Abstract
Agrobacterium tumefaciens is a soil phytopathogen that elicits neoplastic growths on the host plant species. In nature, however, Agrobacterium also may encounter organisms belonging to other kingdoms such as insects and animals that feed on the infected plants. Can Agrobacterium, then, also infect animal cells? Here, we report that Agrobacterium attaches to and genetically transforms several types of human cells. In stably transformed HeLa cells, the integration event occurred at the right border of the tumor-inducing plasmid's transferred-DNA (T-DNA), suggesting bona fide T-DNA transfer and lending support to the notion that Agrobacterium transforms human cells by a mechanism similar to that which it uses for transformation of plants cells. Collectively, our results suggest that Agrobacterium can transport its T-DNA to human cells and integrate it into their genome.
Genetic transformation of plant cells by Agrobacterium tumefaciens is the only known natural example of trans-kingdom DNA transfer. In nature, Agrobacterium introduces several oncogenic genes into the host plant, leading to formation of tumors (1), and in the laboratory this microorganism is used widely for plant genetic engineering (2, 3). Agrobacterium infection requires the presence of two genetic components located on the bacterial tumor-inducing (Ti) plasmid: the transferred DNA (T-DNA), which is introduced into the plant cell genome, and the virulence (vir) region composed of seven loci—virA, virB, virC, virD, virE, virG, and virH encoding most components of the protein apparatus for T-DNA transfer. In addition, several bacterial chromosomal virulence (chv) genes participate in the early stages of Agrobacterium attachment to the plant cells (reviewed in refs. 4 and 5–7).
The infection process begins by chemotactic attraction of Agrobacterium toward wounded sites on the host plant, attachment of the bacteria to the plant cell surface, and activation of the T-DNA transfer machinery (5, 6). During the attachment, Agrobacterium first loosely binds to the plant surface, and then it produces cellulose fibrils that tighten the binding and allow attachment of additional bacteria (8, 9). To transfer its T-DNA into the host plant cell, the attached Agrobacterium uses a complex transport machinery encoded by the vir region and specifically activated by signal molecules contained in exudates from wounded-plant cells. The best studied inducer of vir-gene expression is acetosyringone (AS), a monocyclic phenolic molecule (10).
Besides its natural plant hosts, Agrobacterium has been shown to transfer DNA to other microorganisms such as yeast (11–13), several species of filamentous fungi (14), and a cultivated mushroom (14). However, whether Agrobacterium infects higher eukaryotes other than plants remains unknown. Here we address this question by demonstrating that Agrobacterium can transform human cells genetically by integrating its T-DNA into the cellular genome. Several aspects of the transformation process characteristic for T-DNA transfer to plants, e.g., bacterial attachment and involvement of virulence proteins (reviewed in ref. 7), also were necessary for T-DNA transfer to human cells.
Materials and Methods
Binary T-DNA Vector.
The pNeo plasmid contained a neomycin resistance gene, encoding neomycin phosphotransferase, under the control of the simian-virus-40 early promoter and Herpes-simplex virus thymidine kinase polyadenylation signal (derived from pEGFP-C1, CLONTECH) subcloned into the Klenow-blunted EcoRI and HindIII sites of the pPZP221-based binary vector (15), between its right and left nopaline-type T-DNA borders.
Bacterial Strains and Host Cell Lines.
A. tumefaciens-strain C58C1, harboring Ti-plasmid pGV3850 (16), was used as host for the pNeo binary vector. Strain C58NT1, which does not contain a Ti plasmid (17), was used as control. Agrobacterium strains carrying Ti plasmids with individual vir loci inactivated by Tn3-HoHo1 transposon insertions were described previously (18). Agrobacterium strains ME42 and ME60, with mutated chromosomal virulence genes chvA and chvB, respectively, were also described (19, 20).
