Abstract
The organization of biological materials into versatile three-dimensional assemblies could be used to build multifunctional therapeutic scaffolds for use in nanomedicine. Here we report a strategy to design three-dimensional nanoscale scaffolds that can be self-assembled from RNA with precise control over their shape, size and composition. These cubic nanoscaffolds are only ~13 nm in diameter and are composed of short oligonucleotides making them amenable to chemical synthesis, point modifications and further functionalization. Nanocube assembly is verified by gel assays, dynamic light scattering and cryogenic electron microscopy. Formation of functional RNA nanocubes is also demonstrated by incorporation of a light-up fluorescent RNA aptamer that is optimally active only upon full RNA assembly. Moreover, we show the RNA nano-scaffolds can self-assemble in isothermal conditions (37°C) during in vitro transcription, which opens a route towards the construction of sensors, programmable packaging and cargo delivery systems for biomedical applications.
It is highly desirable to generate a library of nano-scaffolds that allow precise positioning of various therapeutic agents or sensors in 3D space to guarantee their simultaneous delivery to specific areas of the body. In the past 20 years, Seeman and co-workers have largely contributed to the fabrication of DNA-based nano-cages through molecular self-assembly1–3. Diverse 3D DNA nano-scaffolds with the connectivity of a cube4, tetrahedra5,6, bipyramid7, octahedra8,9, dodecahedra6,10, and buckminster-fullerene6 were constructed. The ability of DNA polyhedra to promote targeted delivery by functioning as nano-capsules for molecular cargo has been shown for rigid tetrahedral11 and icosahedral12 DNA cages. Recently, a powerful DNA “origami” technique13 for the design of 2D addressable DNA shapes was applied towards the construction of nano-boxes14, pyramidal tetrahedrons15, and other 3D objects16,17.
To date, most of the nucleic acid based polyhedral nano-scaffolds designed in the laboratory have diameters greater than 15 nm and employ DNA molecules as building blocks3,16,17. While these DNA structures have revealed the potential to develop programmable scaffolds for nanotechnological applications1–3, the DNA biopolymer might not always be able to mimic all the biological functions of RNA. Despite being more chemically labile than DNA, natural RNA molecules are able to function as therapeutic agents such as small interfering and micro RNAs (siRNAs and miRNAs) which do not have DNA analogs18. Furthermore, natural RNA molecules comprise a wide range of working components of biologically essential molecular machines including ribozymes19,20, regulatory aptamers19,21 and nano-motors22.
As a result of greater natural versatility and biologically relevant functionality, RNA might offer building blocks and functional components that have no counterparts in the present day DNA world for building multifunctional therapeutic nano-scaffolds for nano-medicine23,24. Previous works have demonstrated the designs of modular RNA units forming small multimeric particles of various sizes23–30, as well as programmable filaments25,31,32 and 2D nano-arrays and nanogrids consisting of RNA squares26,30. While previous achievements demonstrate that reliable prediction and design of the tertiary structure of RNA can be achieved to build supra-molecular architectures28,30, the structural potential of RNA self-assembly for nano-construction of 3D nano-cages33 and 3D RNA networks has not yet been fully exploited.
We therefore present a strategy to rationally design and construct 3D RNA nano-scaffolds, composed of six (A6–F6) or ten (A10–J10) strands assembled in the shape of a cube. Due to its geometrical simplicity and the relatively large number of participating modules, the cube is an attractive shape for a multifunctional nano-scaffold. The small size of the engineered nano-scaffold (one helical turn per side) allows for the use of relatively short RNA sequences (28–54 nts). This makes the sequences suitable for chemical synthesis, functionalization and/or selective point modifications. Furthermore, the number of possible functions within each scaffold is at least as large as the number of addressable nucleic acid units present in its composition.
Nanocubes rational design
Computational 3D models were generated using the NanoTiler software34 as well as Accelrys Discovery Studio. The computational sequence optimization consists of sequence randomization and Monte Carlo optimization algorithms (Figure S1, Supporting Information (SI)). The objective function of the optimization is the weighted sum of three scores: (i) a rule-based score35,36, (ii) a score comparing the target secondary structure with RNAcofold37–39 predictions of all sequence pairs as well as with RNAfold37,40 predictions of all individual sequences, and (iii) a score evaluating a multi-sequence secondary structure prediction based on a trivial energy model34. Three different cube types were engineered: two cubes with and without dangling ends, each containing six strands of equal length, and a 10-stranded cube with dangling ends containing two different strand lengths (Figure 1). These dangling ends can be modified into functional units as demonstrated below. The sizes of all cubes are 10 bps per side.
Figure 1.
