different members of the cytochrome P450 super-family [37]. This family contains a set of conserved core structures responsible for the heme-binding region of these molecules. The heme-binding region is essential for the normal function of every P450 molecule. Between the hinge region and the conserved core structure lies a less conserved substrate-binding region. The cytochrome P450 protein functions like any classical enzyme molecule [32, 33]. Mutations affecting such enzymes generally produce recessive phenotypes because in heterozygous subjects the normal allele is capable of compensating for the mutant allele. Mutations in the CYP1B1 protein interfere with the integrity of the CYP1B1 protein as well as its ability to adopt a normal conformation and to bind heme; for example, induced mutations in the hinge region have previously been reported to interfere with the heme-binding properties of the cytochrome P450 molecules [38].
First report of mutations in CYP1B1 as a cause for Rieger Anomaly was published in 2006 by Chavarria-Soley et al. [39]. Three reports exist of CYP1B1 mutations in Peters Anomaly; patients are either compound heterozygotes [11, 13] or homozygous in consanguineous families [12].
Segregation of both ARS and Peters' anomaly in the same family has been reported previously [40–43]. Therefore, ARS and Peters' anomaly are discussed to be allelic variants and part of the single disease spectrum of anterior segment dysgenesis (ASD) rather than representing distinct entities. This hypothesis is supported by clinical and genetic findings as well as by the embryonic pathogenesis of ASD. First, several clinical features like corectopia or angle anomalies are common both in ARS and in Peters' anomaly. Second, mutations in FKLH7/FOXC1 genes (as well as mutations in other genes implicated in ASD) are associated with clinically variable manifestations like ARS [44, 45], primary congenital glaucoma [24–26], and Peters' anomaly [40]. Third, ASD is genetically heterogeneous.
Peters' anomaly, for instance, has been associated with mutations in PAX6 [42, 46–48], CYP1B1 [11–13, 49], and FOXC1 [43]. Fourth, there is growing evidence that the ASDs are neural crestopathies that result from a developmental arrest of specific anterior segment tissues derived from neural crest cells [3]. Mutations in CYP1B1 could be contributory in up to 20% of cases of Peters anomaly. Since this case was negative for pathogenic PITX2 mutations, therefore, results of this study support the role of CYP1B1 as a causative gene in ASD as suggested by previous studies [11–13, 39, 49]. Furthermore, this emphasizes the broad range of phenotypic expression for CYP1B1 mutations, and its role in eye development.
7. Conclusion
This is the first study from India reporting CYP1B1 mutations in Axenfeld-Rieger syndrome with bilateral buphthalmos and early onset glaucoma. Result of this study supports the role of CYP1B1 as a causative gene in ASD disorders [11–13, 39, 49] and it emphasizes the broad range of phenotypic expression for CYP1B1 mutations, and its role in oculogenesis.
Acknowledgments
This work was financially supported by the Department of Biotechnology, Govt. of India. The authors would like to thank the family of the patient for their cooperation. The author M. Tanwar thanks University Grants Commission (UGC), Govt. of India for providing Senior Research Fellowship (SRF).
References
- 1.Alkemade PPH. Dysgenesis Mesodermalis of the Iris and Cornea. Assen, The Netherlands: Van Gorcum ad Comp NV; 1969. [Google Scholar]
- 2.Alward WLM. Axenfeld-Rieger syndrome in the age of molecular genetics. American Journal of Ophthalmology. 2000;130(1):107–115. doi: 10.1016/s0002-9394(00)00525-0. [DOI] [PubMed] [Google Scholar]
- 3.Shields MB. Axenfeld-Rieger syndrome: a theory of mechanism and distinctions from the iridocorneal endothelial syndrome. Transactions of the American Ophthalmological Society. 1983;81:736–784. [PMC free article] [PubMed] [Google Scholar]
- 4.Jorgenson RJ, Levin LS, Cross HE, Yoder F, Kelly TE. The Rieger syndrome. The American Journal of Medicine. 1978;2:307–318. doi: 10.1002/ajmg.1320020310. [DOI] [PubMed] [Google Scholar]
- 5.Jena AK, Kharbanda OP. Axenfeld-Rieger syndrome: report on dental and craniofacial findings. Journal of Clinical Pediatric Dentistry. 2005;30(1):83–88. doi: 10.17796/jcpd.30.1.v1732398454r0244. [DOI] [PubMed] [Google Scholar]
- 6.Lubin JR. Oculocutaneous albinism associated with corneal mesodermal dysgenesis. American Journal of Ophthalmology. 1981;91(3):347–350. doi: 10.1016/0002-9394(81)90288-9. [DOI] [PubMed] [Google Scholar]
- 7.Brooks BP, Moroi SE, Downs CA, et al. A novel mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome. Ophthalmic Genetics. 2004;25(1):57–62. doi: 10.1076/opge.25.1.57.29002. [DOI] [PubMed] [Google Scholar]
