Abstract
Chronux is an open-source software package developed for the analysis of neural data. The current version of Chronux includes software for signal processing of neural time-series data including several specialized mini-packages for spike sorting, local regression, audio segmentation, and other data-analysis tasks typically encountered by a neuroscientist. Chronux is freely available along with user tutorials, sample data, and extensive documentation from http://chronux.org/.
Keywords: Software, Time-series Analysis, Matlab
1. Introduction
Neuroscientists are increasingly gathering large time-series data in the form of multichannel electrophysiological recordings, EEG, MEG, fMRI, and optical image time-series. The availability of such data has brought with it new challenges for analysis, and has created a pressing need for the development of software tools for storing and analyzing neural signals. While sophisticated methods for analyzing multichannel time-series have been developed over the past several decades in statistics and signal processing, the lack of a unified user-friendly platform that implements these methods is a critical bottleneck in mining large neuroscientific data. Chronux is an open-source software project that aims to fill this void by providing a comprehensive software platform for the analysis of neural signals. It is a collaborative effort that draws on a number of previous research projects (Fee et al., 1996; Llinas et al., 1999; Mitra and Pesaran, 1999; Tchernichovski et al., 2000; Pesaran et al., 2002; Bokil et al., 2006a,b).
This paper provides a brief description of Chronux. We begin by discussing the architecture of Chronux and related online resources. This is followed by a description of the key routines in Chronux along with examples of their use. While the examples are drawn from our own work, we also provide extensive references to the use of Chronux in the literature. We end with a summary and a discussion of planned enhancements.
2. Materials and Methods
2.1. Software
The Chronux website at http://chronux.org/ is the central location for information about the current and all previous releases of Chronux. The home page contains links to the source code, documentation, tutorials and the discussion forum. Most of the code is written in the Matlab® (The Mathworks, Natick, MA) scripting language, with some compiled C code integrated into Matlab using mex functions. Chronux has been tested in Matlab (releases R13 to the 2007a) under the Windows, Macintosh, and Linux operating systems. Extensive online and within-Matlab help is available. The code along with sample data is available as a zip file. A smaller zip file containing just the code is also available.
The core component of Chronux is a Matlab toolbox for signal processing of neural time-series data. In addition, Chronux contains several mini-packages for spike sorting, local regression, audio segmentation, and other data-analysis tasks typically encountered by a neuroscientist. Chronux is designed to work with univariate and multivariate continuous-valued and point-process data. To install Chronux, first unzip the archive to any location on your computer and set the Matlab path to include the Chronux directory and all its subdirectories (recursively). All Chronux functions and help information will then be available from the Matlab command line. Besides Matlab itself, Chronux requires the Matlab Signal Processing Toolbox. The specscope utility depends upon the Matlab Data Acquisition Toolbox as well. The LOCFIT and Spikesort subpackages utilize Matlab mex functions, which are pre-compiled for common architectures and included with Chronux.
2.2. Analysis
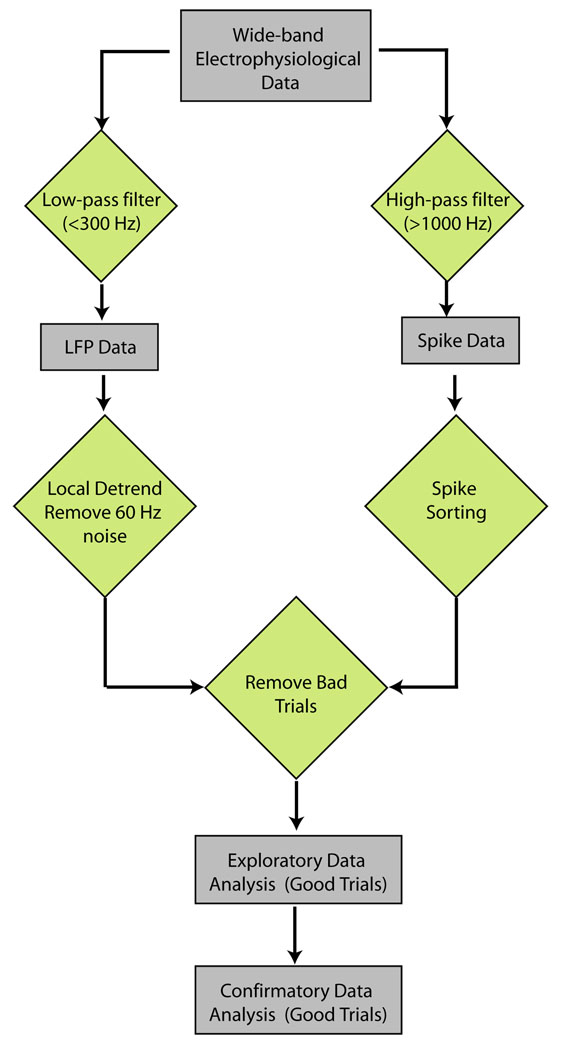
In this section we illustrate the application of Chronux to data obtained using extracellular microelectrodes, such as is the case in an electrophysiology experiment. The typical work-flow for the analysis of such data is shown in Fig. 1. The raw data consists of voltage traces obtained from one or more recording sites. The first step in the analysis is to separate the raw data into spiking activity of individual cells and background synaptic activity i.e. the local field potential (LFP). To this end, the raw data is first high-pass filtered to obtain spikes, and the spikes are then sorted to obtain single and multi-unit (cell) spiking activity. The LFP is obtained by low-pass filtering the raw data. In the next step of analysis, the LFP is preprocessed to remove local trends and line-noise at 50 or 60Hz. Bad trials, as determined by the experimental paradigm, are removed from the spiking data and the LFP, and the good trials are then used for exploratory and confirmatory data analyses. Chronux has routines to aid in each stage of the work-flow, from filtering, spike-sorting, preprocessing, and exploratory and confirmatory data analysis.
