Abstract
Molecular-level information processing1,2, or computing, is essential for ‘smart’ in vivo nanosystems. Natural molecular computing, such as messenger RNA (mRNA) synthesis regulation by special proteins called transcription factors (TFs)3,4, may inspire engineered systems leading to the next generation of nanobiotechnological and nanomedical applications. Synthetic pathways5–15 have already implemented logical control of mRNA levels by certain TF combinations. Here we show an alternative approach toward general-purpose control of mRNA and protein levels by logic integration of transcription factor input signals in mammalian cells. The factors regulate synthetic genes coding for small regulatory RNAs – microRNAs – that in turn control mRNA of interest (i.e., output) via RNA interference pathway. Simple nature of these modular interactions allows in theory to implement any arbitrary logic relation between the TFs and the output16. We construct, test, and optimize increasingly complex circuits with up to three TF inputs, establishing a platform for in-vivo molecular computing.
Our circuits contain sensory, computational and actuation modules resembling those found in engineered control systems (Fig. 1a). In the sensory module, individual TF inputs are ‘transduced’ into microRNA (miR) molecules by directly regulating the expression of engineered, non-native miR genes17 from simple TF-responsive promoters. In the computational module, miRs expressed from these genes down-regulate the output protein-coding transcripts via RNA interference (RNAi), thereby implementing a complex and robust logic control16. The output protein, a fluorescent reporter in our experiments, could also be an actuator that triggers a physiological response.
Figure 1. Design elements of synthetic circuits.
a, An example of a logic circuit with multiple transcription factor inputs and a fluorescent ZsYellow protein output. Three different system modules are shown. Transcription factor inputs A through F, promoters PA through PF, miRs miR-a to miR-f and output-encoding mRNA transcripts containing miR targets Ta to Tf are indicated. Pointed arrows denote activation, and blunt arrows represent repression. Elements in gray denote potential directions for circuit scale-up. b, Detailed structure and shorthand notation for the constructs used in this report. ‘Ex’ denotes exons, and other structural elements are as indicated.
The circuit utilizes the following output dependency on the miR levels:
where miR-a, -b, … target the first output-encoding transcript, c, d,… target the second, etc. The dependency holds because a high level of at least one output-encoding transcript is required to generate the output protein, and none of the miRs that target this transcript can be present16. Linking miR expression to a transcriptional activator or repressor requires the corresponding TF to be absent or present, respectively, for the miR to be absent. In Fig. 1a, TF-A activates miR-a (pointed arrow), TF-B represses miR-b (blunt arrow), TF-D represses miR-d and TF-E activates miR-e. The overall circuit logic becomes
This approach can in principle lead to a fully-predictable regulatory program, conditional on the availability of appropriately-controlled single-input promoters. The specific form of the relation between the inputs and the output, i.e., groups of AND operators connected by OR operators, is known to be universal. Therefore, any digital molecular computation with multiple TF inputs and a single protein output can be expressed using this universal form and implemented using our circuits18.
We experimentally tested our approach using the well-characterized TFs rtTA, Rheo and LacI-Krab as inputs. Known promoters (pTRE, pRheo and pCAGop) regulated by these TFs were used to drive the expression of miR genes (FF3, FF4 and FF6) whose active miR products were identical to active strands of previously tested small interfering RNA (siRNA)16. These miRs target established sequences16 fused into the three prime untranslated region (3’-UTRs) of the mRNA coding for the ZsYellow fluorescent reporter output (Fig. 1, Supplementary Text), causing mRNA degradation and concomitant decrease in the ZsYellow level. Using these well-defined components allowed us to focus on the proof-of-principle of our method.
We developed three different circuits with two or three TF inputs and measured their response to all possible combinations of On and Off input states. The experiments were performed in immortalized human embryonic kidney cells HEK293. Cells were transiently transfected with the invariant part of the circuit, that is, the output and miR genes (Supplementary Table 1). Depending on the desired state (On or Off) of a TF input in a particular measurement, we added, or withheld, the DNA plasmid that constitutively expresses this factor. Each experiment with a particular input combination and appropriately-modified output mRNAs (circuit) was accompanied by a control experiment (control) with the same inputs but with either an unrelated sequence replacing the correct miR target or an absent miR target in the 3’-UTR of the output mRNAs. All non-specific output changes caused by differences in transfection efficiency, non-specific binding of TFs to circuit components, off-target RNAi, and/or other factors are accounted for in the observed output change in the control experiments. Accordingly, On/Off ratios were normalized by the corresponding ratios in the controls to extract the underlying circuit behaviour. Note that our circuits exhibit cell-to-cell variability typical of transient transfections, forming a Poisson distribution with an estimated mean ranging upward of 3–4 plasmid copies per cell19. The strongest signal (or lack thereof) comes from the cells at the high end of this distribution, where the plasmid copy number is three to four times higher than the mean and all the plasmids are likely to be co-transfected.
