Abstract
Background
Peroxiredoxins are redox-sensing enzymes with multiple cellular functions. Previously, we reported on the potent antioxidant function of Drosophila peroxiredoxin 5 (dPrx5). Studies with mammalian and human cells suggest that peroxiredoxins can modulate immune-related signaling.
Methods
Survivorship studies and bacteriological analysis were used to determine resistance of flies to fungal and bacterial infections. RT-PCR and immunoblot analyses determined expression of dPrx5 and immunity factors in response to bacterial challenge. Double mutants for dprx5 gene and genes comprising the Imd/Relish and dTak1/Basket branches of the immune signaling pathways were used in epistatic analysis.
Results
The dprx5 mutant flies were more resistant to bacterial infection than controls, while flies overexpressing dPrx5 were more susceptible. The enhanced resistance to bacteria was accompanied by rapid induction of the Imd-dependent antimicrobial peptides, phosphorylation of the JNK kinase Basket and altered transcriptional profiling of the transient response genes, puckered, ets21C and relish, while the opposite effects were observed in flies over-expressing dPrx5. Epistatic analysis of double mutants, using attacin D and Puckered as read outs of activation of the Imd and JNK pathways, implicated dPrx5 function in the control of the dTak1-JNK arm of immune signaling.
Conclusions
Differential effects on fly survivorship suggested a trade-off between the antioxidant and immune functions of dPrx5. Molecular and epistatic analyses identified dPrx5 as a negative regulator in the dTak1-JNK arm of immune signaling.
General significance
Our findings suggest that peroxiredoxins play an important modulatory role in the Drosophila immune response.
Keywords: Peroxiredoxin, Drosophila, infection, immune, signaling
1. Introduction
Peroxiredoxins (Prx) comprise a family of antioxidant enzymes that also play an important role in signal transduction. Studies with mammalian and human cells suggest that peroxiredoxins can modulate immune-related signaling. Thus it was determined that both Prx 1 and Prx 2 can inhibit NF-κB activation, while Prx1 can also inhibit c-Jun N-terminal kinase (JNK) signaling, and thereby mediate cellular responses to pro-inflammatory molecules [1–4]. In contrast, Prx 4 can activate both NF-κB and c-Jun N-terminal kinase and induce inflammation [5]. Prx 1 also interacts with the macrophage migration inhibitory factor, an important cytokine in the regulation of host immune and inflammatory responses [6]. The human Prx 2 regulates TNF signaling pathways via mitogen-activated protein kinases subsequent to TNF-alpha stimulation [7] and its orthologue from Schizosaccharomyces pombe is required for the peroxide-induced activation of the p38/JNK homolog, Sty1, which mediates stress-response signaling [8]. Thus, there are several lines of evidence suggesting that peroxiredoxins are involved in regulating pro-inflammatory stress responses and innate immunity.
Innate immunity is an ancient mechanism of defense against invading microorganisms and is evolutionary conserved from invertebrates to humans. Drosophila, like other insects, lacks an adaptive immune system and depends solely on the innate immune system to combat pathogens. In response to infection, Drosophila initiates a massive production of numerous antimicrobial peptides (AMPs), comprising the humoral immune reaction. The synthesis of AMPs is induced by the NF-κB-like transcription factors, Dorsal, Dif and Relish, which are activated via two distinct NF-κB signaling pathways, Toll and Imd [9, 10]. Regulation of the immunity effector genes is also influenced by the JNK and JAK/STAT signaling pathways, which can act in a competing or cooperative mode [11–15]. To prevent detrimental effects of prolonged and excessive immune response, flies have developed mechanisms to repress immune signaling. Recently, several factors involved in the negative control of the immune system have been identified, including Caspar, which suppresses nuclear translocation of Relish [16], and POSH, which is involved in the ubiquitination and proteosomal degradation of dTak1 [17]. Immune response signaling can also be turned-off via the JNK and JAK/STAT pathways, which appear to inhibit activation of the Relish-dependent genes [12, 18].
Here, we report that the negative regulation of the Drosophila immune response can be mediated by a redox-sensing enzyme, peroxiredoxin 5 (dPrx5). Prx 5 subtype differs from other 2-Cys peroxiredoxins by the mode of resolution of reactive intermediates and, unlike other `traditional' 2-cys peroxiredoxins, forms intramolecular S-S bonds. Previously, we reported that dPrx5 plays an important role in protecting flies from oxidative stress and apoptosis and promotes longevity in Drosophila [19]. The dprx5 null mutant exhibited a dramatically decreased survivorship relative to controls upon exposure to H2O2 or paraquat and a significantly reduced mean life span under normal conditions [19]. In the present study, we demonstrate that the dprx5 mutant has greater resistance to bacterial infection and provide evidence that dPrx5 acts as a negative regulator of the immune response by modulating JNK-mediated signaling.
2. Materials and Methods
2.1. Fly strains and procedures
The y w strain has been maintained in this laboratory for >15 years. The Actin 5C-GAL4, Da-GAL4 and Tub-GAL4 driver lines were supplied by Dr. Blanka Rogina (University of Connecticut Health Science Center). P{Switch1}106 driver (stock # 8151) was obtained from the Bloomington Stock Center. A strain containing a P-element insertion in the coding region of ird5EY02434 (stock # 19825), relE20 mutant (stock # 9457) and the P{UAS-bsk.DN}2 transgenic line (stock # 6409) were obtained from the Bloomington Stock Center. We received the mutant strain key1 (deficient for Kenny) from Dr. Neal Silverman, University of Massachusetts Medical School, and the mutant strain dtak1 from Dr. Dirk Bohmann, University of Rochester Medical Center. A deficiency strain Df(1)dreddD3 lacking dredd caspase was obtained from Dr. John Abrams, UT Southwestern Medical Center. The key1, dtak1, relE20 and Df(1)dreddD3 alleles are described elsewhere [20–23]. The dprx5 mutant and the UAS-dPrx5 transgenic lines are described in [19]. To exclude background effects on survivorship and AMP expression levels, P-element mutants, UAS transgenes or GAL4 driver lines were backcrossed with y w for 6–8 generations to obtain genetically homogeneous stocks. Df(1)dreddD3, key1, rel20 and dtak1 alleles were crossed to y w or dprx5 flies using a combination of appropriate balancers. In these cases, the experimental double mutants with dprx5 background were derived in the same way as corresponding controls. For experiments flies were collected within 1–2 days after hatching and reared on a standard sucrose-cornmeal medium at 25°C for 8–10 days. The induction of P Switch system was elicited by feeding flies regular food containing 0.1 mg/ml mifepristone for at least 48 hours.
2.2. Generation of the UAS-dPrx5RN transgenic strains
To generate a construct producing redox-negative (RN) peroxiredoxin 5, a conserved Cys79, which is essential for peroxidase activity of the peroxiredoxin 5 subtype, was replaced with a serine residue. A mutagenesis was performed utilizing Splicing by Overlapping Extension PCR (SOE-PCR) using primers containing a single nucleotide mismatch, where a tgc triplet encoding for a cysteine was converted to a tcc triplet coding for a serine. Then the fragment of the UAS-dPrx5 construct, obtained by digestion with Nar1 endonuclease, was substituted with the mutated fragment. Transgenics were obtained by injection into germ cells utilizing the services of the TheBestGene company (www.thebestgene.com).