Figure 1. Typical workflow to analyze electrophysiological data.
The data is filtered to obtain LFP and spiking activity, which are in turn pre-processed to remove artifacts and to exclude bad trials. The cleaned data-set is used for exploratory and confirmatory data analysis.
2.2.1. Spike-sorting
Neuronal noise-sources and systematic variability in the shape of a spike limit the ability to sort multiple-unit waveforms recorded from nervous tissue into their single-neuron constituents. To determine such single-unit activity from electrophysiological data, Chronux implements the spike-sorting algorithm as described in Fee et al. (1996). By implementing a hierarchical clustering scheme, the algorithm efficiently sorts spikes in the presence of anisotropic noise, i.e., noise dominated by particular frequencies, and whose amplitude distribution may be non-Gaussian, which occurs when spike waveforms are a function of interspike interval.
The input to the spike-sorting functions in Chronux is a Matlab structure with the following fields, Waveforms, SpikeTimes, Fs, ThreshV, and ThreshT. Waveforms is an N * M matrix of spike data, where each row contains the voltage values from a single spike (this data can be obtained either be from a single electrode or the concatenated data from a tetrode), SpikeTimes is a row-vector of spike times (in seconds) with N elements, Fs is the sampling rate of the recording (in Hz), ThreshV is a 2-element array corresponding to the high and low voltage thresholds used to extract the spikes and ThreshT is the column index where threshold crossing occurred1.
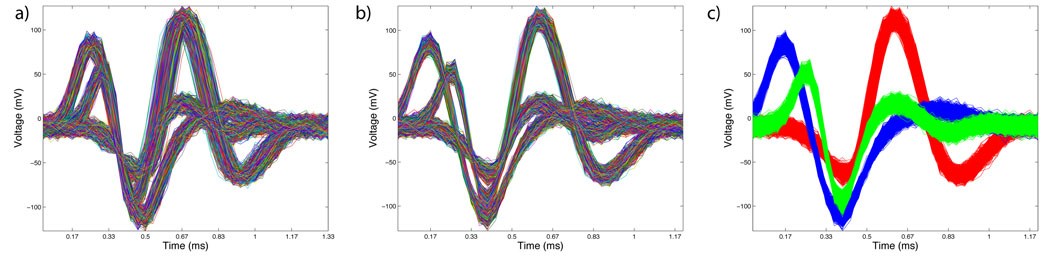
Fig. 3 shows a plot of raw simulated voltage-traces. Typically, noise on the electrode can jitter the exact time at which threshold crossing occurs and can be a significant source of variability for spikes from the same neuron. Also, density estimation techniques used in spike-sorting routines are sensitive to outliers. The spike-sorting package has routines to remove jitter and outliers. Once the data has been pre-processed, the spikes from different cells can be grouped in clusters. The algorithm in Fee et al. (1996) deals with possibly non-Gaussian data (e.g., bursting neurons) by sorting in two steps. The first step fits many local Gaussian spheres to the data to identify groups of spikes with similar shapes; which are subsequently combined into spike assignments in the second step.
Figure 3. Spikesort Package.