3D models for six and ten stranded cubes with corresponding 2D schematics of sequence interactions. Note that 5' start sequences (in black) are base paired in (a) and single-stranded in (b) and (c). The diagrams are drawn to emphasize the symmetry of 3` and 5` positions. Note that the six RNA strands for the 6-stranded cube and the 10 RNA strands for the 10-stranded cube have different length and sequences (Table S1).
In contrast to the step-wise formation of covalently closed DNA cubes reported by Chen and Seeman4, our approach to synthesizing RNA cubes is a one pot self-assembly process. The self-assembly protocol is optimized to be used for all nano-cubes investigated in this project (Materials and Methods). Several different techniques such as native poly-acrylamide gel electrophoresis (PAGE) experiments, dynamic light scattering (DLS), and cryogenic electron microscopy (cryo-EM) were employed to confirm the formation of closed RNA cubes of defined sizes. Hybrid RNA/DNA and DNA cubes were also analyzed.
Nanocubes self-assembly
Native-PAGE results presented in Figure 2 demonstrate the reproducible self-assembly of six (A6–F6) RNA, RNA/DNA, or DNA strands, into finite hexameric structures. Quantification of the bands (Materials and Methods) reveals that the average yields of the RNA or DNA hexamers are greater than 90%. To confirm the formation of the closed hexameric species, sequence A6 was mutated (A6m) to prevent complementary base pairings with strands E6, D6, and F6 (SI, Table S1). Thus, mixing strands A6m, B6, C6, D6, E6, and F6 only leads to the formation of an “open” hexamer. PAGE experiments in Figure 2a demonstrate different electrophoretic mobilities between “open” (lane #6) and “closed” (lane #7) hexamers. By adjusting the porosity of native gels, optimal separation between tetramers, pentamers and hexamers can be achieved. However, the retention factors (Rf) of tetrameric and pentameric assemblies can vary depending on their strand composition, which might favor either circular or linear molecular species (data not shown).
Figure 2.
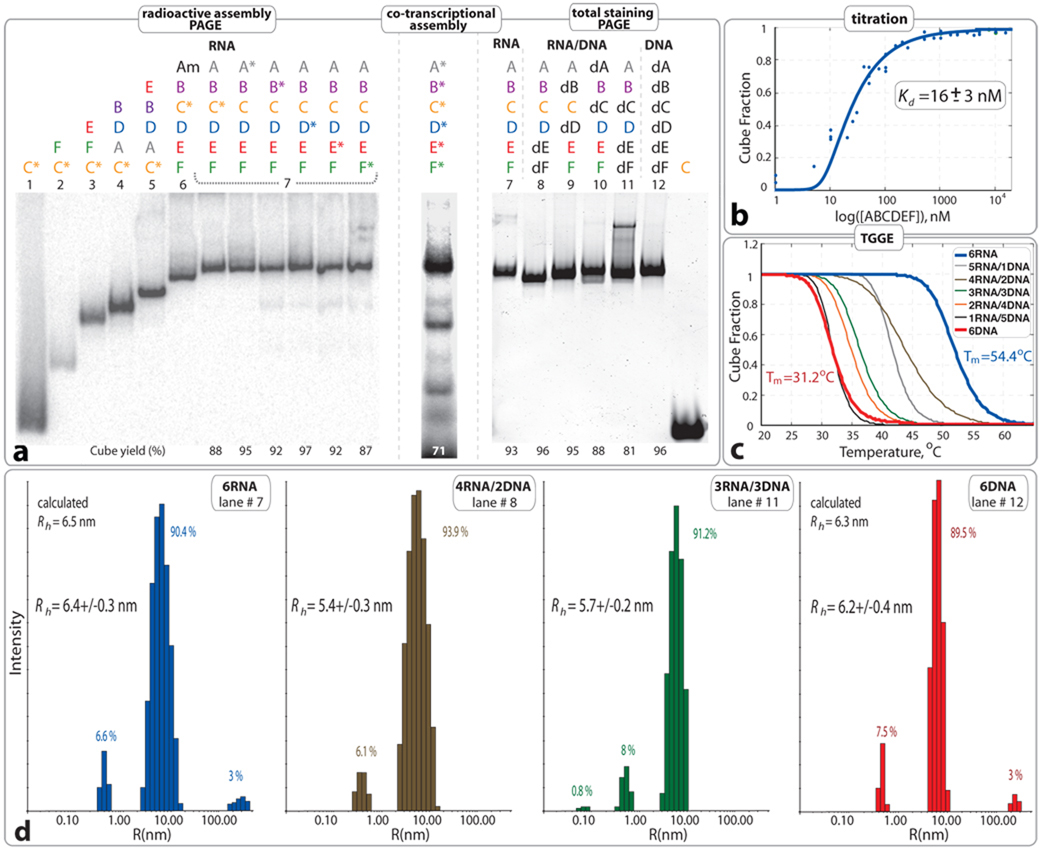
Characterization of 6 stranded cube assemblies (without dangling ends). a. Native PAGE assembly experiments: (left) radioactive assemblies with 32P radiolabeled RNA molecules indicated with asterisks. Am (or A6m) was designed to assemble with B–F (or B6–F6) to form an open hexamer. (middle) Co-transcriptional self-assembly of body-labeled RNA cube strands. (right) Native PAGE assembly experiments with RNA visualization by total SYBR Green II staining. Estimated yields of the hexamers (in %) are shown at the bottom of corresponding lanes. All lanes are numbered to distinguish between twelve different compositions of RNA, RNA/DNA and DNA complexes. b. Titration curve fitting data collected from three independent experiments of RNA cube assembly. c. Thermal melting curves of RNA, DNA, and RNA/DNA hybrid cubes. Corresponding Tm`s are shown Figure S3 in SI. d. Size histograms of six stranded cubes measured by DLS. Compositions are specified for each measurement. Color code is consistent with b and c. Relative assembly yields are calculated from each histogram. All RNA complexes used in a, c, and d experiments were assembled as described in Materials and Methods at 1 µM concentrations.