Human HeLa R19 cells, human embryonic kidney (HEK) 293 cells, and clonal pheochromocytoma PC12 neuronal cells, supplied by the Stony Brook Tissue Culture Facility (Stony Brook, NY), were cultured at 37°C in a humidified atmosphere of 5% CO2/95% air in DMEM (GIBCO/BRL) supplemented with 10% (vol/vol) FBS (GIBCO/BRL). Plant-cell suspension culture of Petunia hybrida (line 3704, Hook Vilm, kindly provided by S. Izhar, Volcani Center, Bet-Dagan, Israel; ref. 21) was grown at 25°C and 110-rpm shaking in UM medium (21, 58) under a 12-h daylight cycle. Petunia protoplasts were prepared from the cell cultures as described (22).
T-DNA Transfer.
A total of 3 × 105 to 5 × 105 human cells was plated on 22 × 22-mm coverslips (105 cells per coverslip), which were then placed in a 90-mm Petri dish and cultured for 1 day in the presence of 100 μg/ml penicillin/streptomycin mixture (GIBCO/BRL) to avoid contamination. Then, 2 to 5 h before the experiment, the medium was replaced with 10 ml of the fresh medium without antibiotics.
Agrobacterium was grown for 16 h at 28°C and 250-rpm shaking in 5-ml yeast extract/peptone (YEP) medium (23) supplemented with appropriate antibiotics to maintain plasmid vectors (100 μg/ml spectinomycin for pNeo and 100 μg/ml carbenicillin for insertion mutants of Ti plasmid). Then, 100 μl of these bacterial cultures was used to inoculate 5 ml of YEP and was grown for 4 to 6 h at 28°C in the presence of 100 μm of AS [from a 100-mM stock solution in DMSO (Aldrich)] to induce vir-gene expression. Note that although minimal medium is more conducive to the induction process than the rich YEP medium, rich media are often used to pregrow Agrobacterium during genetic transformation of different plant species (24, 25). Because for HeLa-cell transformation we wished to employ the conditions most close to those used for plant transformation, YEP rather than a minimal medium was used.
For transformation, 100 μl of preinduced bacterial culture (105–106 bacteria) was added to each tissue-culture plate with human cells, followed by incubation for 48 h at 37°C and 5% CO2 with 0.1 μM AS. After coincubation, cells were washed twice with 10 ml PBS and grown in 10 ml DMEM supplemented with 10% FBS, 0.2 mM cefotaxime to kill Agrobacterium, and 600 μg/ml geneticin (G418, GIBCO/BRL) to select for transformants. The selective medium was changed every 2 days; when no bacteria were detected by microscopic examination of the cell cultures, cefotaxime treatment was stopped, and cell cultures were maintained in the presence of geneticin to select for stable transformants. The transformed cell foci were grown to confluence and used for further analyses. Each transformation experiment was performed 2–6 times and usually included 5–10 culture plates per system. Microsoft EXCEL software was used to calculate standard deviations between the results of individual experiments.
Bacterial Attachment.
HeLa cells (105) were cultured for 16 h on a 22 × 22-mm coverslip in a 6-well dish and transferred to the fresh medium. Then, 2 h later, bacterial cultures were added and coincubated with HeLa cells for 16 h at 37°C as described above for the T-DNA transfer experiments. For attachment to petunia protoplasts, the coincubation was performed at 28°C in suspension cultures. After incubation, the samples were examined and recorded by using a Zeiss Axioskop microscope equipped with MTI CCD-300T-RC video camera (for HeLa cells) or Axioplan 2 microscope equipped with Nikon Culitix 950 camera (for petunia protoplasts).
Southern Blot Analysis of T-DNA Transgenes in HeLa Cells.
Genomic DNA was extracted from six 90-mm plates of confluent cell cultures derived from a single stably transformed cell line by using the High Pure PCR Template Preparation kit (Roche Molecular Biochemicals) according to the manufacturer's instructions. The purified DNA (20 μg) was digested with Eco0109I, HindIII, EcoRI, BamHI, PstI, or SalI restriction enzymes. In the T-DNA region of pNeo binary vector, which is expected to be transferred and integrated into the HeLa-cell genome, there are two cleavage sites for Eco0109I, a single cleavage site for HindIII, EcoRI, BamHI, and PstI, and no sites for SalI. The digested DNA was analyzed by Southern blot hybridization by using standard techniques (26) and a 740-bp radiolabeled probe corresponding to the neomycin resistance gene of pNeo; this probe fragment did not contain the recognition sites for restriction enzymes used to digest the genomic DNA.