(a) Figure shows simulated raw voltage traces of spikes collected from a tetrode. Routines in the Spikesort mini-package allows users to sort such voltage traces to obtain spiking activity. (b) Noise on the electrode can jitter the exact time at which threshold crossing occurs and can be a significant source of variability for spikes from the same neuron. Routines in the Spikesort package allows users to remove such jitter and outliers for better clustering. (c) The algorithm implemented in Chronux deals with possibly non-Gaussian data (e.g., bursting neurons) by performing the sorting in two steps. The first step fits many local Gaussian spheres to the data to identify groups of spikes with similar shapes; which are combined into spike assignments in the second step.
2.2.2. Local Regression and Likelihood based Analysis
Regression refers to the problem of fitting functions to data. Thus, given a set of data points (xi, yi), regression attempts to find a function y = f(x) that best fits the data. These techniques may be classified as parametric or non-parametric. Parametric techniques (e.g. linear regression) assume a particular function form for f(x) and the parameters in that function are determined so as to best match the data. Therefore, the usefulness of these methods depends critically on the data being well-described by the chosen parametric form. Non-parametric techniques on the other hand do not assume any particular form globally, but instead just assume certain smoothness in the underlying function. For example, in Kernel smoothing, f(x0) is estimated as the weighted average of yi for xi within some neighborhood of x0 namely,
| (1) |
where wi denote the normalized weights, f̂(x0) denotes the estimate and I(x0) denotes the chosen neighborhood. While kernel smoothers are easy to use, they implicitly assume that the data is locally constant. In addition, they suffer from bias at the boundaries (Loader (1999)).
Local regression methods are motivated by the need to maintain the simplicity and the non-parametric approach of kernel smoothing without associated problems (Loader, 1999; Parikh, 2009; Hayden et al., 2009). In local regression, one assumes only that f(x) can be described in a local neighborhood of any point x by a low order polynomial. The coefficients of this polynomial are determined by applying the method of least squares only in that neighborhood. This procedure is usually implemented using a local weighting function, an approach that has been referred to by the acronym LOWESS (Locally Weighted Sum of Squares). Local likelihood is an adaptation of local regression to the problem of fitting probability distribution rather than functions to the data.
It is possible to show that local regression reduces the bias at the boundaries that is present in kernel smoothing methods. It is also possible to perform local regression adaptively so that the choice of the smoothing window is guided by the variations in the data. Finally, local regression and likelihood methods can be used to estimate confidence intervals on the first using cross-validation based methods.
The LOCFIT package (Loader, 1999) is included in Chronux. LOCFIT can be used for local regression, local likelihood estimation, local smoothing, density estimation, conditional hazard-rate estimation, classification and censored likelihood models. LOCFIT is written in C and is available in Chronux through a mex interface to Matlab. From a neuroscientist’s standpoint, the most important routines in LOCFIT are,
locfit.m: computes fits for regression as well as density estimation,
lfband.m: computes confidence bands, and
lfplot.m: generate plots of the fit.
The calling sequence for the locfit function to perform local regression is of the form fit=locfit(x,y,opt) where x and y denote the independent and dependent variables, respectively, and opt denotes a sequence of optional name, value pairs. When x is absent, the problem is treated as a density (or rate) estimation problem.
Here, we present two examples to demonstrate the LOCFIT package. In the first example, shown in Fig.4, we determine a local regression fit, along with confidence intervals, where the independent variable is time and the dependent variable is voltage. Such a situation would be encountered, for example, when the user wants to compute a smooth evoked response to a certain stimulus.
Figure 4. LOCFIT package.
(a) Regression fit using LOCFIT. A local regression fit, along with confidence intervals, where the independent variable was time and the dependent variable was simulated voltage from a cell. (b) Firing-rate estimate: LOCFIT can be used to estimate firing-rate given a sequence of spike times based on a local Poisson likelihood. Also shown are the LOCFIT computed 95% local confidence bands around the smoothed rate estimate. For comparison a histogram is also shown.
The second example, shown in Fig. 4, demonstrates the use of LOCFIT to estimate firing-rate given a sequence of spike times. In this case, there is only one variable, the spike-times, and LOCFIT was used to estimate the rate based on a local Poisson likelihood. Also shown are the 95% local confidence-bands around the smoothed rate-estimate. For comparison a histogram is also shown. In this case the calling sequence was fit=locfit(t,‘deg’, 3, ‘nn’,0.35), where deg controls the polynomial degree (the default is 2) and ‘nn’ denotes the nearest-neighbour bandwidth. In this example, this parameter is chosen so as to include 35% of the total number of points. The nearest-neighbour bandwidth is adaptive in that larger windows are used in regions where there are few points and smaller windows are used in regions where there is a high density of points. LOCFIT also allows the user to specify a fixed bandwidth. This is specified by an additional input argument called ‘h’. If both nearest-neighbour and fixed bandwidths are specified LOCFIT uses the larger of the two.