Prior to testing multi-input circuits, we calibrated their single-input ‘branches’ using Cytomegalovirus (CMV) immediate-early promoter-driven output (Supplementary Text). To facilitate calibration, miR-encoding RNAs were embedded as introns between two exons coding for a fluorescent protein, such that miR expression was accompanied by the generation of a fluorescent protein serving as a miR level reporter. Following the initial adjustment of the transfection conditions, we implemented circuits with two TF inputs. The first one used the activator rtTA and the repressor LacI-Krab as inputs. LacI-Krab repressed a pCAGop-driven AmCyan-FF4 while rtTA activated pTRE-driven DsRed-FF3. The level of the fluorescent protein ZsYellow is expected to be high when rtTA is Off and LacI-Krab is On, or using logic notation, ‘ZsYellow = NOT(rtTA) AND LacI-Krab’. This circuit behaved as intended (Fig. 2a, Supplementary Fig. 5). The corrected On/Off ratios with different input combinations varied from ~ 5 to ~2, with an average of ~4.2 (Supplementary Table 3). To further confirm that these changes were RNAi-specific, we used flow cytometry to measure miR reporters and output levels in individual cells (Fig. 2b, Supplementary Methods). The data show that increasing miR levels lead to a decrease in the output protein, after correction for non-specific effects. Consistent with this result, inspection of microscopy images shows minimal overlap between the miR reporters and the output in the circuit experiments, as opposed to significant overlap in the controls.
Figure 2. Experimental implementation of two-input regulatory programs.
Plasmid amounts are given in Supplementary Table 1. Red and green bars (mean ± s.d.) correspond to the predicted Off and On states, respectively. a–c, Regulatory program ‘ZsYellow = NOT(rtTA) AND LacI-Krab’. a, From left to right: circuit schematics; representative microscopy snapshots and quantitative performance of the circuit with CMV-driven output. b, Surface plot of control-corrected ZsYellow output as a function of miR-FF3 and FF4 levels judged by the levels of DsRed and AmCyan, respectively. c, Images and quantitative analysis of the circuit with EF1a-driven output. d, Regulatory program ‘ZsYellow = NOT(rtTA) AND NOT (Rheo)’ implemented with EF1a-driven output. Left: circuit schematics; right: anticipated circuit behavior, microscopy images and quantitative analysis.
Modularity is the key attribute of our design approach, where only a few well-defined changes are required to alter the regulatory program. To demonstrate this feature, we replaced the pCAGop promoter of the AmCyan-FF4 fusion with a pRheo promoter leading to a new program: ‘ZsYellow = NOT(rtTA) AND NOT(Rheo)’. The system behaved as expected, generating On/Off ratios ranging from 2- to 6-fold (Supplementary Fig. 6, Supplementary Table 3) and showing a correct relation between miR and output levels (Supplementary Fig. 7).
While these data qualitatively fitted expectations, the On/Off ratios of the circuits required quantitative improvement. Low ratios were caused by high output levels in the Off state (leakage) and other non-specific effects. To improve performance we placed the output under the control of alternative constitutive promoters, and found that the promoter EF1a significantly reduced leakage in the Off state (Supplementary Text). We attribute this improvement to a transcriptional delay of the output caused by an intron in the EF1a promoter as well as to weaker absolute expression from this promoter (Supplementary Fig. 9). Therefore we used EF1a-driven output in two-input circuits without changing the remaining components. For the ‘ZsYellow = NOT(rtTA) AND LacI-Krab’ regulatory program using the EF1a promoter, the average On/Off ratio was ~27 (Fig. 2c, Supplementary Fig. 10). The average ratio was ~13 in the regulatory program ‘ZsYellow = NOT (rtTA) AND NOT (Rheo)’, a significant improvement over the CMV-driven version (Fig. 2d, Supplementary Fig. 11).