To verify that all 6 RNA strands participate in self-assembly of the hexamer, radio-assay PAGE experiments were carried out (Figure 2a). In these experiments, each of the six radio-labeled molecules (marked with “*”) was individually mixed with five other non-labeled molecules followed by the assembly protocol. The results show identical gel shifts for all 6 cubes with different labeled strands, suggesting the participation of all strands in the formation of a closed species (Figure 2a, lanes #7). Likewise, the formation of DNA hexamers was confirmed by using three different labeled DNA strands (SI, Figure S2).
The assemblies of the 6- and 10-stranded cubes with 5' dangling ends were also confirmed by PAGE experiments and the yields for both nano-constructs were estimated to be greater than 90% (SI, Figures S4, S5, and S11). For instance, SYBR Green II total RNA staining as well as assembly reactions performed with body-labeled RNA strands demonstrate that all the RNA strands expected to enter into cube composition are localized in a unique band product on native PAGE gels. In the case of the 6-stranded cube, the sequence composition or the presence of the dangling ends can alter the relative gel shifts for the hexamers.
All the assembly protocols used in this project involve stepwise incubation at several temperatures, while the production and folding of naturally transcribed RNAs is an isothermal process. To demonstrate the potential feasibility of cube assembly in conditions mimicking an intracellular environment, we monitored its formation throughout an in vitro transcription reaction (Material and Methods). Equimolar concentrations of DNA templates for all RNA strands were simultaneously added to the α[P32]-ATP body-labeling transcription mixture and the final products were characterized with PAGE. Co-transcriptional assembly results reveal the ability of all three nano-scaffolds to self-assemble isothermally (37°C) during in vitro transcription with yields greater than 70 % (Figure 2a; SI, Figures S4 and S5).
Comparison of RNA, DNA, and RNA/DNA nanocubes
Because the concentration of the RNA strands is a key factor in self-assembly processes, we measured the apparent dissociation equilibrium constants (Kd) for the RNA and DNA hexamers (see Materials and Methods). For the RNA hexamer Kd was found to be ~16 nM (Figure 2b), while the DNA hexamer Kd was measured to be at least 10 times higher (~170 nM, data not shown).
Nanostructures of hybrid (RNA/DNA) composition are of great interest due to their ability to maintain the diverse functionality of RNA, while incorporating the chemical stability of DNA. To test for hybrid cube viability, some of the RNA/DNA hybrids of the 6-stranded cube without dangling ends were characterized by total staining PAGE assembly experiments. The results (Figure 2) demonstrated slight differences in the gel shifts for the major bands which can be attributed to the differences in shape and hydrodynamic radii of the cubes based on the number of A-form (RNA/RNA, RNA/DNA) and B-form (DNA/DNA) helices (see also Table S2).
Melting temperatures (Tm’s) were determined by TGGE experiments30 for the six stranded (Figure 2c; SI, Figures S3 and S4) and ten stranded (SI, Figure S5) cubes by measuring the decrease in the yield of cubes versus temperature. Relative thermal stabilities of assembled RNA, DNA and RNA/DNA hybrid cubes were also compared by temperature gradient gel electrophoresis (TGGE) and are summarized in Table S2 (SI). The RNA cubes have Tm's about 15–20 °C higher than those of DNA cubes which can be explained by the higher thermal stability of A-form RNA duplexes versus B-form DNA duplexes41–43. However, the Tm values of RNA/DNA hybrids nanocubes can significantly vary in function of the number of strands of RNA and DNA entering into their composition as well as the location and orientation of the RNA strands with respect to DNA strands (Table S2). These results suggest that the thermal stability of nano-scaffolds can be potentially tuned by altering the ratio of RNA to DNA strands and their location within the assembly.