2.3. Infection experiments
Septic infection was caused by pricking adult flies with a needle dipped in a 24 h culture of bacteria. Natural infection was elicited by feeding flies with a bacterial suspension in a Luria broth containing 1 % sucrose. Natural infection with B. bassiana was obtained by shaking flies for 1–2 min with a sporulating fungal culture. The B. subtilis strain BGSC1A1, the strains of M. luteus, S. marcescens (ATCC#8100) and P. aeruginosa were a kind gift of Dr. Christine Buchanan, SMU. Beauveria bassiana and Pectobacterium carotovorum were obtained from the ATCC (#9453 and #17799) and the E. coli strain used was an ampicillin-resistant transformant of the laboratory DH5-a strain.
2.4. Immunoblotting
Protein concentrations were determined by the Bio-Rad Protein Assay (Bio-Rad). 5–10 μg of protein extracts were resolved by 10% SDS-PAGE followed by transfer to PVDF membrane (Millipore) and immunoblots were performed as described previously [24]. The dPrx5 protein was detected with antibodies raised against a recombinant dPrx5 protein. To control for loading, anti-actin antibodies (MP Biomedicals) were used. Phosphorylated JNK (Basket) was analyzed with anti-phospho-JNK antibodies (Cell Signaling Technology); anti-JNK antibodies were obtained from Santa Cruz Biotechnology.
2.5. RT-PCR
Forward and reverse primers specific for different genes are indicated in the Table I. Real-time quantitative PCR was performed with cDNA aliquots obtained as described previously [25] and pairs of gene-specific fluorescent primers. Primers for the housekeeping gene (Actin) and AMP genes were labeled differentially (ea JOE-labeled versus FAM-labeled). Consequently, fluorescent signals obtained during co-amplification of a target gene and a house-keeping gene were detected using different channels. Alternatively, PCR reactions were performed with SYBR Green (Molecular Probes) added to PCR reaction mixtures, and signals obtained for a target gene were standardized against signals obtained for a house-keeping gene (rp49 or actin) in parallel sets of reactions. Fluorescent primers for real-time PCR were designed using D-LUX™ Designer (Invitrogen) and quantitative real-time PCR analysis was performed using Rotor-Gene™ RG-3000 (Corbett Research) and software.
Table I.
Primers used for the real time RT-PCR analysis of gene expression.
| Gene | Primer name and structure. | |
|---|---|---|
| Basket | Forward | 5' CCGCTTACGATACTATCACC |
| Reverse | 5' GACCAGGTAGACATCCTGA | |
| Immune response deficient 5 | Forward | 5' GGCTAGACATCTGCCCAAGTC |
| Reverse | 5' CCATCGCTGCGCTTAGCTC | |
| Puckered | Forward | 5' GCAGAATTTGCGCAAGAGCGG |
| Reverse | 5' GAGCAGTTACTACCCGCCAG | |
| Relish | Forward | 5' ATCCGGAGAAGGGAGAACTGG |
| Reverse | 5' CAGTAGGGCGTAAGCCTGTG | |
| Ets21C | Forward | 5' ATCCAGCTGTGGCAGTTTCT |
| Reverse | 5' TGCACCTTGGTCATGATGTT | |
| Attacin A+B | Forward | 5' ACCAGAGATTAGCACGTCCATCC |
| Reverse | 5' FAM-CGTTGACTGGATTACTCCCACATC | |
| Attacin C | Forward | 5' GTCAGTTCCAGGCCGTGTCC |
| Reverse | 5' FAM-CGCTCCACTCCCGTAACCAG | |
| Attacin D | Forward | 5' CGTTGAGGTTGAGATTGCCACT |
| Reverse | 5' FAM-CGGTCCCTCAGTTCGGCATGAC | |
| Cecropin C | Forward | 5' CCAATGCGCTCGATTCTCTTG |
| Reverse | 5' FAM-CGATTCCATCAGCATTGGACAAT | |
| Diptericin | Forward | 5' CCACCGCAGTACCCACTCAAT |
| Reverse | 5' FAM-CGATGACTGCAAAGCCAAAACCA | |
| Drosocin | Forward | 5' AGTGAGGCGCGAGGCACTG |
| Reverse | 5' CATCAGATGGGTGGTGGCCTGA | |
| Defensin | Forward | 5' CGCATAGAAGCGAGCCACATG |
| Reverse | 5' GCAGTAGCCGCCTTTGAACC | |
| Drosomycin | Forward | 5' GCTGTCCTGATGCTGGTGGT |
| Reverse | 5' FAM-CGGAAAGGGACCCTTGTATCTTC | |
| Metchnikowin | Forward | 5' CAGTGCTGGCAGAGCCTCAT |
| Reverse | 5' FAM-CGTCGGTTAGGATTGAAGGGCGA | |
2.6. Statistical analysis
All analyses were performed using Prism 4.0a for Macintosh or Microsoft EXCEL software.
3. Results
3.1. The dprx5 mutant is more resistant to infection
To investigate resistance to various microbes, we used at least 10 da old flies, when the increased mortality rate in the dprx5 mutant is stabilized and becomes similar to that of the y w control [19]. Another reason for using flies of this age is that 10 da old flies are fully developed, which minimizes potential effects of variations in the levels of hormones, such as juvenile hormone and ecdysone, which may tremendously affect immunocompetence [26].
As indicated in the Introduction, the innate immune response in Drosophila entails at least two distinct signaling pathways: Toll and Imd. It is well known that Gram-negative bacteria induce humoral immunity mainly via the Imd pathway, while Gram-positive bacteria and fungi primarily activate the Toll pathway (reviewed in [27]), although the activation of either pathway is not always strictly selective and the immune response is more complex [28]. For instance, some Gram-positive bacteria, such as B. subtilis, are also sensed through the Imd pathway [10] and vice versa, Gram-negative bacteria, such as P. aeruginosa activates both Toll and Imd pathways to combat the infection [29].
When flies were infected with B. bassiana, survivorship in both controls and mutant flies was almost equivalent reaching approximately 30% by day 10 after infection (Fig. 1 A). We also investigated survivorship of flies infected with Gram-positive bacteria M. luteus; however no significant mortality was determined with this pathogen, since ~ 90% of both control and mutant flies survived after 4 days post-infection (data not shown). Similarly, we were not able to discern differences in survivorship of flies that were naturally infected with P. carotovorum, as only minor mortality was observed (data not shown). However, we were able to determine differences in recovery from septic injury with B. subtilis, which is known to activate the Imd pathway [10]. Infection with this bacterium caused approximately 80% death of control after 72 h post-infection, while almost 60% of dprx5 flies survived infection (Fig. 1 B). The dprx5 mutant had also slightly enhanced resistance to natural infection with P aeruginosa that was particularly evident during 24–72 hours post-infection (Fig. 1 C). Septic injury with E. coli caused only moderate mortality (less than 30%) within 12 hours followed by recovery in both control and mutant flies (data not shown). However, bacteriologic analysis revealed that the mutant flies were cleared from infection significantly faster relative to the controls (Fig. 1 E). The opposite effects on resistance to bacteria were observed in flies expressing dPrx5 from the UAS-dPrx5 transgene using global Act-GAL4 or Da-GAL4 drivers in flies with the dprx5 null background. Expression of dPrx5 at high levels severely affected survivorship of flies infected with P. aeruginosa, where median survival time of experimental flies was about one third of that in driver and transgene controls (Fig. 1 D). Together, these data indicate that the dprx5 mutant has enhanced resistance to bacterial infection compared to its isogenic control. In contrast, high levels of dPrx5 reduced survival of infected flies. The data also suggest that the immune response triggered via the Imd pathway may be a cause of the observed enhanced resistance to bacteria.
Fig. 1. Survivorship of dprx5 mutant after infection and clearance from bacteria.