Thermal Asymmetric Interlaced (TAIL) PCR Amplification of the Integration Junction.
TAIL PCR was performed as described for mapping of genomic sequences flanking T-DNA insertions in Arabidopsis (27). Briefly, genomic DNA from the same stably transformed HeLa cell line used for the Southern blot analysis was purified (see above), and the putative T-DNA integration junction was amplified by using three consecutive rounds of PCR. The first PCR was carried out in a 50-μl volume containing 20 ng DNA, 0.1 nM dNTP, 2.5 mM of the T-DNA-specific sense primer TR1 (5′-GCACTGGCCGTCGTTTTACAACG-3′), 2.5 mM of a degenerate antisense primer AD2 (5′-NTCGASTWTSGWGTT-3′, in which N is A or C or G or T, S is C or G, and W is A or T), and 2 units of TKARA EX-Taq polymerase (Panvera , Madison WI) with 5 μl of EX-TaqI0X reaction buffer. After 5 min of denaturation at 92°C, the junction fragment was amplified for 5 cycles of 60 sec at 94°C, 60 sec at 62°C, and 2.5 min at 72°C followed by 22 cycles of 30 sec at 94°C, 60 sec at 68°C, 2.5 min at 72°C, 30 sec at 94°C, 60 sec at 44°C, and 2.5 min at 72°C. The resulting mixture was diluted 10 times and subjected to the second PCR under the same conditions but by using a downstream-nested T-DNA-specific sense primer TR2 (5′-GGGAAAACCCTGGCGTTACCC-3′). Then 10 times-diluted products of the second reaction were amplified the third time by using another downstream-nested T-DNA-specific sense primer TR3 (5′-CCTTTCGCCAGCTGGCGTAATAGCG-3′). The resulting amplified T-DNA–HeLa-DNA junction fragment was subcloned into a pBluescript SK(+) (Stratagene) plasmid, and its DNA sequence was determined.
Results
Agrobacterium Attachment to HeLa Cells.
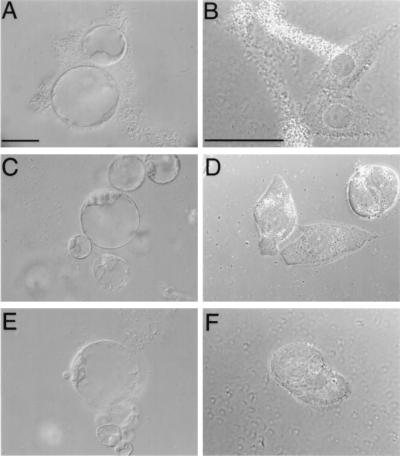
Infection of host cells by Agrobacterium must begin from attachment of the bacteria to the host cell. Thus, we examined the ability of Agrobacterium to associate with HeLa cells and compared it to the well characterized Agrobacterium plant-cell binding (9, 28, 29). Fig. 1A shows that Agrobacterium attaches to petunia protoplasts, enmeshing itself and the plant cells into aggregates (9, 28, 29). Fig. 1B demonstrates a similar attachment pattern with characteristic bacterial aggregates at and around the host cells observed after incubation of Agrobacterium with HeLa cells.
Figure 1.
Agrobacterium attachment to petunia protoplasts and HeLa cells. (A and B) Wild-type Agrobacterium incubated with petunia protoplasts and HeLa cells, respectively. (C and D) chvA mutant of Agrobacterium incubated with petunia protoplasts and HeLa cells, respectively. (E and F) chvB mutant of Agrobacterium incubated with petunia protoplasts and HeLa cells, respectively. (Bars = 50 μm.)
To investigate whether Agrobacterium binding to HeLa cells requires the same bacterial factors as its attachment to plant cells, we used two avirulent Agrobacterium mutants, chvA and chvB. These mutations, which are located on the bacterial chromosome, result in inability of the bacteria to bind to host cells (19, 20). Neither chvA nor chvB mutants (Fig. 1 C and E, respectively) attached to plant protoplasts or HeLa cells (Fig. 1 D and F, respectively).