Structural characterization by DLS and cryo-EM
Using dynamic light scattering (DLS), the hydrodynamic radii (Rh) for pre-assembled hexameric RNA and DNA particles were determined to be 6.4 and 6.2 nm, respectively (Figure 2). These values are in good agreement with the predicted radii of circumscribed spheres around the RNA and DNA cube models of 6.5 and 6.3 nm, respectively (Materials and Methods, Eq. 2). The hydrodynamic radii of the 10-stranded and 6-stranded cubes with dangling ends are 6.9 and 7.1 nm, respectively (SI, Figures S3 and S4). The larger radii can be attributed to the presence of 6nt dangling ends and are consistent with estimated radii of 6.8 nm each and with PAGE results (SI, Figure S6).
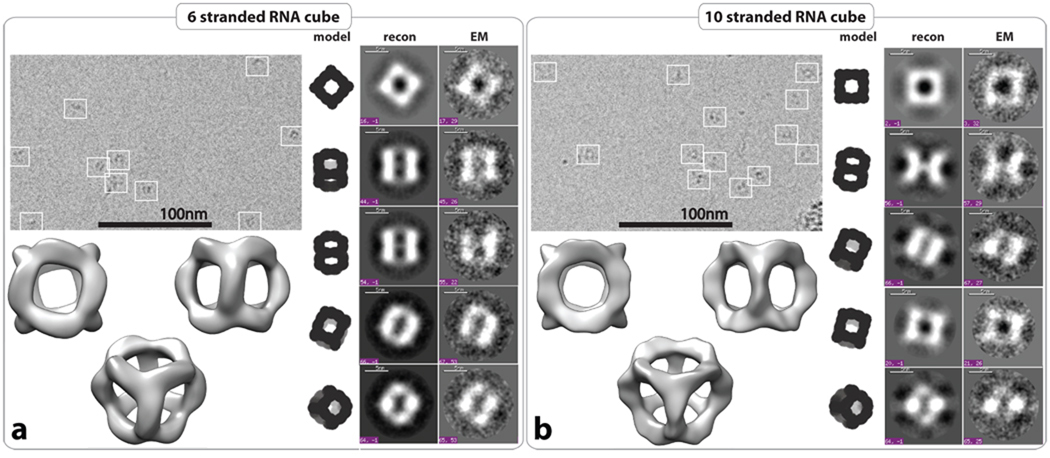
Overall, PAGE results and DLS data strongly suggest the formation of closed, compact molecular complexes composed either of six or ten strands. However, one of the most convincing evidences for the formation of RNA cubes comes from cryogenic electron microscopy (cryo-EM) imaging and further single particle reconstruction6,9,44. The cryo-EM images show that most RNA particles have the expected size (Figure 3) and are equally distributed in the entire imaging field (SI, Figure S7). Using the EMAN reconstruction packages45, the 3D structures of the 6-stranded and 10-stranded cubes were obtained at resolutions of 8.9 Å (from 2,038 particles) and 11.7 Å (from 1,677 particles), respectively (SI, Figure S7, and Materials and Methods). The computed projections from these 3D reconstructions match well with the class averages of observed particles with similar views (Figure 3). RNA cube reconstructions have structural features in good agreement with the predicted 3D cubic model displayed in Figure 1. Structural variations observed between the 6- and 10-stranded cubes can be explained by the different structures of their 3-way junction corners (Figure 1).
Figure 3.
Structural characterization of RNA cubes by cryo-EM with single particle image reconstruction. Panels a and b correspond to the characterization of 6 and 10 stranded RNA cubes respectively. Each panel on the top left represents typical cryo-EM images of the RNA particles. On the right side, class averages for each RNA cube as observed by cryo-EM (EM) with corresponding projections of the reconstructed 3D structure and theoretical RNA cube model. Reconstructed 3D models of the six and 10-stranded RNA cubes have been obtained at 8.9 Å and 11.7 Å resolution, respectively. All RNA complexes used in cryo-EM experiments were assembled at 1 µM of each RNA strand as described in the Materials and Methods.