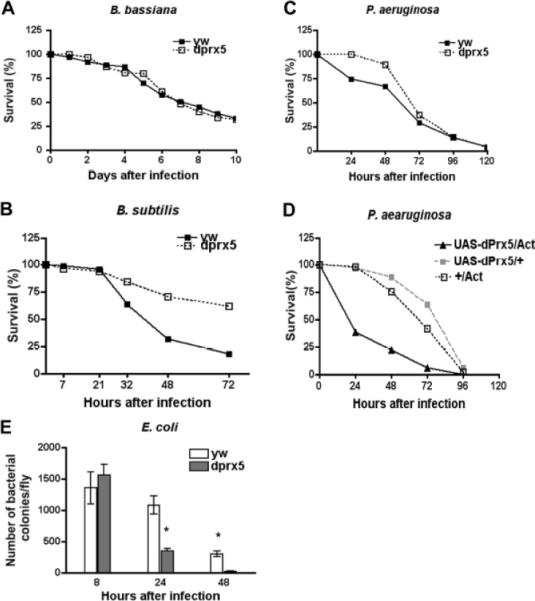
A minimum one hundred flies was used for each cohort. All flies were aged until day 10 followed by infection. The results of survivorship are pooled data of three-four independent experiments. A, B, C and D, a percentage of live flies is shown on the Y axis. A, dprx5 mutant and control (yw) flies were exposed to a sporulating culture of B. bassiana followed by incubation at 25°C. B, dprx5 mutant and control (yw) flies were septically injured with B. subtilis and incubated at 25°C. Paired t test showed statistically significant differences in mean survivorship for y w versus dprx5 infected with B. subtilis (P=0.049). C and D, dprx5 mutant and control (yw) flies (C) or flies expressing dPrx5 from the UAS-dPrx5 transgene (D) were naturally infected with P. aeruginosa and incubated at 25°C. UAS-dPrx5 transgene was expressed in flies with dprx5 null background using ubiquitous Act-GAL4 driver (UAS-dPrx5/Act). The resulting genotype was yw; UAS-dPrx5/Act; dprx5/dprx5 and the genotypes of corresponding controls were yw; UAS-dPrx5/+; dprx5/dprx5 (UAS-dPrx5/+) and yw; +/Act; dprx5/dprx5 (+/Act). Paired t test showed statistically significant differences in survivorship between infected flies expressing dPrx5 from transgene (UAS-dPrx5) and corresponding driver and transgene controls (P=0.030 and P=0.027) and marginally significant differences in mean survivorship between dprx5 mutant and y w control (P=0.055). E, flies were septically injured with ampicillin-resistant strain of E. coli followed by bacteriological analysis of fly lysates at various time intervals after infection. At least ten flies were taken for each sample. The number of colonies per fly is indicated on the Y axis. The results are mean ± SD from two independent experiments and the statistically significant differences (P<0.05) are denoted by asterisks.
3.2. AMP levels
To determine whether the observed resistance to bacteria in the dprx5 mutant relies, at least in part, upon activation of the specific immune pathway(s), we measured levels of the key effectors of humoral immunity, the anti-microbial peptides (AMP). We used infection with B. bassiana to trigger the Toll pathway and injury with E. coli to trigger the Imd pathway and evaluated the levels of different AMPs.
The basal AMP levels in mutant and control flies were almost undetectable except drosomycin, which was expressed in flies under normal conditions at approximately the same levels in both control and mutant flies (not shown). Infection with B. bassiana caused elevation in the levels of the anti-fungal AMPs, drosomycin (50–60 fold induction) and metchnikowin (8–12 fold induction), while other AMPs were not induced (Fig. 2 A and not shown data). Both drosomycin (Drs) and metchnikovin (Mtk) are induced primarily via the Toll pathway; however, there were no differences in the response between control and mutant flies after 24 h post-infection (Fig. 2 A). Septic injury with E. coli caused up-regulation of several tested AMPs after 8 h post-infection, although the induction of attacins (Att) and to a lesser extent diptericin A (Dipt) and cecropin C (CecC) was notably pronounced in the dprx5 mutant relative to control. Furthermore, the differences in the induction of attacin D (AttD) and cecropin C (CecC) between the dprx5 mutant and y w control were statistically significant (Fig. 2 A). It is well known that the induction of attacins, cecropins and diptericins is associated primarily with the Imd/Relish pathway; moreover, the activation of AttD and DiptA is controlled only by Relish [30]. The effects on the levels of AMPs, normally up-regulated in the dprx5 mutant (Fig. 2 B), were reverted by ubiquitous (by 2.5–6 fold) over-expression of UAS-dPrx5, resulting in ~ 2 fold decrease in the AMP levels. Taken together, the observed differences in the induction of AMPs suggest that dPrx5 may modulate the immune response mediated via the Imd pathway.
Fig. 2. AMP levels in flies infected with B bassiana and E. coli.
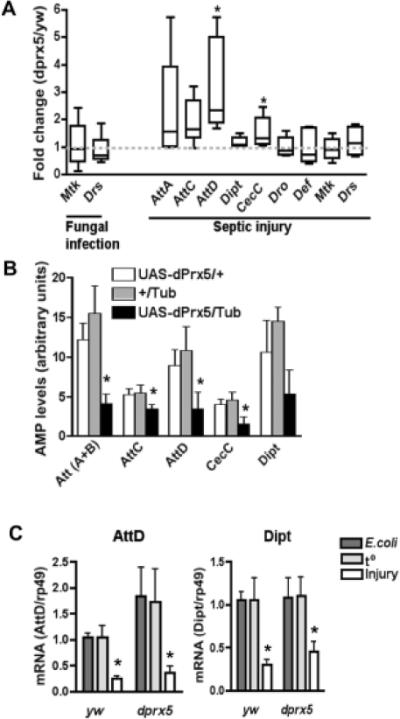
A, Control and mutant 10 days old flies were exposed to sporulating fungi and maintained for 24 h at 25°C or they were injured with a needle dipped in a E. coli culture followed held for 8 h at 25°C. RNA was extracted and AMP levels were then determined by real-time RT-PCR. The signals obtained for AMP genes were normalized with respect to signals obtained for house-keeping gene (rp49). Plotted are fold changes in the AMP levels in dprx5 mutant compared to y w control. At least four independent determinations were performed for each AMP. More than one fold significant differences between the dprx5 mutant relative to y w control were determined for AttD (P=0.0455) and CecC (P=0.0148), as denoted by asterisks. B, 10 da old flies over-expressing dPrx5 (UAS-dPrx5/Act) and corresponding driver and transgene controls (UAS-dPrx5/+ and +/Act) were infected with E. coli by pricking. RNA was extracted and AMP levels were then determined by real-time RT-PCR. The signals obtained for AMP genes were normalized with respect to signals obtained for house-keeping gene (actin). The results are mean ± SD from three independent experiments and the statistically significant differences (P<0.05) are denoted by asterisks. C, RNA was isolated from flies that were pricked with a needle dipped in E. coli suspension (E.coli) or the same suspension inactivated for 5 min by boiling (t°), as well as from flies pricked with a sterile needle (Injury). Relative to a housekeeping gene mRNA ratios are plotted on the Y axis.
We also investigated whether the observed AMP induction is a general response to potentially elevated oxidative stress in the dprx5 mutant or a response to proliferating bacteria. To test this, we injected flies with live and killed bacteria and compared AMP levels. No differences in AttD or Dipt levels were found between flies pricked with live or thermo-inactivated bacteria, while significant differences in the AttD or Dipt levels were found between samples isolated from flies injured with a sterile needle and flies pricked with bacterial suspensions. In the latter, the response was significantly stronger (Fig. 2 C). Strong induction of diptericin or attacin was also not observed in flies treated with paraquat for the periods from 1 h to 8 h (data not shown). Finally, we did not find any differences in the levels of H2O2 in the whole body lysates or mitochondria prepared from mutant and control flies (data not shown). Altogether, the data suggest that the observed early response in the dprx5 mutant is not due to bacterial proliferation or elevated ROS levels but rather due to specific effects of dPrx5 on factors involved in regulating immunity.