2.2.3. Spectral Analysis
The Spikesort and LOCFIT packages aid primarily in the preprocessing of neural data. The spectral analysis package, on the other hand, along with the associated statistical routines, help in exploratory and confirmatory data analyses.
In conventional spectral analysis, the spectrum of a time-series x(t) observed in a time-window of duration T is estimated as the square of the absolute value of its tapered fourier transform namely,
where h(t) is a taper or windowing function. Some standard choices for the windowing function are the constant, the Hanning window, the Hamming window and the Parzen window (Percival and Walden, 1993; Bokil et al., 2006c).
The conventional estimate suffers from three significant problems. First, the choice of windowing function is arbitrary. Second, for any choice of the windowing function, the estimate is biased i.e. the spectral estimate differs from the population spectrum. Third, the estimate has high variance. The last problem can be ameliorated by breaking up the observation window into shorter segments and averaging spectral estimates computed over those segments. But this is typically impossible for neural time-series which are stationary only over relatively short durations (a few hundred milliseconds). The multi-taper method provides a principled framework for addressing all these shortcomings.
In the multi-taper method, rather than choosing an arbitrary windowing function, one decides on a half-bandwidth W and asks for windowing functions that are maximally concentrated within [−W,W]. For duration T approximately K = 2TW −1 such functions, known as Slepian sequences, can be found. Thus, for W > 1/T there are multiple such functions that are well-concentrated in frequency and therefore have bias reducing characteristics. These functions are orthogonal to each other and the multi-taper estimate of the spectrum is given by using each of these tapers as windowing function and averaging the resulting spectral estimates namely,
where are the Slepian sequences or tapers. Since the functions are orthogonal, they give independent estimates of the spectrum and therefore lead to estimates that have reduced variance. In addition, by dropping one taper in turn, it is possible to compute robust, Jackknife based confidence intervals on all quantities, even for a single trial.
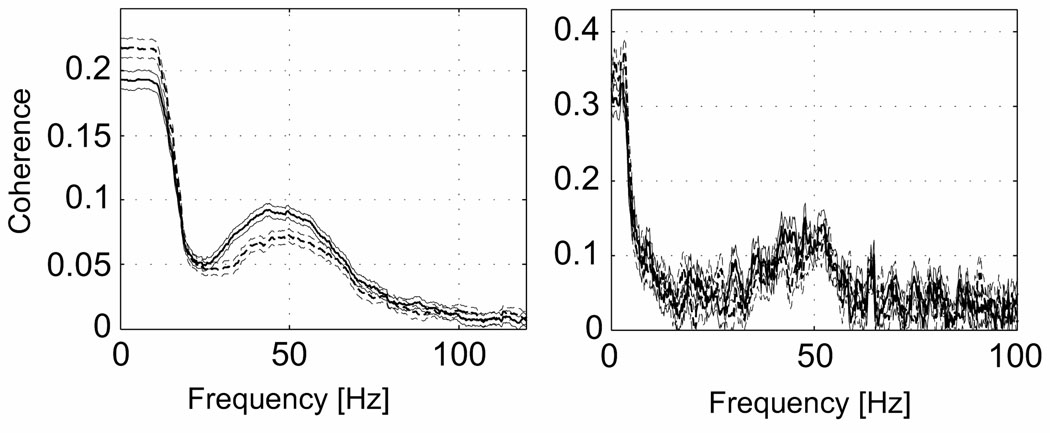
The framework discussed above can be extended to include multiple time-series and measures of their association. Figure xxx shows the multi-taper estimate and the conventional estimate of the coherence between the spike and the local field potential in area V4 of an awake macaque monkey during an attention guided task.
Chronux provides routines to estimate time-frequency spectrograms and spectral derivatives as well as measures-of-association such as cross-spectra and coherences. Univariate and multivariate signals may be analyzed as appropriate. Specialized computations such as the space-frequency singular value decomposition are available for multivariate signals. Where possible, Chronux provides confidence intervals on estimated quantities using both asymptotic formulae based on appropriate probabilistic models, and nonparametric bands based on the Jackknife method. Finally, Chronux includes statistical tests such as the two-group tests for the spectrum and the coherence, a non-stationarity test based on quadratic inverse theory, and the F-test to detect periodic signals in a colored background. The latter is particularly useful for removing 50 or 60 Hz line noise from recorded neural data (Viswanathan and Freeman, 2007). Chronux can handle both continuous-valued data as well as point-process data. The point-process data may be a sequence of values, such as times of occurrence of spikes, or a sequence of counts in successive time-bins (Maier and Ghazanfar, 2007; Maier et al., 2008; Michels et al., 2008; Harvey et al., 2009; Lehmkuhle et al., 2009; Turner et al., 2008; Zhang et al., 2009; van der Meer and Redish, 2009).