T-DNA Transfer from Agrobacterium to Human Cells.
To establish whether Agrobacterium transfers T-DNA to human cells, we constructed the binary vector pNeo. This plasmid carries a T-DNA that contains a neomycin resistance gene driven by the Simian-virus-40 early promoter, which is specific for mammalian cells. pNeo was introduced then into the Agrobacterium helper-strain C58C1, harboring the Ti plasmid pGV3850 (16), which serves as a source of vir-gene products in a binary transformation system. For control experiments, pNeo was introduced into the Agrobacterium C58NT1 strain cured of its Ti plasmid (17).
Coincubation with AS-induced Agrobacterium resulted in genetic transformation of HeLa cells as evidenced by formation of geneticin-resistant cell lines (Table 1). The transformation frequency was comparable to that of calcium phosphate stable transfection of mammalian cells (Table 1) but lower than the highly efficient lipofectin-mediated transformation method (30). The resistance was retained through many cell duplications (usually the resistant lines were maintained for 2 to 6 weeks and then discarded), suggesting stable integration of the transgene. Incubation with Agrobacterium lacking the Ti plasmid but harboring pNeo never gave rise to antibiotic-resistant HeLa cells (Table 1), indicating that presence of the vir genes was essential for genetic transformation. The requirement for the Ti plasmid also indicates that potentially spontaneous uptake by HeLa cells of pNeo DNA released from bacteria was not responsible for transformation. Also, the expression of neomycin resistance could not be attributed to contaminating bacteria, because this gene in the pNeo vector is driven by the simian-virus-40 early promoter; such viral promoters are specific strictly for animal cells (31) and are not expressed in bacteria. As expected, no transformants were obtained when the cells were incubated with the purified pNeo plasmid alone, in the absence of Agrobacterium (Table 1). Surprisingly, however, Agrobacterium stably transformed HeLa cells, albeit with lower efficiency, even in the absence of exogenously added vir-inducer AS (Table 1), suggesting that human cells and/or their growth medium may have a certain capacity for vir-gene induction. Alternatively, two different mechanisms may be responsible for transformation of HeLa cells by Agrobacterium: vir dependent and vir independent.
Table 1.
Genetic transformation of human cells by Agrobacterium
| Plasmids | Host-cell type | Total number of geneticin-resistant transformants | Efficiency of transformation per 106 cells, ±SD* |
|---|---|---|---|
| pTi–pNeo in Agrobacterium | HeLa | 140 | 17 ± 4 |
| pTi–pNeo in Agrobacterium without preinduction | HeLa | 74 | 9 ± 2 |
| pNeo in Agrobacterium | HeLa | 0 | <0.1 |
| pNeo alone | HeLa | 0 | <0.1 |
| pNeo Ca-Phosphate | HeLa | 179 | 19 ± 6 |
| pTi–pNeo in Agrobacterium | HEK393 (kidney) | 167 | 18 ± 6 |
| pNeo in Agrobacterium | HEK393 (kidney) | 0 | <0.1 |
| pNeo alone | HEK393 (kidney) | 0 | <0.1 |
| pTi–pNeo in Agrobacterium | PC12 (neuronal) | 112 | 12 ± 3 |
| pNeo in Agrobacterium | PC12 (neuronal) | 0 | <0.1 |
| pNeo alone | PC12 (neuronal) | 0 | <0.1 |
Microsoft excel software was used to calculate standard deviations between two to six individual experiments, each of which included 5–10 culture plates per system.
Next, we examined the ability of Agrobacterium to infect other human cell lines. Table 1 shows that HEK393 cells and neuronal PC12 cells also were susceptible to genetic transformation by Agrobacterium, forming geneticin-resistant transformants. Similarly to HeLa cells, transformation of HEK393 and PC12 depended on the presence of the Ti plasmid (Table 1).
Evidence for T-DNA Integration into the HeLa-Cell Genome.