3.3. Epistatic analysis
To position dPrx5 regulatory effects within specific immune pathway(s), we generated a series of double-mutant lines involving dprx5 in combination with alleles that are known to reside in the pathways regulating Drosophila immunity. Since the induction of AMPs suggested that dPrx5 might modulate the Imd-dependent immune response, a phenotypic analysis was performed using flies that are mutant for genes of the Imd pathway. The immune response, triggered via the Imd pathway, is initiated after stimulation of pattern recognition receptors [31, 32] and requires a receptor proximal complex, which includes Imd and dFADD [33, 34]. The signal is subsequently transmitted through activation of the kinase complex DmIKK [35], by means of the kinase kinase dTak1 [21]. Activation of DmIKK is necessary for phosphorylation of the NF-κB protein Relish, which is further processed by Dredd caspase [36]. The liberated RHD domain of Relish is then transported into the nucleus where it activates transcription of the AMP effector genes. We generated a series of double-mutant lines involving kenny, ird5, dtak1, rel or dredd alleles in combination with dprx5. It has been previously reported that AMPs, triggered primarily by the Imd/Relish module, have significantly reduced levels in kenny, dtak1, rel or dredd mutants [22, 35]. Indeed, we found that the levels of AttD, which was used as a read out gene for the Imd pathway, were almost undetectable in dredd, rel and kenny or ird5 ten days old mutants infected with E. coli and were significantly reduced in the dtak1 mutant (Fig. 3). The reduction in AttD levels was partially compensated by the absence of dPrx5 expression in the dprx5;dtak1 double mutant, but not in the double mutants for dPrx5 and the IKK complex (kenny and ird5) nor the dredd;dprx5 and rel,dprx5 double mutants (Fig. 3).
Fig. 3. The immune response induced by bacterial infection is altered in mutants.
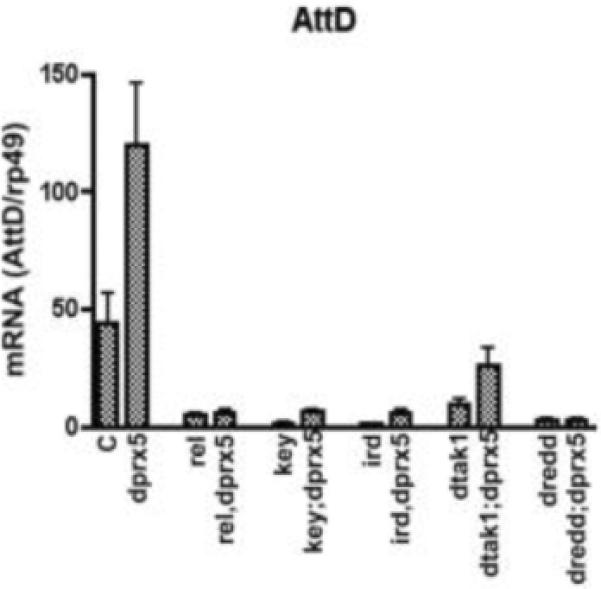
Ten day old y w control (C), dprx5, tak1, relE20, kenny (key1), ird5 and dredd mutant flies and double mutants underexpressing dTak1, Relish, genes comprising IKK complex (Kenny and Ird5), Dredd caspase and dPrx5 were infected with E. coli and incubated at 25°C for 6 hours. RNA was extracted and the transcriptional levels of attacin D as a read-out gene for the Imd/Relish pathway were determined. The results are mean ± SD from three independent experiments. Statistically significant differences (P<0.05) were between y w control and the dprx5 mutant, as well as mutants of the genes comprising the IMD/Relish axis, and double mutants. There were also significant differences between dtak1 mutant and dtak1;dprx5 double mutant.
The absence of compensatory effects in the IKK-Relish module suggests that the activity of Relish is still required for the induction of Imd-dependent AMPs in either normal or dprx5 background. However, partial up-regulation of the Imd-dependent antimicrobial peptides, which occurs in the absence of dTak1 activity in the dprx5 null background, suggested the presence of an alternate route for activation.
3.4. dPrx5 modulates the JNK pathway
We examined whether dPrx5 may affect AMP expression via the Imd/JNK branch of the immune signaling pathways. It has been shown that Imd-dependent activation of Drosophila innate immunity bifurcates downstream of dTak1, leading to the activation of both NF-κB and JNK pathways [11, 13, 37]. We evaluated activation of the JNK pathway in the dprx5 mutant background by assessing phosphorylation patterns of JNK kinase (Basket) following infection. Activation of Basket occurred rapidly in the y w control and was sustained for at least 1 h after infection before dropping off. In the dprx5 mutant, activated Basket was already observed prior to infection, showed no further activation in response to infection, and, in fact, declined significantly by 2 h post-infection (Fig. 4). In contrast, phosphorylation of Basket in flies over-expressing dPrx5 (UAS-dPrx5/Act) had a temporal profile, similar to that in the control but less pronounced (Fig. 4). Unlike phosphorylation, the expression levels of Basket (JNK) did not change in response to infection (data not shown).
Fig. 4. dPrx5 affects JNK signaling by signal-dependent phosphorylation of Basket.
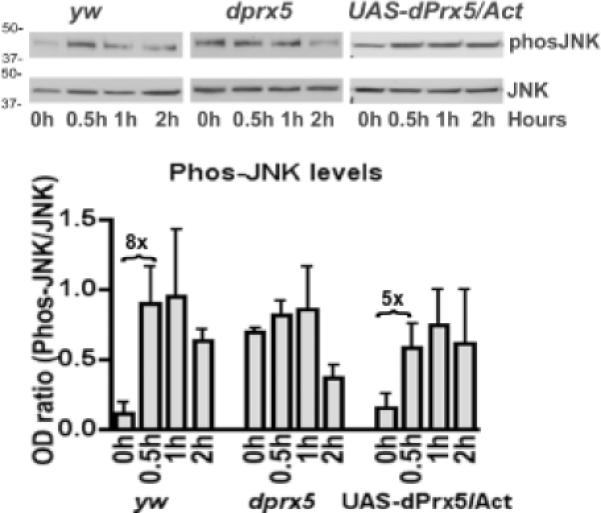
Proteins were isolated from control (y w), dprx5 mutant and flies over-expressing dPrx5 (UAS-dPrx5) at different time intervals after infection with E.coli and resolved by PAGE electrophoresis. Immunoblot analysis was performed with anti-phos-JNK antibodies to detect phosphorylated JNK (Basket) and with anti-JNK antibodies as a control. Total protein levels were equal in each sample, as was determined after re-probing blots with anti-Actin antibodies (not shown). Protein molecular weight markers (kDa) are indicated on the left. Intensity of the phos-JNK signals standardized against signals obtained for JNK is plotted on the graphs. The results are mean ± SD, n=3.
The altered phosphorylation of Basket in flies under- or over-expressing dPrx5 resulted in compromised induction of transient response genes puckered, ets21C and relish, which was evaluated by measuring their mRNA levels. As determined by RT-PCR analysis, induction of the gene coding for the dual specificity phosphatase Puckered, a target gene of the JNK pathway, was observed in both the dprx5 mutant and control flies, although the response was more robust in the dprx5 mutant and less pronounced in flies over-expressing dPrx5 (Fig. 5 A). We also determined changes in the response of another putative target of JNK signaling, transcription factor Ets21C [15, 38]. As noted in Fig. 5 B, induction of Ets21C was found to occur earlier in the dprx5 mutant and was dampened in the dPrx5 over-expressor (Fig. 5 B). Similarly, the transcription factor Relish was found to exhibit a much more robust response in the dprx5 mutant while that of the overexpressor was reduced (Fig. 5 C).