Space constraints preclude covering all aspects of the spectral analysis package here, but the functions generally have a uniform function calling signature. We illustrate three canonical routines below.
-
mtspectrumc:
As a first example, we show how to estimate the multi-taper spectrum (Humphries et al., 2006; Sarnthein and Jeanmonod, 2008) of an LFP obtained from a single-tetrode placed in the macaque lateral intraparietal area (LIP) during a memory saccade experiment. Fig. 5 shows the spectrum estimated by the Chronux function mtspectrumc. For comparison we also display the ordinary periodogram. The Matlab calling signature of the mtspectrumc function is as follows:
[S,f,Serr] = mtspectrumc(data,params);
The first argument is the data matrix in the form of times * trials or channels, while the second argument params is a structure defining the sampling frequency2, the time-bandwidth variable used to compute the tapers, and the amount of zero padding to use. It also contains flags controlling the averaging of the data over trials and the computation of the error. The time-bandwidth variable in its simplest form, is a two-dimensional vector whose first element, TW, is the time-bandwidth product, where T is the duration and W is the desired bandwidth. The second element gives the number of tapers to be used. For a given TW, the number of tapers that can be used can be at most 2TW−1. Higher order taper functions will not be sufficiently concentrated in frequency and may lead to increased broadband bias if used. For further details on the params argument, please refer to the online documentation.
The three variables returned by mtspectrumc are the estimated spectrum S, the frequencies at which the spectrum has been estimated f, and the confidence bands Serr. The spectrum is in general two-dimensional, with the first dimension being the power as a function of frequency and the second dimension being the trial or channel. The second dimension is 1 when the spectrum is averaged over the trials or channels. The confidence bands are provided as a lower and upper confidence band at a p-value set by the user. As indicated by the last letter c in its name, the routine mtspectrumc is applicable to continuous-valued data such as the local field potential or EEG. The corresponding routines for point-processes stored as a sequence of times is mtspectrumpt and for binned point-processes is mtspectrumpb.
-
mtspecgramc: The second example is a moving-window version of mtspectrumc called mtspecgramc (Friedman et al., 2006; Ghazanfar et al., 2008; Sirotin and Das, 2009). This function, mtspecgrampt, and mtspecgrampb, calculate the multitaper spectrum over a moving window with user adjustable time-width and step-size. The calling signature of this function is:
[S,t,f,Serr] = mtspecgramc(data,movingwin,params);
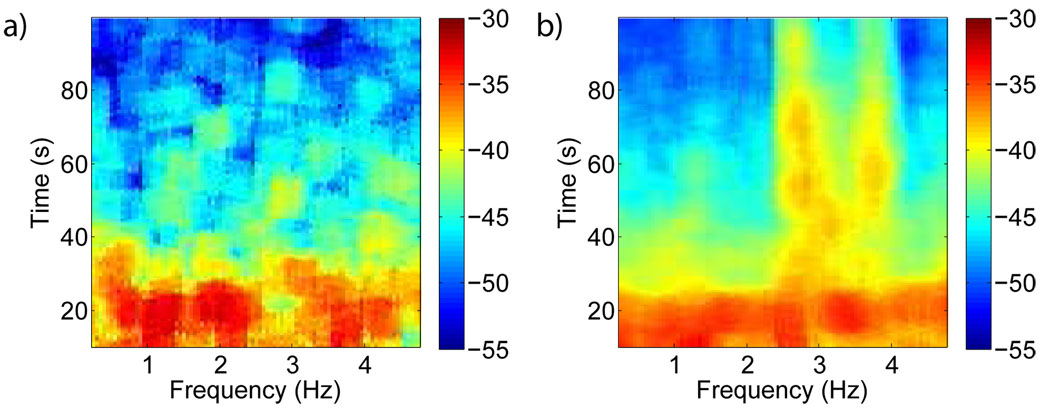
The movingwin argument is supplied as [winsize winstep] in units consistent with the sampling frequency. The returned spectrum here is in general three dimensional: times * frequency * channel or trial. Fig. 6 shows the spectrogram of the LFP (similar to Pesaran et al. (2002)) obtained during a memory saccade experiment from the macaque lateral intraparietal area (LIP). Compared to baseline activity the spectrogram reveals elevated high-frequency activity during the memory period of the task.