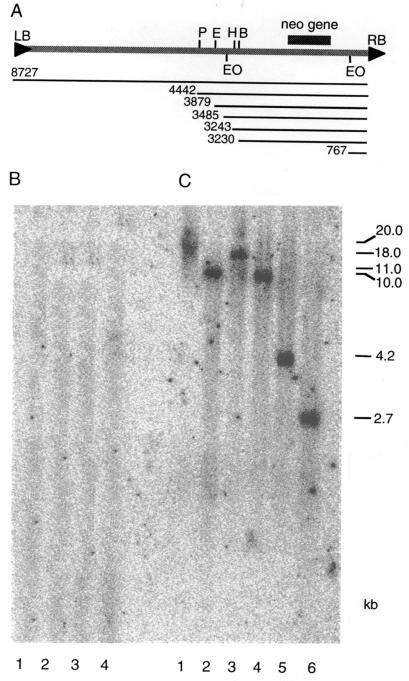
Genomic DNA was isolated from one of the stable geneticin-resistant HeLa cell lines (transformant 7–14), digested with SalI, PstI, BamHI, EcoRI, HindIII, or Eco0109I restriction enzymes, and analyzed by Southern blot hybridization by using the neomycin resistance gene as probe (Fig. 2A). For control experiments, DNA isolated from a wild-type untransformed HeLa-cell line was used. Fig. 2 shows that alhough no signal was detected in the DNA from wild-type cells (B), a neomycin-specific signal was present in the transformed cell genome (C). Digestion with SalI, which has no recognition sites within the entire T-DNA region of pNeo, produced a single high-molecular-weight band (Fig. 2B, lane 1), indicating a single integration site for the T-DNA.
Figure 2.
Southern blot analysis of a geneticin-resistant HeLa cell line stably transformed by Agrobacterium. (A) Organization of the pNeo plasmid. The T-DNA region of pNeo with the restriction sites used for the Southern blot analysis is shown. P, H, E, B, and EO indicate PstI, BamHI, EcoRI, HindIII, or Eco0109I restriction enzyme sites, respectively. LB and RB indicate left and right T-DNA borders, respectively. Thin lines indicate the length of the entire T-DNA and distances of each restriction site from RB in base pairs. The location of the neomycin resistance (neo) gene is shown. The pNeo backbone (6 kb, not shown) outside of the T-DNA region is that of the parental pPZP221 vector (ref. 15; GenBank accession no. U10490). (B) Wild-type untransformed cell line. (C) Stably transformed cell line. Lanes 1 to 6 show digestions with SalI, PstI, BamHI, EcoRI, HindIII, or Eco0109I restriction enzymes, respectively. The blots were hybridized with a 740-bp radiolabeled probe corresponding to the neomycin resistance gene of pNeo (see A). The sizes of the restriction-fragment bands were calculated based on standards and are indicated on right in kilobase pairs (kb).
To estimate the number of T-DNA inserts within this integration site, we performed digestion with PstI, BamHI, EcoRI, and HindIII, each of which has a single recognition site in the T-DNA but outside of the neomycin resistance gene. The resulting single bands with variable sizes (Fig. 2B, lanes 2–5, respectively) suggested integration of a single T-DNA copy. Also, that the size of these fragments differed from that predicted for restriction digestion of the complete pNeo plasmid (14,727 bp) or its free unintegrated T-DNA region (8,727 bp; Fig. 2A), suggests that these fragments represent the neomycin resistance gene linked to the chromosomal sequences. In the undigested samples, the signal coincided with very high-molecular-weight DNA (data not shown), confirming integration of the T-DNA into the chromosomal DNA. In control digestion with Eco0109I, which has two recognition sites bracketing the neomycin resistance gene, a fragment corresponding to the predicted T-DNA fragment (Fig. 2A) was released (Fig. 2B, lane 6).