RNA nanocubes functionalization
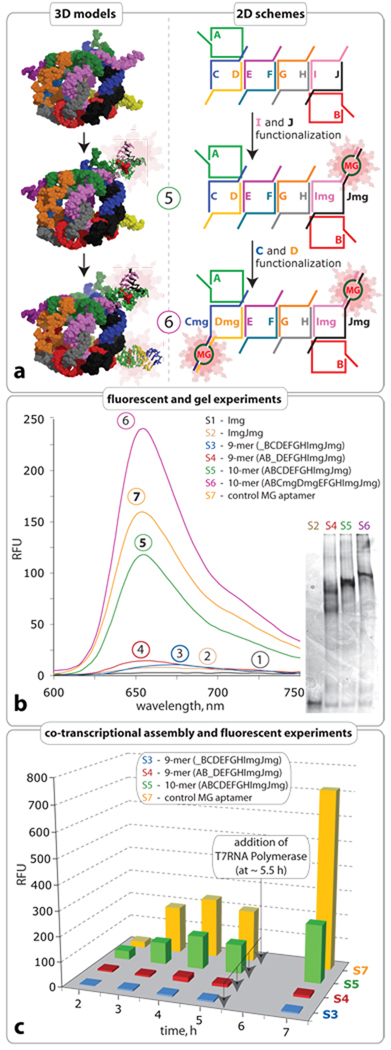
To demonstrate the concept of functional activation through nano-scaffold assembly, Malachite Green (MG) aptamers (PDB ID: 1f1t)46 were integrated into the dangling ends of the 10-stranded cube sequences. The triphenylmethane dye, MG was chosen as a signaling agent due to its fluorescent properties47. In an unbound state, the MG molecule exhibits no fluorescence, however, upon binding to an RNA aptamer a large increase in fluorescence is observed48. The MG aptamer was separated into two strands, each of which was incorporated into the flanking sequence of two different strands of the cube (Figure 4). The MG aptamer sequences were designed to have low mutual affinity, such that dimers would not form an active aptamer (Figure S8). As seen in Figure 4, emission remains relatively low for the monomer, dimer and all eight possible nonamers (Figure 4, spectra 1–3; for all nonamers see Figure S9 in SI). Analysis of the decamer (cube) spectrum (Figure 4, spectrum 5) indicates a sharp increase in fluorescence due to full cube formation. Therefore, only when the cube is formed are the aptameric flanking sequences brought into close enough proximity to form the active MG binding aptamer. To reveal the potential multifunctionality of the nano-scaffold, a second aptamer was introduced into the same cube, resulting in a two-fold increase in the MG emission signal (spectrum 6). The successful formation of functionalized nano-cubes was confirmed by native PAGE experiments (Figure 4). A single RNA oligonucleotide containing the MG aptamer sequence within a helical region was used as a positive control (Figure 4, sample S7, supporting Table S1). Based on the emission signal of the control molecule, the yield of the functionally active cube S5 was estimated to be 77.3 % at RNA concentration of 1µM.
Figure 4.
Functionalization of RNA nano-cube scaffold with Malachite Green (MG) aptamer. a. Scheme showing the functionalization. b. Emission spectra representing binding of MG to RNA aptamer and native PAGE demonstrating the formation of the constructs. Monomer, dimer and nonamer samples (S1, S2, S3, S4) are unable to bring the aptamers into close enough proximity necessary for fluorescent emission in presence of MG. The functionalized cube sample (S5) shows an increase in fluorescence demonstrating correct formation of the MG binding pocket. The cube sample (S6) shows two-fold increase in fluorescence demonstrating simultaneous correct formation of its two MG binding pockets. All RNA complexes used in the fluorescent experiments were assembled at RNA strand concentration of 1 µM as described in Materials and Methods. Based on the emission signal of the control molecule, the yield of the functionally active cube (S5) was estimated to be 77.3 %. c. Comparison of co-transcriptional self-assembly of nonamers S3 and S4 with 10-stranded RNA nano-cube (S5) functionalized with one MG aptamer at 37°C. Aliquots of the transcription mixture were taken after 2, 3, 4, 5, and 7 hours, followed by the addition of DNAse to stop the reaction. MG was added just prior to fluorescent data acquisition. Note that after 5h, more T7 RNA polymerase was added to each transcription mix. Control S7 corresponds to a MG aptamer molecule.