-
coherencycpt: In the final example (Fig. 7), we compute spike-field coherence (van der Meer and Redish, 2009; Chandrasekaran et al., 2009) for data recorded from the primary visual cortex of a monkey during an attention modulation task (Womelsdorf et al., 2006). The coherence was computed using coherencycpt. The function is called with two time-series as arguments, the continuous LFP data and the corresponding spikes, which are stored as event times (hence the letters ‘cpt’ at the end of the function name). A typical call to this function would take the form:
[C,phi,f,S12,S1,S2] = coherencycpt(data1,data2,params);
The output arguments above are the magnitude C and phase phi of the coherency, the frequencies f, the cross-spectrum S12 and individual spectra S1,S2 from which the coherence is computed. The phase convention is that the phase is positive/negative when data1 leads/lags data2. As with the spectra, confidence intervals on the magnitude and phase of the coherency can also be obtained. These quantities will be returned as additional output variables. Additional input arguments include a flag for incorporating a finite-size correction to the degree of freedom used to compute confidence intervals. This is necessary when the number of spikes is small. For the full calling signature of this and other related routines, see the online documentation.
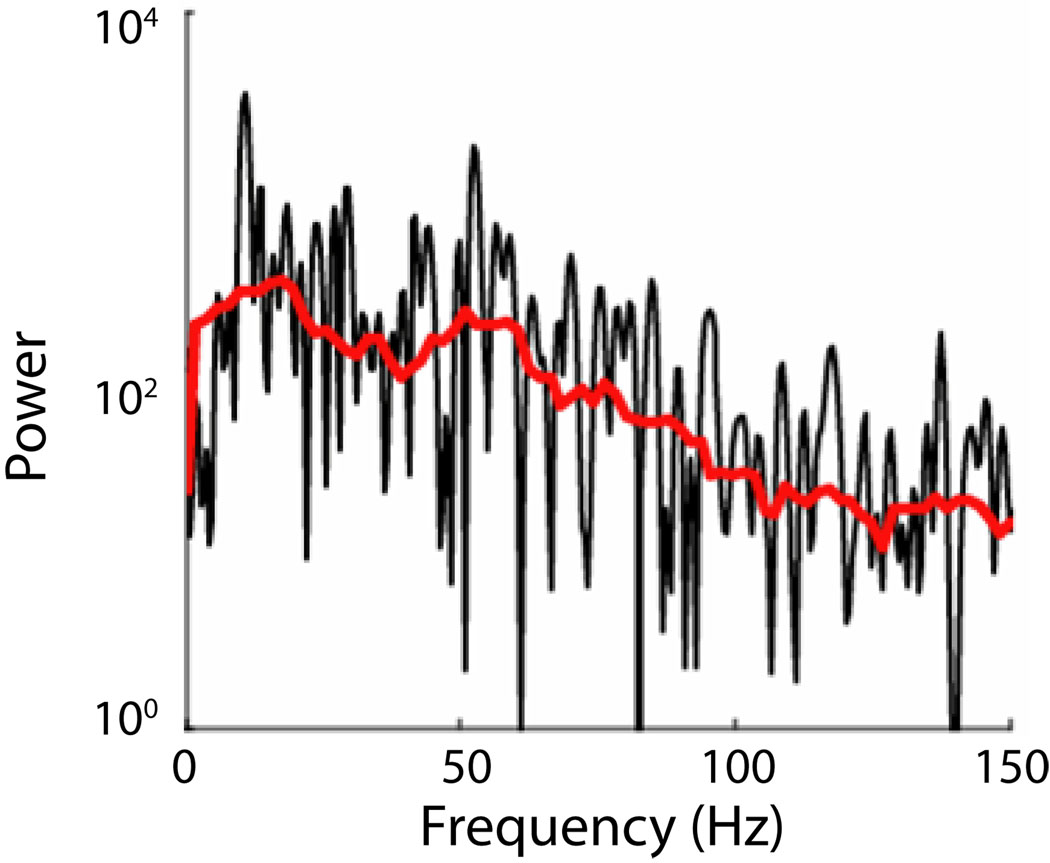
Figure 5. Spectrum.
Comparison of a periodogram (black) and multitaper estimate (red or light) of a single trial local field potential measurement from macaque during a working memory task. This estimate used 9 tapers.
Figure 6. Spectrogram.
Data from macaque monkey performing a working memory task: (a) Basline spectrogram of the LFP and (b) Spectrogram of the LFP during the memory period, which begins when the target is made visible at 2.5s, is plotted on a logarithmic scale. Sharp enhancement in high frequency power occurs during the working memory period.
Figure 7. Coherence.