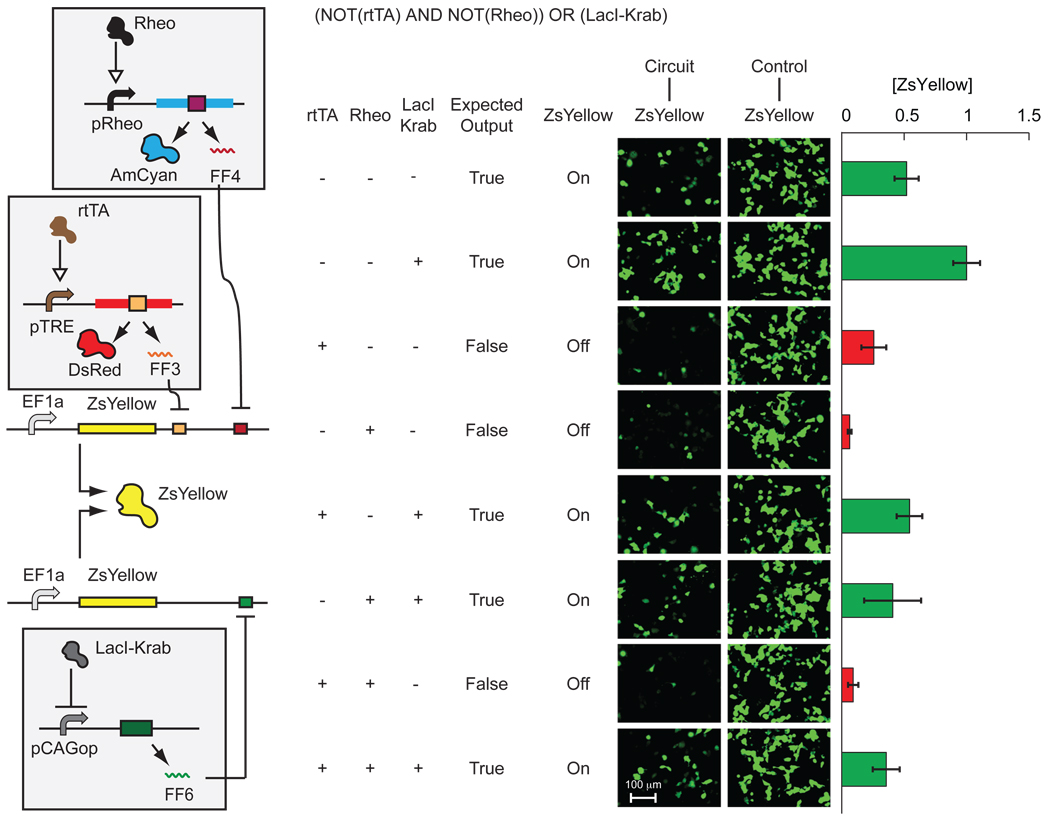
Next, we created a three-input circuit to implement the program ‘ZsYellow = (NOT(rtTA) AND NOT(Rheo)) OR (LacI-Krab)’. In our measurements (Fig. 3, Supplementary Table 4, Supplementary Fig. 12), seven out of eight possible input combinations behaved as predicted. One expected Off output (rtTA is On) was relatively high yet it was still 30% lower than the lowest measured On output. This result is consistent with the relatively low On/Off ratio of the pTRE-DsRed-FF3 construct observed in single- and two-input circuits (Fig. 2, Supplementary Fig. 8). The average ZsYellow level, normalized to the highest measured output, was ~0.46 among the On states and ~0.14 among the Off states, roughly as expected (0.5 and 0, respectively, because the highest output is generated by two separate transcripts). The same circuit implemented with CMV-driven output showed greatly deteriorated performance, as expected from two-input circuit comparison (Supplementary Fig. 13). Indeed, some of the Off states are higher than some of the On states and the average On level was ~0.5 of the maximum compared to 0.3 in the Off state.
Figure 3. Experimental implementation of a three-input regulatory program.
Plasmid amounts are given in Supplementary Table 1. From left to right: circuit schematics; table of TF input states, the expected outputs and fluorescent output levels of ZsYellow; microscopy images of ZsYellow output; and quantitative output intensity as obtained by FACS analysis. Red and green bars (mean ± s.d.) indicate anticipated Off and On states, respectively.
While our circuits’ primary purpose is to respond to changing TF levels, in some cases those proteins can be controlled by molecular cofactors. The activators used in our experiments require cofactors (Dox for rtTA and RSL1 for Rheo) that we used at saturating concentrations. One might consider the TFs and the cofactors as separate inputs, in which case the circuit logic would include both these species. For example, the ‘NOT(rtTA) AND NOT(Rheo)’ program would become ‘(NOT(Dox) OR NOT(rtTA)) AND (NOT(RSL1) OR NOT(Rheo))’. There are a total of 24 = 16 input combinations in this formula and setting some of them permanently to False (or True) allows us to operate with a part of the truth table (Supplementary Table 5). When the TFs are constitutively expressed, the circuits could be used as logic integrators of their cognate cofactors, as shown experimentally in Supplementary Fig. 14. Independent control of TF and cofactor inputs further expands the complexity of our circuits and suggests their use as tuneable gene-expression platforms or sophisticated sensors of small molecule expression profiles in cells.