Fig. 5. Expression analysis of transient response genes puckered (A), ets21C (B) and relish (C), induced by bacterial infection.
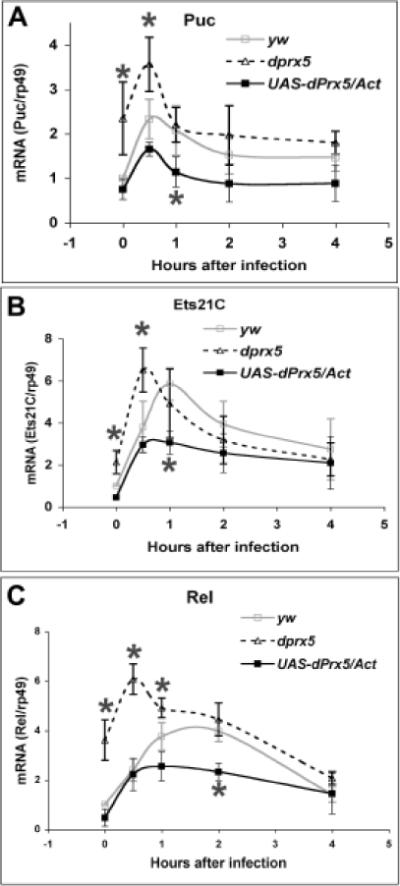
Total RNA was isolated from dprx5 mutant, control (y w) flies and flies over-expressing dPrx5 (UAS-dPrx5/Act) infected with E. coli at different time intervals. RT-PCR was performed with primers specific for Puckered, Ets21C and Relish, and signals obtained were standardized against signal obtained for the housekeeping gene rp49. Statistically significant (P<0.05) were differences in the levels of Puc, Ets21C and Rel at 0 h and 30 min post-infection and also Rel at 1 h post-infection, as denoted by asterisks. Significant were also differences between control and flies over-expressing dPrx5 at 30 min post-infection (Puc), 1 h (Puc, Ets21C and Rel) and 2 h (Rel). The results are mean ± SD from three independent experiments.
We also investigated whether peroxiredoxin 5 effects on the JNK pathway are due to its peroxidatic activity. To test this, we generated a UAS construct carrying a redox-negative Prx5 variant (UAS-dPrx5RN), where catalytic cysteine-79 was replaced with redox-inactive serine-79 (see Materials and Methods). This type of amino acid substitution compromises only peroxidase activity of peroxiredoxin 5, and not other properties of this protein [39].
The catalytically inactive UAS-dPrx5RN variant and normal UAS-dPrx5 were expressed with the Da-GAL4 driver in flies having a dprx5 null background. For these experiments, transgenic lines expressing comparable levels of dPrx5 and dPrx5RN proteins were selected (Fig. 6 C). Unlike UAS-dPrx5, which rescued the dprx5 mutant phenotype from increased susceptibility to H2O2 or paraquat [19], redox negative UAS-dPrx5RN failed to protect flies from oxidants (data not shown), thus confirming that the mutated form of peroxiredoxin 5 lost its antioxidant activity. At the same time, the effects on the activation of the JNK pathway, observed in flies expressing normal dPrx5, were absent in flies expressing the redox-negative peroxiredoxin 5, whose phenotype was largely similar to that of the dprx5 mutant. For instance, flies expressing dPrx5RN exhibited induction of the downstream response genes Puckered and Ets21C, effects that were dampened in flies expressing wild type dPrx5 (Fig. 6 A and B). Similarly, the phosphorylated Basket, observed in the dprx5 mutant prior to infection, was also observed in flies expressing the redox-negative peroxiredoxin 5 (Fig. 6 C). In contrast, kinase Basket was not phosphorylated in the absence of infection in yw flies and flies expressing normal dPrx5 (Fig. 6 C). Taken together, these data show that peroxiredoxin 5 modulates the JNK pathway by virtue of its peroxidase activity.
Fig. 6. Effects of the wild type and redox-negative forms of dPrx5 on JNK signaling.
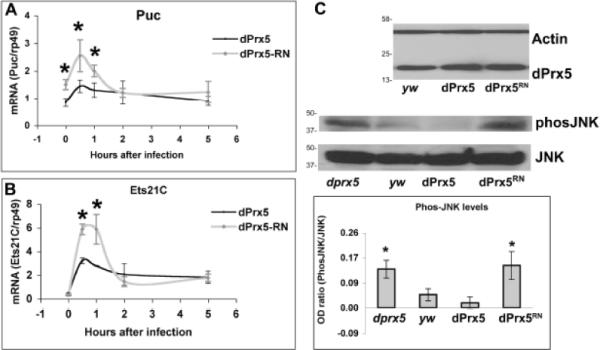
The expression of dPrx5 or dPrx5RN was achieved using corresponding UAS transgenes and dprx5 or recombinant Da-GAL4, dprx5 alleles. The genotype of the generated flies was yw; UAS-dPrx5(or dPrx5RN)/+; dprx5/Da-GAL4,dprx5. A and B, expression analysis of puckered and Ets21C, induced by bacterial infection. Total RNA was isolated from flies infected with E. coli at different time intervals. RT-PCR was performed with primers specific for Puckered and Ets21C and signals obtained were standardized against signals obtained for the housekeeping gene rp49. Statistically significant (P<0.05) differences are denoted by asterisks. C, Expression levels of wild type and redox-negative forms of dPrx5 protein are shown on the upper image. Phosphorylation of Basket in flies expressing wild type and redox-negative forms of dPrx5 are shown on the lower image. Proteins were isolated from control (y w), dprx5 mutant (dprx5) and flies expressing UAS-dPrx5 or UAS-dPrx5RN in dprx5 null background (indicated as dPrx5 and dPrx5RN) and resolved by PAGE electrophoresis. Immunoblot analysis was performed with anti-dPrx5 and anti-Actin antibodies to control for loading. Phosphorylated JNK (Basket) was detected with anti-phos-JNK antibodies and with anti-JNK antibodies as a control. Protein molecular weight markers (kDa) are indicated on the left. Intensity of the phos-JNK signals standardized against signals obtained for JNK is plotted on the graphs. The results are mean ± SD, n=3. Asterisks indicate statistically significant differences between the dprx5 mutant or flies expressing dPrx5RN and yw control and dPrx5-expressing flies.