The assembly of the functionalized 10-stranded cube with MG aptamer was also monitored during in vitro transcription by following an increase in MG emission signal (Figure 4c). Equimolar concentrations of 10 DNA templates for A10–J10mg RNA strands (Figure 4c, Sample S2) were simultaneously added to the transcription mixture as described above and aliquots of the transcription mixture were taken after 2, 3, 4, 5, and 7 hours for fluorescence measurements in presence of MG. Amplification of the emission signal over the course of the reaction confirms the proper folding of the MG aptamer into its active conformation upon cube assembly during transcription at 37°C (Figure 4c, sample S5; and supporting Figure S10a). After 5 hours, a slight decrease in emission signal occurs due to the partial or entire inactivation of the T7RNA polymerase and simultaneous RNA degradation. Additional T7RNA polymerase was therefore added to the transcription mixture at ~5.5 hours. By contrast to the transcription mixture containing the ten DNA templates coding for the 10-stranded cube, removal of one of the templates essentially resulted in partial RNA assemblies unable to trigger the fluorescent signal emission in the presence of MG (Figure 4c, samples S3, S4 and SI, Figure S10b–c). These results are consistent with the idea that the MG aptamer is optimally active only once the full RNA assembly is achieved. The 10-stranded cube has a lower emission signal than the control MG aptamer molecule (Figure 4c, sample S7). This is expected as less MG aptamers are obtained in the 10-stranded RNA mix than in the MG aptamer control mix for an even amount of RNA produced during transcription.
As suggested previously, co-transcriptional assembly of functionalized nano-scaffolds confirm their ability to self-assemble isothermally (37°C) during in vitro transcription, in conditions mimicking intracellular context.
Conclusion
In summary, we have demonstrated a strategy to design and engineer programmable, 3D RNA self-assembling nano-scaffolds with radii not exceeding 6.5 nm. By contrast to other strategies taking advantage of pre-folded RNAs26,30,33, nanocube strand components are short enough to be amenable to chemical synthesis. This allows (i) introduction of chemically stable RNA nucleotide analogs at specific sequence position to enhance their chemical stability, (ii) chemical functionalization important for therapeutic delivery and (iii) high yield of synthesis. Thermal stabilities of these nano-scaffolds can be tuned by altering their strand compositions. Functionalization can be introduced through modification of the core strands and triggered by the full assembly of the nano-scaffold, thus providing vast potential for biomedical applications. In addition, the ability of these cubic RNA scaffolds to self-assemble isothermally at 37°C during in vitro transcription opens a completely new route towards the in vivo construction of detection sensors, programmable packaging and cargo delivery systems.
Materials and Methods
RNA preparation
RNA molecules were prepared by transcription of PCR amplified DNA templates. Synthetic DNA molecules coding for the antisense sequence of the designed RNA were purchased from IDT DNA and amplified by PCR using primers containing the T7 RNA polymerase promoter. PCR products were purified using the QiaQuick PCR purification kit and RNA molecules were prepared by in vitro transcription using home-made T7 RNA polymerase and purified on denaturing urea gel (PAGE) (10% acrylamide, 8M urea). The RNA was eluted from gel slices overnight at 4°C into buffer containing 300 mM NaCl, 10 mM Tris pH 7.5, 0.5 mM EDTA then ethanol precipitated, and rinsed twice with 90% ethanol, vacuum dried and dissolved in TE buffer.
pCp labeling of RNA molecules
T4 RNA ligase was used to label the 3`-ends of RNA molecules by attaching [32P]Cp. Labeled material was purified on denaturing polyacylamide gels (10% acrylamide, 8M urea).
ATP labeling of DNA
T4 polynucleotide kinase was used to label the 5`-ends of DNA molecules by moving [32P] from the gamma position of ATP. Labeled material was purified on denaturing polyacrylamide gels (10% acrylamide, 8M urea).
RNA and DNA cubes self-assembly
All assembly experiments reported in this study were analyzed on 7% (37.5:1) non-denaturing polyacrylamide native gels containing 2 mM Mg(OAc)2 and 50 mM KCl and run at 4°C with running buffer (89 mM Tris-borate, pH 8.3/ 15 or 2 mM Mg(OAc)2). Prior to the addition of the buffer and Mg(OAc)2, the RNA(DNA) samples containing cognate RNA(DNA) molecules at concentrations 1 µM were heated to 95°C for two minutes and immediately snap cooled at 45°C followed by assembly buffer addition (tris-borate buffer (89 mM, pH 8.3), 2 mM Mg(OAc)2, and 50 mM KCl) and incubation for 30 minutes at 45°C.
Non-denaturing PAGE, TGGE experiments, and Kd measurements
All constructs were assembled as described above and an equal volume of loading buffer (same buffer with 0.01% bromphenol blue, 0.01% xylene cyanol, 50% glycerol) was added to each sample before loading on native gel. Gels were run for 4 hours, at 25 W with temperature set to be below 10°C, dried under vacuum, exposed to a phosphoimager screen for 16 hours, and scanned using a Typhoon phosphoimager. For total gel staining, SYBR® Green II RNA gel stain was used to visualize RNA or DNA bands using Typhoon phosphoimager with the emission of SYBR Green II centered at 520 nm. The stained RNA or DNA bands appear as black bands on the white background.