Our study has demonstrated that bottom-up construction can lead to complex systems with predictable behaviour. At the same time we observed that the different components are not strictly modular and that non-specific background processes can lead to deviations from expectation, a well-recognized challenge facing the construction of synthetic biological networks10,20. More efficient decoupling of the synthetic components from the endogenous processes21 and from each other could reduce these deviations. Fluctuations may also limit scalability, and they need to be contained by proper tuning of individual interactions such that the input effect becomes efficiently saturated at the low end of its naturally-occurring distribution in the cell population. For example, RNAi knock-down could be augmented with transcriptional repression22. In addition, one could use auxiliary circuitry such as bistable switches23, feedback, and ultrasensitive response24. Resolving these challenges will enable larger circuits, as well as cascades in which the output of an upstream computation serves as an input to a downstream circuit to engineer increasingly complex regulatory programs.
Future applications of our systems with exogenous TFs include tuneable gene expression systems. Extending the inputs to endogenous TFs will lead to reporters and modulators of cell states, ‘intelligent’ therapeutic agents and new nanotechnology applications. In particular, our tools for in vivo molecular computation could become essential components of smart nanobiomaterials that interact with the biological environment1. In this context, we envision distributed and non-distributed modes of system operation. In the distributed mode the TF sensors are delivered to individual cells while the computational and the actuation modules are encapsulated in artificial nanovesicles supporting RNAi and protein translation25. The miRs generated by the sensors would be shuffled to the nanovesicles using naturally-secreted exosomes26, and the encapsulated components would respond by producing a desired protein that would either be secreted or delivered to specific cells via targeted endocytosis27. In a non-distributed mode, the entire network could be compartmentalized in channel-equipped nanocarriers28 with cell-free extracts supporting transcription/translation/RNAi in order to detect secreted inputs such as TFs or their ligands, and generate appropriate response species.
Methods
Cell culture
Maintenance of human embryonic kidney cells (293-H) and plasmid transfection were performed as described16. The summary of plasmid amounts can be found in Supplementary Table 1. All transfections were performed in 12-well plates using 1 mL of growth medium per well. To increase transfection efficiency 25 µM Chloroquine (Sigma) was added 1 h prior to transfection. 3 hours after transfection the ligands RSL1 and/or Doxycycline were added at 1 µM and 1 µg/mL final concentrations, respectively. For fluorescence-activated cell sorting (FACS) analysis, cells were either prepared as described16 or trypsinized with 0.1 ml 0.25% trypsin-EDTA. After addition of 0.5 ml DMEM (Invitrogen) the cell suspension was collected and used directly for FACS analysis. The output was assayed 48 h post-transfection and quantified using population-based readouts (Supplementary Methods).
siRNA molecules
SiRNAs were obtained from Dharmacon and processed as described previously16.
Recombinant DNA constructs
The details of the cloning strategy are described in Supplementary Methods. Most of the constructs were made using standard restriction-ligation methods, and the exon-intron constructs were made using seamless cloning approaches29 with SII-type SapI restriction enzyme (New England Biolabs). MiR stem-loop templates were designed based on published siRNA sequences16 following the guidelines of the pPRIME method30.
Microscopy measurements
All microscopy images were taken from live cells grown in glass-bottom wells (Mattek) in the transfection medium supplemented with 10% FBS. We used Zeiss Axiovert 200 microscope equipped with shutter filter wheels, Prior mechanized stage and an environmental chamber (Solent) held at 37 °C during measurements. The images were collected by an Orca ERII camera cooled to −60 °C, in the high precision (14 bit) mode using a 20× PlanApochromat NA 0.8, PH2 objective. The collection settings for the fluorophores were S500/20× (excitation) and S535/30m (emission) filters for ZsYellow, S430/25× (excitation) and S470/30m (emission) filters for AmCyan and S565/25× (excitation) and S650/70m (emission) for DsRed. A dichroic mirror 86004v2bs (Chroma) was used for ZsYellow and AmCyan. The dichroic mirror 86021bs (Chroma) was used for DsRed. Data collection and processing were performed by the Metamorph 7.0 software (Molecular Devices). All images illustrating a given circuit and its control underwent the same processing.