3.5. Expression of Imd- and JNK target genes is affected by dominant-negative Basket
To further test involvement of the JNK pathway in the induction of Imd-dependent AMPs, we generated flies with reduced JNK activity by expressing the dominant-negative form of Basket (BskDN), which is nonactivatable due to a mutation in the phosphorylation site T181A [40]. BskDN-expressing flies were generated in normal (y w) or dprx5 null background using the mifepristone-inducible P{Switch1}106 driver that targets responder gene expression to the Drosophila fat body. Flies were fed regular food containing mifepristone for two days prior to infection, resulting in a 5–7 fold increase in the levels of BskDN compared those of the endogenous Basket (Fig. 7 A). As anticipated, activation of the JNK pathway was significantly dampened in flies over-expressing the dominant-negative Basket protein, as determined by measuring mRNA levels of Puckered, a target gene of the JNK pathway (Fig. 7 B). While the induction of Puc was completely abrogated in the BskDN-expressing flies, the effects on the induction of Ets21C were less pronounced but still significant (Fig. 7 C), which confirmed that Ets21C is at least partially regulated via the JNK pathway. The induction of Puckered or Ets21C was not restored in the absence of dPrx5, which suggests that dPrx5 acts upstream of Basket. To investigate the effects of the dPrx5 mutation in flies with normal and Basket-DN backgrounds on the Imd-triggered immune response, we determined the transcript levels of Relish and the Relish-dependent AMPs, diptericin A, cecropin C and attacin D, as well as drosomycin, which is induced primarily via the Toll pathway. We found that JNK activity is required for the early activation of Relish, since no induction was observed at 1 h post-infection in flies over-expressing dominant-negative Basket (Fig. 7 D). In contrast, JNK was found to have dampening effects on subsequent Relish up-regulation later, since Relish levels continued to rise after 4 h in the dprx5 mutant background (Fig. 7 D). Consistent with previous results (Fig. 2 A), the induction of CecC and AttD, but not Dipt or Drs, was more pronounced in the dprx5 mutant (Fig. 7 E–H). The effects were further enhanced by over-expression of BskDN, although the dominant-negative Basket alone did not have any effects on the expression of these AMPs. We also found that JNK signaling is needed for more robust induction of Drs (Fig. 7 H).
Fig.7. Effects of dPrx5 and JNK signaling on the immune response genes.
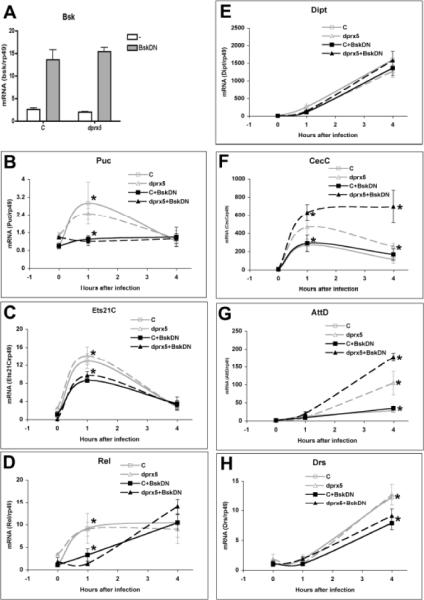
The genotype of the control (C) was UAS-BskDN; S106/+;+/+ and the genotype of flies under-expressing dPrx5 (dprx5) was UAS-BskDN; S106/+;dprx5/dprx5. Expression of dominant-negative Basket was achieved by feeding one week-old flies food containing 0.1 mg/ml mifeprisone dissolved in ethanol (BskDN) or ethanol-only added food as a control (−). Food with mifepristone was changed daily. A, Analysis of Basket expression. After two days of feeding mifepristone, RNA was isolated followed by real time RT-PCR analysis with primers specific for Basket. B–H, After feeding for two days food with or without mifepristone, flies were infected with 24 h broth culture of P. aeruginosa by pricking and incubated at 25°C for 0 h, 1 h and 4 h followed by RNA extraction. RT-PCR was performed with primers specific for Puckered (B), Ets21C (C), Relish (D), Dipt (E), CecC (F), AttD (G) and Drs (H) and the gene-specific signals were standardized against signals obtained for the housekeeping gene rp49. Statistically significant (P<0.05) were differences in the levels of Puc, Ets21C and Rel between control flies (no mifepristone) and flies expressing BskDN in normal (C) or dprx5 background at 1 h after infection, as denoted by asterisks. Significant differences were also in the levels of CecC between dprx5 mutant and control, expressing dominant-negative Basket at 1 h and 4 h post-infection; AttD at 4 h post-infection between control and the dprx5 mutant expressing or not expressing BskDN, as well as Drs between flies with wild type or dominant-negative Basket, as denoted by asterisks. All RT-PCR results are mean ± SD from three-four determinations for each of two independent experiments.
3.6. dPrx5 is up-regulated in response to septic injury
The above data indicated that dPrx5 acts as a negative regulator of the immune response to infection. It is plausible then that activation of pathway(s) involved in regulating immunity could serve to induce dPrx5 expression, which through feedback inhibition could result in a more controlled immune response. Indeed we found that there was an increase in dPrx5 protein and mRNA levels (not shown) after septic injury with different bacteria, which became particularly evident after 5 h post-infection (Fig. 8).
Fig. 8. dPrx5 response to infection.
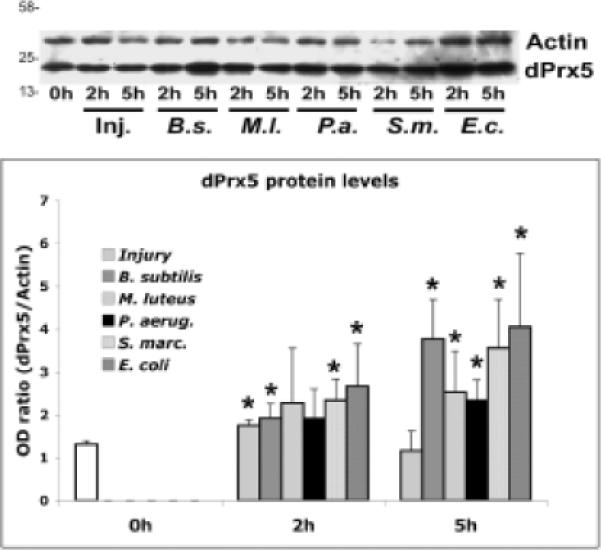
Control and dprx5 mutant flies were septically injured with B. subtilis, M. luteus, P. aeruginosa, S. marcescens or E. coli suspensions or with a sterile needle and kept for 2 h and 5 h at 25°C. At least ten flies were taken for each sample. Proteins were extracted, loaded in the amounts of 5 μg and resolved by PAGE, followed by immunoblot analysis using anti-dPrx5 antibodies and anti-actin antibodies to control for a loading. Protein molecular weight markers (kDa) are indicated on the left. Ratios of dPrx5 to actin signals are plotted on the Y axis. The results are mean ± SD from three determinations. Significant were differences between non-infected flies and flies infected with bacteria, as indicated by asterisks.
4. Discussion
In this study we showed that Drosophila peroxiredoxin 5 is involved in negative control of the immune response, and furthermore its action has been tracked to the JNK signaling pathway.
The dprx5 mutant was found to exhibit enhanced resistance to bacteria that primarily activate the Imd immune pathway. Several lines of evidence support this conclusion: i) downstream effectors of the immune response associated with the Imd pathway, particularly attacins and cecropins were clearly up-regulated in the absence of dPrx5 (Figs. 2 A and 7 F–G); ii) the dprx5 mutant was cleared from the E. coli sepsis significantly faster (Fig. 1 E); and iii) the mortality of the mutant, infected with B. subtilis or P. aeruginosa, was reduced. The reduced mortality rate was particularly evident during the first 1–2 days after infection (Fig. 1 B and C). At the same time, production of AMPs in the mutant was enhanced and likely prevented bacterial proliferation during early stages of infection. To evade host defense, some bacteria, such as P. aeruginosa, employ suppression of AMP production early during infection [41]. The initial AMP deficit appeared to be compensated in the dprx5 mutant. Furthermore, the elevated levels of attacins, cecropins and diptericins, which display antimicrobial activity to a wide spectrum of both Gram-positive and Gram-negative bacteria [42–44], seem to underlie the enhanced resistance in the dprx5 mutant.
While being relatively resistant to microbial challenge, the dprx5 mutant is highly susceptible to oxidative stress caused by hydrogen peroxide or paraquat, as was reported previously [19]. Similarly, dPrx5 overexpression conferred greater resistance to oxidative stress but enhanced significantly susceptibility to bacterial infection (Fig. 1 D), suggesting a trade-off between dPrx5 immune and antioxidant functions.