- 8.Vieira V, David G, Roche O, et al. Identification of four new PITX2 gene mutations in patients with Axenfeld-Rieger syndrome. Molecular Vision. 2006;12:1448–1460. [PubMed] [Google Scholar]
- 9.Reese AB, Ellsworth RM. The anterior chamber cleavage syndrome. Archives of Ophthalmology. 1966;75(3):307–318. doi: 10.1001/archopht.1966.00970050309003. [DOI] [PubMed] [Google Scholar]
- 10.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Human Molecular Genetics. 1997;6(4):641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 11.Vincent A, Billingsley G, Priston M, et al. Phenotypic heterogeneity of CYP1B1: mutations in a patient with Peters’ anomaly. Journal of Medical Genetics. 2001;38(5):324–326. doi: 10.1136/jmg.38.5.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edward DP, Al Rajhi A, Lewis RA, Curry SA, Wang Z, Bejjani BA. Molecular basis of Peters anomaly in Saudi Arabia. Ophthalmic Genetics. 2004;25(4):257–270. doi: 10.1080/13816810490902648. [DOI] [PubMed] [Google Scholar]
- 13.Churchill AJ, Yeung A. A compound heterozygous change found in Peters’ anomaly. Molecular Vision. 2005;11:66–70. [PubMed] [Google Scholar]
- 14.Vincent AL, Billingsley G, Buys Y, et al. Digenic inheritance of early-onset glaucoma: CYP1B1, a potential modifier gene. American Journal of Human Genetics. 2002;70(2):448–460. doi: 10.1086/338709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanwar M, Dada T, Sihota R, Dada R. Identification of four novel cytochrome P4501B1 mutations (p.I94X, p.H279D, p.Q340H, and p.K433K) in primary congenital glaucoma patients. Molecular Vision. 2009;15:2926–2937. [PMC free article] [PubMed] [Google Scholar]
- 16.Sunyaev S, Ramensky V, Koch I, Lathe W, III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Human Molecular Genetics. 2001;10(6):591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 17.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Research. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends in Genetics. 2000;16(5):198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 19.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4(7):1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George Priya Doss C, Rajasekaran R, Sudandiradoss C, Ramanathan K, Purohit R, Sethumadhavan R. A novel computational and structural analysis of nsSNPs in CFTR gene. Genomic Medicine. 2008;2(1-2):23–32. doi: 10.1007/s11568-008-9019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gale CP, Grant PJ. The characterisation and functional analysis of the human glyoxalase-1 gene using methods of bioinformatics. Gene. 2004;340(2):251–260. doi: 10.1016/j.gene.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Vaux C, Sheffield L, Keith CG, Voullaire L. Evidence that Rieger syndrome maps to 4q25 or 4q27. Journal of Medical Genetics. 1992;29(4):256–258. doi: 10.1136/jmg.29.4.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy ABM, Panicker SG, Mandal AK, Hasnain SE, Balasubramanian D. Identification of R368H as a predominant CYP1B1 allele causing primary congenital glaucoma in Indian patients. Investigative Ophthalmology and Visual Science. 2003;44(10):4200–4203. doi: 10.1167/iovs.02-0945. [DOI] [PubMed] [Google Scholar]
- 25.Tanwar M, Dada T, Sihota R, Das TK, Yadav U, Dada R. Mutation spectrum of CYP1B1 in North Indian congenital glaucoma patients. Molecular Vision. 2009;15:1200–1209. [PMC free article] [PubMed] [Google Scholar]
- 26.Stoilov IR, Costa VP, Vasconcellos JPC, et al. Molecular genetics of primary congenital glaucoma in Brazil. Investigative Ophthalmology and Visual Science. 2002;43(6):1820–1827. [PubMed] [Google Scholar]
- 27.Mashima Y, Suzuki Y, Sergeev Y, et al. Novel cytochrome P4501B1 (CYP1B1) gene mutations in Japanese patients with primary congenital glaucoma. Investigative Ophthalmology and Visual Science. 2001;42(10):2211–2216. [PubMed] [Google Scholar]
- 28.Bejjani BA, Lewis RA, Tomey KF, et al. Mutations in CYP1B1, the gene for cytochrome P4501B1, are the predominant cause of primary congenital glaucoma in Saudi Arabia. American Journal of Human Genetics. 1998;62(2):325–333. doi: 10.1086/301725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plášilová M, Stoilov I, Sarfarazi M, Kádasi L, Feráková E, Ferák V. Identification of a single ancestral CYP1B1 mutation in Slovak Gypsies (Roms) affected with primary congenital glaucoma. Journal of Medical Genetics. 1999;36(4):290–294. [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazaki H, Inui Y, Yun C-H, Guengerich FP, Shimada T. Cytochrome P450 2E1 and 2A6 enzymes as major catalysts for metabolic activation of N-nitrosodialkylamines and tobacco-related nitrosamines in human liver microsomes. Carcinogenesis. 1992;13(10):1789–1794. doi: 10.1093/carcin/13.10.1789. [DOI] [PubMed] [Google Scholar]