FACS measurement and data analysis
50,000–150,000 cells from each transfected well of 3 biological replicas were analyzed on a BD LSRII flow analyzer. ZsYellow was measured using a 488 nm laser, a 505 nm longpass filter and a 530/30 emission filter. AmCyan was measured with a 405 nm laser, a 460 nm longpass filter and a 480/40 emission filter. DsRed was measured with 561 nm laser and a 585/20 emission filter. The data were compensated and analyzed as described in Supplementary Methods. To account for non-specific changes in the output, we corrected the data as follows: the mean values of the ZsYellow output for the data set obtained by flow cytometry for ‘circuit’ experiments were divided by the mean ZsYellow outputs for the corresponding control experiments prior to averaging over three biological replicas and normalizing by the highest measured ‘On’ state.
Supplementary Material
Acknowledgements
We thank anonymous reviewers for thoughtful comments. M. L. is a recipient of Deutsche Forschungs Gemeinschaft scholarship. The research was funded by the Bauer Fellows program and by the NIGMS grant GM068763 for National Centres of Systems Biology. M.L. acknowledges her father R. Leisner who passed away as this manuscript was prepared for submission: One learns most from those one loves (Goethe).
Footnotes
Author contribution. Y.B. designed research and supervised the project. M. L., L. B., J. L., Z. X. and Y.B. performed research. M. L., Y.B., J. L. and L.B. wrote the manuscript.
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Shapiro E, Benenson Y. Bringing DNA computers to life. Sci. Amer. 2006;294:44–51. [PubMed] [Google Scholar]
- 2.Jungmann R, Renner S, Simmel FC. From DNA nanotechnology to synthetic biology. Hfsp Journal. 2008;2:99–109. doi: 10.2976/1.2896331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genetics. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 4.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 5.Weiss R, Homsy GE, Knight TF. In: Evolution as Computation: DIMACS Workshop. Landweber LF, Winfree E, editors. Springer; 1999. pp. 275–295. [Google Scholar]
- 6.Guet CC, Elowitz MB, Hsing WH, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 7.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 8.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 2007;3 doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronson JE, Mazur WW, Cornish VW. Transcription factor logic using chemical complementation. Mol. Biosystems. 2008;4:56–58. doi: 10.1039/b713852k. [DOI] [PubMed] [Google Scholar]
- 10.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nature Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 11.Sayut DJ, Niu Y, Sun LH. Construction and enhancement of a minimal genetic AND logic gate. Applied and Environ. Microbiol. 2009;75:637–642. doi: 10.1128/AEM.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nature Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechn. Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 14.Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U. Plasticity of the cis-regulatory input function of a gene. Plos Biology. 2006;4:555–561. doi: 10.1371/journal.pbio.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox RS, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 2007;3:11. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinaudo K, et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda Y, Kawasaki H, Taira K. Construction of microRNA-containing vectors for expression in mammalian cells. Meth. Mol. Biol. 2006;338:167–173. doi: 10.1385/1-59745-097-9:167. [DOI] [PubMed] [Google Scholar]
- 18.Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc. Natl Acad. Sci. USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwake G, et al. Predictive Modeling of Non-Viral Gene Transfer. Biotech. Bioeng. 2010;105:805–813. doi: 10.1002/bit.22604. [DOI] [PubMed] [Google Scholar]
- 20.Marguet P, Balagadde F, Tan CM, You LC. Biology by design: reduction and synthesis of cellular components and behaviour. J. Royal Soc. Interface. 2007;4:607–623. doi: 10.1098/rsif.2006.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An WL, Chin JW. Synthesis of orthogonal transcription-translation networks. Proc. Natl Acad. Sci. USA. 2009;106:8477–8482. doi: 10.1073/pnas.0900267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 23.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 24.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nature Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Liu SJ, Bleris L, Benenson Y. Logic integration of mRNA signals by an RNAi-based molecular computer. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.Gullotti E, Yeo Y. Extracellularly activated nanocarriers: a new paradigm of tumor targeted drug delivery. Mol. Pharmaceutics. 2009;6:1041–1051. doi: 10.1021/mp900090z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broz P, et al. Toward intelligent nanosize bioreactors: A pH-switchable, channel-equipped, functional polymer nanocontainer. Nano Letters. 2006;6:2349–2353. doi: 10.1021/nl0619305. [DOI] [PubMed] [Google Scholar]
- 29.Lu Q. Seamless cloning and gene fusion. Trends in Biotechnology. 2005;23:199–207. doi: 10.1016/j.tibtech.2005.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.