The spike-field coherence recorded from visual cortex of monkey, showing significant differences between the attended and unattended conditions. In addition to the coherence in the two conditions, we also show the 95% confidence bands computed using the Jackknife.
3. Discussion
We have presented Chronux, a comprehensive software platform for the analysis of neural signals. The current version of Chronux includes a Matlab toolbox for signal processing of neural time-series data as well as several specialized mini-packages for spike-sorting, local regression, audio segmentation, and other tasks. Future developments will include improvements and extensions to the spectral analysis and spike sorting libraries, the addition of a machine-learning toolbox and the development of a modular graphical user interface to aid analysis.
As an open source project released under the GNU Public License (GPL v2), we welcome development, code contributions, bug reports, and discussion from the community. To date, Chronux has been downloaded over 10,000 times and has been used in several publications, some of which we have referenced in this paper. Questions or comments about Chronux can be posted on the discussion forum at http://chronux.org/forum/.
Figure 2. Spectrum computed using conventional methods and multi-taper methods.
Spike-LFP coherence in area V4 in awake macaque during an attention guided saccade task. Shown in each graph are the coherences for two different conditions differing only in selective attention, along with jackknife estimates of the errors. The differences can only be seen in the Multitaper estimate on the left, smoothed with a 15 Hz bandwidth. The unsmoothed conventional estimate in the right panel, with a frequency resolution of 1 Hz, is not able to differentiate between the two conditions. Note also that the multi-taper method provides Jackknife based confidence intervals (figure courtesy Pascal Fries).
Acknowledgements
The Chronux platform was initially supported by grant R01MH071744 from NIH to PPM. We thank David Kleinfeld, Murray Jarvis, Bijan Pesaran, Hiren Maniar, and David Thomson for contributions. We would also like to specially acknowledge Catherine Loader for her contributions to the Chronux project, and specifically for the LOCFIT package.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Candidate spike waveforms are typically obtained by identifying epochs in time where the voltage crosses certain thresholds
The current version of Chronux assumes continuous-valued data to be uniformly sampled
Disclosure Statement
Dr. Mitra is on the board of directors of Medametrics, LLC.
Contributor Information
Hemant Bokil, Email: hemantbokil@gmail.com.
Peter Andrews, Email: andrewsp@cshl.edu.
Jayant E. Kulkarni, Email: kulkarni@cshl.edu.
Samar Mehta, Email: sbmehta@gmail.com.
Partha Mitra, Email: mitra@cshl.edu.
References
- Bokil H, Pesaran B, Andersen RA, Mitra PP. A method for detection and classification of events in neural activity. IEEE Transactions on Biomedical Engineering. 2006a;53:1678–1687. doi: 10.1109/TBME.2006.877802. [DOI] [PubMed] [Google Scholar]
- Bokil H, Purpura K, Schofflen J-M, Thompson D, Pesaran B, Mitra PP. Comparing spectra and coherences for groups of unequal size. Journal of Neuroscience Methods. 2006b;159:337–345. doi: 10.1016/j.jneumeth.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Bokil H, Tchernichovsky O, Mitra P. Dynamic phenotypes. Neuroinformatics. 2006c;4(1):119–128. doi: 10.1385/NI:4:1:119. URL http://dx.doi.org/10.1385/NI:4:1:119. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran C, Trubanova A, Stillittano S, Caplier A, Ghazanfar AA. The natural statistics of audiovisual speech. PLoS Comput Biol. 2009 Jul;5(7):e1000436. doi: 10.1371/journal.pcbi.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee MS, Mitra PP, Kleinfeld D. Automatic sorting of multiple unit neuronal signals in the presence of anisotropic and non-gaussian variability. Journal of Neuroscience Methods. 1996;69:175–188. doi: 10.1016/S0165-0270(96)00050-7. [DOI] [PubMed] [Google Scholar]