For TGGE, analysis performed at 2 mM Mg(OAc)2, a linear temperature gradient, typically from 30 to 60 °C, was applied perpendicular to the electric field. Cube concentration was typically 1 µM. Gels were run for 1 hour, at 30 W.
Dissociation constants (Kd`s) were calculated by plotting the fraction of a cube (f) versus the total concentration (CT) of the RNA strands corresponding to this particular fraction. The combined data collected from three independent measurements was subjected to nonlinear curve fitting with the equation:
| (Eq. 1) |
which was solved for f and fit non-linearly to obtain Kd49.
All gels were quantified using ImageQuant software. Equally sized boxes were drawn around the bands corresponding to the hexamers (cubes). The yield for each hexamer-forming complex was calculated by dividing the corresponding quantified value for hexamer by the total sum of the values for all complexes present in the corresponding lane.
Assembly of RNA cubes during transcription
Equimolar concentrations of PCR amplified DNA templates (containing the T7 RNA polymerase promoter) for all six (A6–F6) or ten (A10–J10) RNA strands were simultaneously added all together to the transcription mixture (diH2O, 50 mM Tris pH 7.5, 10 mM MgCl2, 2 mM spermidine, 2.5 mM NTPs, 10 mM DTT) containing α[P32]-ATP (10 mCi/ml) for body-labeling. Transcription was initiated with the addition of home-made T7 RNA polymerase and stopped after 4 hours with RQ1 RNase-free DNase followed by the direct characterization of the transcription mixture with native PAGE as described above. To compensate the difference in transcription time for varying lengths of 10-stranded cube sequences (A10–J10) the initial concentrations of longer DNA templates for A10 and B10 were doubled.
Dynamic Light Scattering
For DLS, 10 µL of sample solution containing preassembled RNA/DNA cubes were measured by DynaPro99 (Protein Solution/Wyatt) with the laser wavelength equal to 824 nm at 24°C. The theoretical hydrodynamic radii (Rh) were calculated using the equation:
| (Eq. 2) |
where L is the length (nm) of the cube side. Assuming a 0.23 nm rise per basepair and a 2.6 nm diameter of the RNA duplex, the side length of the RNA cube is calculated to be 7.5 nm. The DNA cube side length is calculated to be 7.3 nm using 0.33 and 2.0 nm for the rise per basepair and duplex diameter, respectively. For the cubes with dangling ends, the theoretical Rh`s were calculated by measuring the distance between the center of mass and the furthest atom of the cube 3D CPK model.
Cryo-EM imaging
Samples containing the six-stranded RNA cube with dangling ends (6-cube) and the 10-stranded RNA cube (10-cube) for cryo-EM were prepared as described above. Micrographs were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 kV, a nominal magnification of 80,000X, and a dose of ~30 e-/Å2. 413 images for 6-cube and 335 images for 10-cube and were automatically collected by the Leginon system50 and recorded with a Tietz F415 4k × 4k pixel CCD camera. Experimental data were processed using the Appion software package51. 2,038 particles for 6-cube and 1,677 particles for 10-cube were manually selected. The 3D reconstruction was carried out using the EMAN reconstruction package45. A resolution of 9.6 Å for 6-cube and 11.5 Å for 10-cube was determined by Fourier Shell Correlation (FSC) at a cutoff of 0.5.
Fluorescent Experiments
The fluorescent experiments were carried out using a NanoDrop 3300 Fluorospectrometer with the following settings: excitation wavelength was set at blue in all experiments. Emission was scanned from 540 to 800 nm. Signal was registered in Relative Fluorescent Units (RFU) at 660 nm. All RNA complexes used in the fluorescent experiments were assembled as described above at 1 µM concentrations. The experiments with MG binding to the decamer (10-stranded cube) resulting in enhanced MG emission were repeated and reproduced (within 10% error) at least five times. The experiments with the nine nonamers and the 10-stranded cube with two embedded aptamers were repeated and reproduced twice (within 10% error).
Supplementary Material
Acknowledgments
Authors thank V. A. Piunova for helping with DLS, K. Kahn for helping with the Kd curve analysis, C. Potter and B. Carragher for their invaluable scientific input regarding Cryo-EM and single particle reconstruction. Cryo-EM imaging was performed at NRAMM, which is supported by NIH though the National Center for Research Resources' P41 program (RR17573). This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (to B.A.S.) and by NIH RO1GM079604 (to L.J.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. K.A. and L.J. wish to dedicate this work to Sts. Cyril and Methodius.