A series of molecular and genetic approaches were used to investigate mechanisms by which dPrx5 controls the immune response. We determined that the activity of Relish was still required for the induction of the Imd-dependent AMPs (Fig. 3). The levels of AttD, an indicator for the Imd/Relish pathway, were significantly reduced in the rel mutant as well as mutants for the IKK complex and Dredd caspase, genes that are required for processing and activation of Relish [36, 37]. When any of these mutants were placed in a dprx5 background the AttD levels remained low. On the other hand, AttD suppression was partially compensated when dtak1 mutation was combined with the dprx5 mutation. dTak1 controls activation of both Imd and JNK-mediated immune response [11, 13, 14, 21], which led us to suggest that dPrx5 could have negative control over activation of Relish-dependent genes by modulating the JNK branch of immune signaling. Indeed, we found that temporal patterns of activation of the JNK pathway, specifically phosphorylation of the JNK kinase Basket and transcriptional profiling of the downstream target genes, puckered and ets21C, were altered accordingly in the dprx5 mutant (Figs. 4 and 5). We have also found that dPrx5 may control local peroxide levels, thus perhaps modulating the redox state of the dTak1-JNK kinase partners and coupled phosphatases, which in turn impacts kinase function. Enhanced phosphorylation of the JNK kinase Basket and the altered transcriptional profile of the JNK downstream targets (Fig. 6) in flies expressing redox-negative dPrx5 strongly suggest that dPrx5 regulates the dTak1-JNK arm of immune signaling by virtue of its peroxidase activity.
Altogether, the data placed dPrx5 downstream or at the level of dTak1 but upstream of Basket. This is supported by recent evidence that both JNK and the Imd pathways integrate to positively control the induction of AMPs [11]. This runs counter to a study conducted by Kim et al., where RNAi and over-expression in S2 cells revealed a negative crosstalk between the JNK and Imd pathways, in which the JNK pathway was required for shutting down activation of AttA, triggered via the Imd pathway [12]. At present we attribute this discrepancy to the greater level of complexity of the immune response in the whole organism, which relies on the fat body in addition to a hemocyte-based subset of the immunity in contrast to the immune response in macrophage-like S2 cells.
There are two current models of Imd-mediated immune signaling in response to infection. An earlier model, initially suggested by Brennan and Anderson [45] places dTak1 within the Imd pathway downstream of IMD and upstream of IKK complex and is based on the finding that the loss of dTak1 activity can block activation of the AMPs, elicited by IMD over-expression [21]. A second model, proposed by Delaney et al. and Kleino [11, 46], places dTak1 upstream of JNK, rather than IKK. According to their data, activation of JNK signaling could rescue AMP expression from the loss of dTak1, while suppression of JNK signaling results in inhibition of Relish-dependent antimicrobial peptides. Our data favor the latter model, where the loss of dPrx5 activity results in activation of the JNK kinase Basket and transient response genes and positively controls the induction of Relish-dependent AMPs.
It is interesting that JNK was also required for the early activation of Relish at the transcriptional level, as Rel induction was significantly abrogated in BskDN flies (Fig. 7 D). However, expression of Rel or over-expression of BskDN did not seem to impact activation of the Rel-dependent AMPs, Dipt, Cec and Att, but did affect the Toll-dependent Drs (Fig. 7 H). It appears that constitutively produced amounts of Relish protein are sufficient for activation of the Imd/Relish-mediated response, where Relish processing and nuclear transport permit induction of the effector AMP genes. Nevertheless, differences in Relish mRNA levels in response to infection, that were also observed between control and dprx5 mutant flies, suggest that Relish protein may play another function in modulating the immune response. Consistent with other reports [11], the patterns of Drs response also suggest that JNK is required for the Toll-mediated response and that dPrx5 acts upstream of the JNK kinase Basket (Fig. 7 H).
The proposed model does not fully explain the observed differences in the activation of Relish-dependent AMPs, nor the synergistic effects on AttD and CecC induction, observed in flies producing the dominant-negative Basket in the dprx5 mutant background (Fig. 7 E–G). The analysis of Dipt, CecC and AttD promoters using AliBaba2 software (www.gene-regulation.com) did not reveal obvious differences that might affect the induction of attacin, cecropin and diptericin. Unlike Dipt, transcription of AttD and CecC could also require additional modifications of Relish, as suggested [47]. On the other hand, alterations in JNK signaling may in its turn cause changes in the transcription factors required for proper control of different AMPs. The differential responses of Dipt, CecC and AttD (Fig. 7) support this hypothesis. We also cannot exclude that, in addition to affecting phosphorylation of JNK (Basket), dPrx5 may also function downstream of JNK and directly affect activity of transcription factors or transcription complexes involved in regulating expression of different AMPs. Our previous finding that dPrx5 has multiple subcellular localizations, including nuclear, supports this notion. Possible interactions of a nuclear-localized form of dPrx5 with transcription complexes have been reported [39].
The dPrx5 itself is up-regulated in response to septic injury by different bacteria, both Gram-positive and Gram-negative. It is known that both injury and bacterial challenges cause release of ROS, which is either a component of cellular immune response to kill invaders or a downstream event of cell receptor-ligand interactions, which facilitates cell signaling. On the other hand, the excess of ROS, released in response to infection, may cause damage to the host cells. Thus, the up-regulation of dPrx5 after bacterial challenge could only be a response to the elevated ROS levels and serve a protective role to host cells during the immune response, at a time when excess reactive oxygen species are released. However, we did not find evidence that dPrx5 is up-regulated in response to oxidants, including paraquat or hydrogen peroxide; nor did we find any evidence for the presence of ARE (antioxidant response element) in the dPrx5 promoter [19]. Besides, adult flies without dPrx5 seem to survive better during infection than controls and transgenic flies that express dPrx5 at higher levels (Fig. 1). Thus, any protective role that dPrx5 plays in the immune response would appear to be minor. Given the role of dPrx5 in suppressing the immune response, its up-regulation by bacterial infection may instead represent a negative feedback mechanism, quenching an excessive immune response. The presence of a κB binding motif in the dprx5 gene promoter [19] supports this hypothesis.
In summary, we have determined that one of the biological functions of peroxiredoxin 5 is to control the immune response. To date, the role of antioxidants, such as peroxiredoxins, during infection has been generally described as protective in nature, where peroxiredoxins scavenge excessive ROS that are released by cells in order to kill microbial invaders. Our findings, however, suggest that peroxiredoxins may function to dampen the activation of the immune response, acting as negative regulators of specific immune pathways. The results of the present research placed dPrx5 at the dTak1-JNK branch of immune signaling, although the data suggest that dPrx5 may also act downstream or in parallel to JNK. The latter conjecture is currently under investigation.
Acknowledgements
We thank Drs. J. Abrams, D. Bohmann, B. Rogina and N. Silverman for providing fly strains, Dr. C. Buchanan for providing bacterial strains and Drs. R. Jones and J. Waddle (SMU) for a critical reading of the manuscript. This work was supported by grant R21 AG025096-01 from the National Institute on Aging/National Institutes of Health.