- Friedman WA, Jones LM, Cramer NP, Kwegyir-Afful EE, Zeigler HP, Keller A. Anticipatory activity of motor cortex in relation to rhythmic whisking. J Neurophysiol. 2006 Feb;95(2):1274–1277. doi: 10.1152/jn.00945.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazanfar AA, Chandrasekaran C, Logothetis NK. Interactions between the superior temporal sulcus and auditory cortex mediate dynamic face/voice integration in rhesus monkeys. J Neurosci. 2008 Apr;28(17):4457–4469. doi: 10.1523/JNEUROSCI.0541-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Collman F, Dombeck DA, Tank DW. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature. 2009 Oct;461(7266):941–946. doi: 10.1038/nature08499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden BY, Smith DV, Platt ML. Electrophysiological correlates of default-mode processing in macaque posterior cingulate cortex. Proc Natl Acad Sci U S A. 2009 Apr;106(14):5948–5953. doi: 10.1073/pnas.0812035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MD, Stewart RD, Gurney KN. A physiologically plausible model of action selection and oscillatory activity in the basal ganglia. J Neurosci. 2006 Dec;26(50):12921–12942. doi: 10.1523/JNEUROSCI.3486-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmkuhle MJ, Thomson KE, Scheerlinck P, Pouliot W, Greger B, Dudek FE. A simple quantitative method for analyzing electrographic status epilepticus in rats. J Neurophysiol. 2009 Mar;101(3):1660–1670. doi: 10.1152/jn.91062.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas RR, Ribary U, Jeanmonod D, Kronberg E, Mitra PP. Thalamocortical dysrhythmia: A neurological and neuropsychiatric syndrome characterized by magnetoencephalography. PNAS. 1999;96:15222–15227. doi: 10.1073/pnas.96.26.15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loader C. Local Regression and Likelihood. Springer; 1999. [Google Scholar]
- Maier A, Wilke M, Aura C, Zhu C, Ye FQ, Leopold DA. Divergence of fmri and neural signals in v1 during perceptual suppression in the awake monkey. Nat Neurosci. 2008 Oct;11(10):1193–1200. doi: 10.1038/nn.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier JX, Ghazanfar AA. Looming biases in monkey auditory cortex. J Neurosci. 2007 Apr;27(15):4093–4100. doi: 10.1523/JNEUROSCI.0330-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels L, Moazami-Goudarzi M, Jeanmonod D, Sarnthein J. Eeg alpha distinguishes between cuneal and precuneal activation in working memory. Neuroimage. 2008 Apr;40(3):1296–1310. doi: 10.1016/j.neuroimage.2007.12.048. [DOI] [PubMed] [Google Scholar]
- Mitra PP, Pesaran B. Analysis of dynamic brain imaging data. Biophysical Journal. 1999;76:691–708. doi: 10.1016/S0006-3495(99)77236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H. Ph.D. thesis. The University of Michigan; 2009. On improving the eectiveness of control signals from chronic microelectrodes for cortical neuroprostheses. [Google Scholar]
- Percival DB, Walden WT. Spectral Analysis for Physical Applications: Multitaper and Conventional Univariate Techniques. Cambridge University Press; 1993. [Google Scholar]
- Pesaran B, Pezaris J, Sahani M, Mitra P, Andersen R. Temporal structure in neuronal activity during working memory in macaque parietal cortex. Nature Neuroscience. 2002;5:805–811. doi: 10.1038/nn890. [DOI] [PubMed] [Google Scholar]
- Sarnthein J, Jeanmonod D. High thalamocortical theta coherence in patients with neurogenic pain. Neuroimage. 2008 Feb;39(4):1910–1917. doi: 10.1016/j.neuroimage.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Sirotin YB, Das A. Anticipatory haemodynamic signals in sensory cortex not predicted by local neuronal activity. Nature. 2009 Jan;457(7228):475–479. doi: 10.1038/nature07664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernichovski O, Nottebohm F, Ho C, Pesaran B, Mitra P. A procedure for an automated measurement of song similarity. Animal Behavior. 2000;59:1167–1176. doi: 10.1006/anbe.1999.1416. [DOI] [PubMed] [Google Scholar]
- Turner GC, Bazhenov M, Laurent G. Olfactory representations by drosophila mushroom body neurons. J Neurophysiol. 2008 Feb;99(2):734–746. doi: 10.1152/jn.01283.2007. [DOI] [PubMed] [Google Scholar]
- van der Meer MAA, Redish AD. Low and high gamma oscillations in rat ventral striatum have distinct relationships to behavior, reward, and spiking activity on a learned spatial decision task. Front Integr Neurosci. 2009;3:9. doi: 10.3389/neuro.07.009.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan A, Freeman RD. Neurometabolic coupling in cerebral cortex reflects synaptic more than spiking activity. Nat Neurosci. 2007 Oct;10(10):1308–1312. doi: 10.1038/nn1977. [DOI] [PubMed] [Google Scholar]
- Womelsdorf T, Fries P, Mitra PP, Desimone R. Gamma-band synchronization in visual cortex predicts speed of change detection. Nature. 2006;439:733–736. doi: 10.1038/nature04258. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lin S-C, Nicolelis MAL. Acquiring local field potential information from amperometric neurochemical recordings. J Neurosci Methods. 2009 May;179(2):191–200. doi: 10.1016/j.jneumeth.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]