Footnotes
Supporting Information
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
Author contributions
K.A.A. and L.J. conceived and designed the experiments: E.B., B.A.S., K.A.A., and L.J. contributed to the sequence design: K.A.A. and A.J.Y performed all the experiments: K.A.A. and L.J. analyzed the data: N.V and E.J. contributed to cryo-EM analysis tools: K.A.A. and L.J. co-wrote the paper.
References
- 1.Seeman NC. An overview of structural DNA nanotechnology. Mol Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 3.Lin C, Liu Y, Yan H. Designer DNA Nanoarchitectures. Biochemistry. 2009;48:1663–1674. doi: 10.1021/bi802324w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen JH, Seeman NC. The electrophoretic properties of a DNA cube and its substructure catenanes. Electrophoresis. 1991;12:607–611. doi: 10.1002/elps.1150120902. [DOI] [PubMed] [Google Scholar]
- 5.Goodman RP, et al. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 6.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 7.Erben CM, Goodman RP, Turberfield AJ. A self-assembled DNA bipyramid. J Am Chem Soc. 2007;129:6992–6993. doi: 10.1021/ja071493b. [DOI] [PubMed] [Google Scholar]
- 8.Andersen FF, et al. Assembly and structural analysis of a covalently closed nano-scale DNA cage. Nucleic Acids Res. 2008;36:1113–1119. doi: 10.1093/nar/gkm1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann J, Cebulla MP, Monninghoff S, von Kiedrowski G. Self-assembly of a DNA dodecahedron from 20 trisoligonucleotides with C(3h) linkers. Angew Chem Int Ed Engl. 2008;47:3626–3630. doi: 10.1002/anie.200702682. [DOI] [PubMed] [Google Scholar]
- 11.Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew Chem Int Ed Engl. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 12.Bhatia D, et al. Icosahedral DNA Nanocapsules by Modular Assembly. Angew Chem Int Ed Engl. 2009;48:4134–4137. doi: 10.1002/anie.200806000. [DOI] [PubMed] [Google Scholar]
- 13.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 14.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 15.Ke Y, et al. Scaffolded DNA Origami of a DNA Tetrahedron Molecular Container. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 16.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 19.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 20.Joyce GF. Directed evolution of nucleic acid enzymes. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 21.Davidson EA, Ellington AD. Engineering regulatory RNAs. Trends Biotechnol. 2005;23:109–112. doi: 10.1016/j.tibtech.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Wendell D, et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo P. RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy. J Nanosci Nanotechnol. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaeger L, Leontis NB. Tecto-RNA: One-Dimensional Self-Assembly through Tertiary Interactions. Angew Chem Int Ed Engl. 2000;39:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 26.Chworos A, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 27.Shu D, Moll W-D, Deng Z, Mao M, Guo P. Bottom-up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr Opin Struct Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J Am Chem Soc. 2008;130:93–102. doi: 10.1021/ja071516m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyfman AY, et al. Controlled spacing of cationic gold nanoparticles by nanocrown RNA. J Am Chem Soc. 2005;127:11886–11887. doi: 10.1021/ja051144m. [DOI] [PubMed] [Google Scholar]
- 32.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res. 2006;34:1381–1392. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Severcan I, et al. A polyhedron made of tRNAs. Nat Chemistry. 2010 doi: 10.1038/nchem.733. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bindewald E, Grunewald C, Boyle B, O'onnor M, Shapiro BA. Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler. J Mol Graph Model. 2008;27:299–308. doi: 10.1016/j.jmgm.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seiffert J, Huhle A. A full-automatic sequence design algorithm for branched DNA structures. J Biomol Struct Dyn. 2008;25:453–466. doi: 10.1080/07391102.2008.10507193. [DOI] [PubMed] [Google Scholar]
- 36.Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 37.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 38.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernhart SH, et al. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol Biol. 2006;1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofacker IL. Fast Folding and Comparison of RNA Secondary Structures. Monatshefte f Chemie. 1994;125:167–188. [Google Scholar]
- 41.Freier SM, et al. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugimoto N, Nakano S, Yoneyama M, Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. High-resolution structuralanalysis of a DNA nanostructure by cryoEM. Nano Lett. 2009;9:2747–2750. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]
- 45.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 46.Baugh C, Grate D, Wilson C. 2.8 A crystal structure of the malachite green aptamer. J Mol Biol. 2000;301:117–128. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 47.Duxbury DF. The photochemistry and photophysics of triphenylmethane dyes in solid and liquid media. Chem. Rev. 1993;93:381–433. [Google Scholar]
- 48.Afonin KA, Danilov EO, Novikova IV, Leontis NB. TokenRNA: a new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. Chembiochem. 2008;9:1902–1905. doi: 10.1002/cbic.200800183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 50.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.