Abbreviations
- Prx
peroxiredoxin
- AMP
antimicrobial peptide
- Att
attacin
- Cec
cecropin
- Dipt
diptericin, Drs, drosomycin
- Mtk
metchnikovin
- Imd
immune deficiency
- JNK
c-Jun N-terminal kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Shau H, Kim AT, Hedrick CC, Lusis AJ, Tompkins C, Finney R, Leung DW, Paglia DE. Endogenous natural killer enhancing factor-B increases cellular resistance to oxidative stresses. Free Radic Biol Med. 1997;22:497–507. doi: 10.1016/s0891-5849(96)00372-3. [DOI] [PubMed] [Google Scholar]
- [2].Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kappaB activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- [3].Hansen JM, Moriarty-Craige S, Jones DP. Nuclear and cytoplasmic peroxiredoxin-1 differentially regulate NF-kappaB activities. Free Radic Biol Med. 2007;43:282–288. doi: 10.1016/j.freeradbiomed.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kim YJ, Lee WS, Ip C, Chae HZ, Park EM, Park YM. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006;66:7136–7142. doi: 10.1158/0008-5472.CAN-05-4446. [DOI] [PubMed] [Google Scholar]
- [5].Haridas V, Ni J, Meager A, Su J, Yu GL, Zhai Y, Kyaw H, Akama KT, Hu J, Van Eldik LJ, Aggarwal BB. TRANK, a novel cytokine that activates NF-kappa B and c-Jun N-terminal kinase. J Immunol. 1998;161:1–6. [PubMed] [Google Scholar]
- [6].Jung H, Kim T, Chae HZ, Kim KT, Ha H. Regulation of macrophage migration inhibitory factor and thiol-specific antioxidant protein PAG by direct interaction. J Biol Chem. 2001;276:15504–15510. doi: 10.1074/jbc.M009620200. [DOI] [PubMed] [Google Scholar]
- [7].Kang SW, Chang TS, Lee TH, Kim ES, Yu DY, Rhee SG. Cytosolic peroxiredoxin attenuates the activation of Jnk and p38 but potentiates that of Erk in Hela cells stimulated with tumor necrosis factor-alpha. J Biol Chem. 2004;279:2535–2543. doi: 10.1074/jbc.M307698200. [DOI] [PubMed] [Google Scholar]
- [8].Veal EA, Findlay VJ, Day AM, Bozonet SM, Evans JM, Quinn J, Morgan BA. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol Cell. 2004;15:129–139. doi: 10.1016/j.molcel.2004.06.021. [DOI] [PubMed] [Google Scholar]
- [9].Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- [10].Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci U S A. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Delaney JR, Stoven S, Uvell H, Anderson KV, Engstrom Y, Mlodzik M. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. Embo J. 2006;25:3068–3077. doi: 10.1038/sj.emboj.7601182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim T, Yoon J, Cho H, Lee WB, Kim J, Song YH, Kim SN, Yoon JH, Kim-Ha J, Kim YJ. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules. Nat Immunol. 2005;6:211–218. doi: 10.1038/ni1159. [DOI] [PubMed] [Google Scholar]
- [13].Silverman N, Zhou R, Erlich RL, Hunter M, Bernstein E, Schneider D, Maniatis T. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J Biol Chem. 2003;278:48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- [14].Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, Ramet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7:811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- [15].Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- [16].Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci U S A. 2006;103:16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsuda M, Langmann C, Harden N, Aigaki T. The RING-finger scaffold protein Plenty of SH3s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 2005;6:1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim LK, Choi UY, Cho HS, Lee JS, Lee WB, Kim J, Jeong K, Shim J, Kim-Ha J, Kim YJ. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 2007;5:e238. doi: 10.1371/journal.pbio.0050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Radyuk SN, Michalak K, Klichko VI, Benes J, Rebrin I, Sohal RS, Orr WC. Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila. Biochem J. 2009;419:437–445. doi: 10.1042/BJ20082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- [21].Vidal S, Khush RS, Leulier F, Tzou P, Nakamura M, Lemaitre B. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 2001;15:1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- [24].Michalak K, Orr WC, Radyuk SN. Drosophila peroxiredoxin 5 is the second gene in a dicistronic operon. Biochem Biophys Res Commun. 2008;368:273–278. doi: 10.1016/j.bbrc.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Radyuk SN, Sohal RS, Orr WC. Thioredoxin peroxidases can foster cytoprotection or cell death in response to different stressors: over- and under-expression of thioredoxin peroxidase in Drosophila cells. Biochem J. 2003;371:743–752. doi: 10.1042/BJ20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, Garbuzov A, Palli SR, Tatar M, Silverman N. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol. 2008;211:2712–2724. doi: 10.1242/jeb.014878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- [28].Hedengren-Olcott M, Olcott MC, Mooney DT, Ekengren S, Geller BL, Taylor BJ. Differential activation of the NF-kappaB-like factors Relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J Biol Chem. 2004;279:21121–21127. doi: 10.1074/jbc.M313856200. [DOI] [PubMed] [Google Scholar]
- [29].Lau GW, Goumnerov BC, Walendziewicz CL, Hewitson J, Xiao W, Mahajan-Miklos S, Tompkins RG, Perkins LA, Rahme LG. The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect Immun. 2003;71:4059–4066. doi: 10.1128/IAI.71.7.4059-4066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. Embo J. 2004;23:4690–4700. doi: 10.1038/sj.emboj.7600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- [34].Naitza S, Rosse C, Kappler C, Georgel P, Belvin M, Gubb D, Camonis J, Hoffmann JA, Reichhart JM. The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity. 2002;17:575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- [35].Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci U S A. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stoven S, Ando I, Kadalayil L, Engstrom Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Johansson KC, Metzendorf C, Soderhall K. Microarray analysis of immune challenged Drosophila hemocytes. Exp Cell Res. 2005;305:145–155. doi: 10.1016/j.yexcr.2004.12.018. [DOI] [PubMed] [Google Scholar]
- [39].Kropotov AV, Grudinkin PS, Pleskach NM, Gavrilov BA, Tomilin NV, Zhivotovsky B. Downregulation of peroxiredoxin V stimulates formation of etoposide-induced double-strand DNA breaks. FEBS Lett. 2004;572:75–79. doi: 10.1016/j.febslet.2004.07.011. [DOI] [PubMed] [Google Scholar]
- [40].Adachi-Yamada T, Nakamura M, Irie K, Tomoyasu Y, Sano Y, Mori E, Goto S, Ueno N, Nishida Y, Matsumoto K. p38 mitogen-activated protein kinase can be involved in transforming growth factor beta superfamily signal transduction in Drosophila wing morphogenesis. Mol Cell Biol. 1999;19:2322–2329. doi: 10.1128/mcb.19.3.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Apidianakis Y, Mindrinos MN, Xiao W, Lau GW, Baldini RL, Davis RW, Rahme LG. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc Natl Acad Sci U S A. 2005;102:2573–2578. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Samakovlis C, Kimbrell DA, Kylsten P, Engstrom A, Hultmark D. The immune response in Drosophila: pattern of cecropin expression and biological activity. EMBO J. 1990;9:2969–2976. doi: 10.1002/j.1460-2075.1990.tb07489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tryselius Y, Samakovlis C, Kimbrell DA, Hultmark D. CecC, a cecropin gene expressed during metamorphosis in Drosophila pupae. Eur J Biochem. 1992;204:395–399. doi: 10.1111/j.1432-1033.1992.tb16648.x. [DOI] [PubMed] [Google Scholar]
- [44].Winans KA, King DS, Rao VR, Bertozzi CR. A chemically synthesized version of the insect antibacterial glycopeptide, diptericin, disrupts bacterial membrane integrity. Biochemistry. 1999;38:11700–11710. doi: 10.1021/bi991247f. [DOI] [PubMed] [Google Scholar]
- [45].Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- [46].Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, Stoven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Ramet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. Embo J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wiklund ML, Steinert S, Junell A, Hultmark D, Stoven S. The N-terminal half of the Drosophila Rel/NF-kappaB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Dev Comp Immunol. 2009;33:690–696. doi: 10.1016/j.dci.2008.12.002. [DOI] [PubMed] [Google Scholar]