- 31.Nebert DW. Proposed role of drug-metabolizing enzymes: regulation of steady state levels of the ligands that effect growth, homeostasis, differentiation, and neuroendocrine functions. Molecular Endocrinology. 1991;5(9):1203–1214. doi: 10.1210/mend-5-9-1203. [DOI] [PubMed] [Google Scholar]
- 32.Swindell EC, Eichele G. Retinoid metabolizing enzymes in development. BioFactors. 1999;10(2-3):85–89. doi: 10.1002/biof.5520100201. [DOI] [PubMed] [Google Scholar]
- 33.Chen H, Howald WN, Juchau MR. Biosynthesis of all-trans-retinoic acid from all-trans-retinol: catalysis of all-trans-retinol oxidation by human P-450 cytochromes. Drug Metabolism and Disposition. 2000;28(3):315–322. [PubMed] [Google Scholar]
- 34.Michels-Rautenstrauss KG, Mardin CY, Zenker M, Jordancand N, Gusek-Schneider G-C, Rautenstrauss BW. Primary congenital glaucoma: three case reports on novel mutations and combinations of mutations in the GLC3A (CYP1B1) gene. Journal of Glaucoma. 2001;10(4):354–357. doi: 10.1097/00061198-200108000-00017. [DOI] [PubMed] [Google Scholar]
- 35.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Human Molecular Genetics. 1997;6(4):641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 36.Choudhary D, Jansson I, Sarfarazi M, Schenkman JB. Characterization of the biochemical and structural phenotypes of four CYP1B1 mutations observed in individuals with primary congenital glaucoma. Pharmacogenetics and Genomics. 2008;18(8):665–676. doi: 10.1097/FPC.0b013e3282ff5a36. [DOI] [PubMed] [Google Scholar]
- 37.Yamazaki S, Sato K, Suhara K, Sakaguchi M, Mihara K, Omura T. Importance of the proline-rich region following signal-anchor sequence in the formation of correct conformation of microsomal cytochrome P-450s. Journal of Biochemistry. 1993;114(5):652–657. doi: 10.1093/oxfordjournals.jbchem.a124232. [DOI] [PubMed] [Google Scholar]
- 38.Stoilov I, Akarsu AN, Alozie I, et al. Sequence analysis and homology modeling suggest that primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. American Journal of Human Genetics. 1998;62(3):573–584. doi: 10.1086/301764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chavarria-Soley G, Michels-Rautenstrauss K, Pasutto F, et al. Primary congenital glaucoma and Rieger's anomaly: extended haplotypes reveal founder effects for eight distinct CYP1B1 mutations. Molecular Vision. 2006;12:523–531. [PubMed] [Google Scholar]
- 40.Phillips JC, del Bono EA, Haines JL, et al. A second locus for Rieger syndrome maps to chromosome 13q14. American Journal of Human Genetics. 1996;59(3):613–619. [PMC free article] [PubMed] [Google Scholar]
- 41.Holmström GE, Reardon WP, Baraitser M, Elston JS, Taylor DS. Heterogeneity in dominant anterior segment malformations. British Journal of Ophthalmology. 1991;75(10):591–597. doi: 10.1136/bjo.75.10.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson IM, Fletcher JM, Jordan T, et al. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peter's anomaly. Nature Genetics. 1994;6(2):168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 43.Honkanen RA, Nishimura DY, Swiderski RE, et al. A family with Axenfeld-Rieger syndrome and Peters Anomaly caused by a point mutation (Phe112Ser) in the FOXC1 gene. American Journal of Ophthalmology. 2003;135(3):368–375. doi: 10.1016/s0002-9394(02)02061-5. [DOI] [PubMed] [Google Scholar]
- 44.Nishimura DY, Swiderski RE, Alward WLM, et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nature Genetics. 1998;19(2):140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 45.Komatireddy S, Chakrabarti S, Mandal AK, et al. Mutation spectrum of FOXC1 and clinical genetic heterogeneity of Axenfeld-Rieger anomaly in India. Molecular Vision. 2003;9:43–48. [PubMed] [Google Scholar]
- 46.Azuma N, Yamaguchi Y, Handa H, Hayakawa M, Kanai A, Yamada M. Missense mutation in the alternative splice region of the PAX6 gene in eye anomalies. American Journal of Human Genetics. 1999;65(3):656–663. doi: 10.1086/302529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanjo Y, Kawasaki S, Mari K, Sotozono C, Inatomi T, Kinoshita S. A novel mutation in the alternative splice region of the PAK6 gene in a patient with Peters' anomaly. British Journal of Ophthalmology. 2004;88(5):720–721. doi: 10.1136/bjo.2003.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dansault A, David G, Schwartz C, et al. Three new PAX6 mutations including one causing an unusual ophthalmic phenotype associated with neurodevelopmental abnormalities. Molecular Vision. 2007;13:511–523. [PMC free article] [PubMed] [Google Scholar]
- 49.Vincent A, Billingsley G, Priston M, et al. Further support of the role of CYP1B1 in patients with Peters anomaly. Molecular Vision. 2006;12:506–510. [PubMed] [Google Scholar]