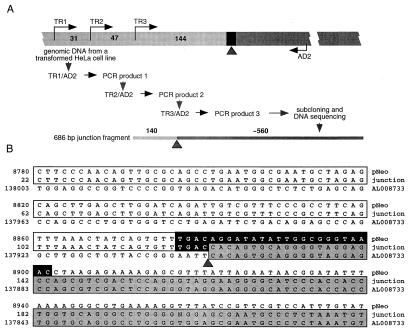
To confirm the integration event further, we used TAIL PCR (ref. 27; outlined in Fig. 3A) to isolate the T-DNA–HeLa DNA junction from the same cell line with a single T-DNA insertion analyzed in Fig. 2. Unlike integration of transposable elements, which is generally precise, T-DNA insertions are imprecise; nevertheless, in plants, the junction points at the right T-DNA border are more consistent and occur close to or within the border sequence, whereas the left border junctions can be spread over much longer segments, often deleting the border itself (reviewed in refs. 32–34). Similar preservation of the right border region relative to the left border region was observed during Agrobacterium-mediated transformation of yeast (13) and filamentous fungi (14). Thus, we focused on isolation of the right border junction site. Our TAIL PCR experiments using DNA from wild-type untransformed HeLa cells as negative control produced no specific fragments (data not shown), whereas the DNA from the transformed HeLa-cell line yielded only one fragment that contained both HeLa and Agrobacterium sequences (Fig. 3A).
Figure 3.
Cloning of the T-DNA–HeLa DNA integration junction. (A) TAIL-PCR cloning strategy. Wide bar illustrates the genomic DNA from a transformed HeLa cell line used as a template, black rectangle indicates the region of the right T-DNA border, and numbers indicate distances between the nested sense PCR primers TR1 and TR2, TR2 and TR3, and TR3 and the right border. AD2 is the degenerate antisense primer expected to anneal within HeLa-cell DNA. Narrow bar indicates the amplified junction fragment in outline, and numbers indicate the size of its corresponding T-DNA and HeLa-DNA components. Light and dark segments of both bars indicate T-DNA and HeLa-cell DNA, respectively, whereas the arrowhead indicates the integration point between these sequences. For primer sequences and description of PCRs, see Materials and Methods. (B) Nucleotide sequence alignment of the right T-DNA-border region of pNeo, the isolated integration junction from an Agrobacterium-transformed HeLa-cell DNA, and the human genomic DNA (GenBank accession no. AL008733). All sequences are shown in the 5′ to 3′ direction. The pNeo sequence is based on the right border region of the parental pPZP221 vector (15) (GenBank accession no. U10490) and the human DNA sequence is from clone RP1–163G9 from chromosome 1p36.2–36.3. The consensus nopaline-type right T-DNA-border sequence, described in refs. 33 and 57, is indicated by a black box. Homology of the junction fragment to pNeo is indicated by open boxes and to the human DNA by shaded boxes. Arrowhead indicates the integration point at which the right T-DNA border is fused to the human DNA. Note also that the DNA sequences upstream of the position 22 and downstream of the position 221 in the 686-bp junction fragment were identical to the corresponding regions of the pNeo and AL008733 DNA, respectively (data not shown).
The DNA sequence of this fragment was determined and compared with the sequence of the right T-DNA-border region of the pNeo binary vector and to the known chromosomal DNA sequences of the human genome (Fig. 3B). This analysis identified a pNeo sequence within the junction fragment (open boxes in Fig. 3B). It also demonstrated that, from the 25-bp right border sequence of pNeo (black box in Fig. 3B), four bases remained at the junction site, consistent with the observations that the nopaline-type right border is cleaved between its third and fourth base during generation of the transferable copy of the T-DNA, the T strand (35) (potentially, the fourth border base of the junction may have been derived from a duplication of the adjacent identical base in the HeLa DNA; see Fig. 3B).
That the cleaved right border sequence was found at the integration junction and, importantly, that no pNeo sequences to the right of the right T-DNA border, i.e., non-T-DNA sequences, were found in the junction site (Fig. 3B) is indicative of the true T-DNA integration rather than nonspecific insertion of plasmid DNA. Fig. 3 also shows that, at the junction point (arrowhead), the T-DNA sequence was linked to a HeLa-DNA sequence virtually identical to the region between bases 137,901 and 137,583 of the human DNA sequence from clone RP1–163G9 on chromosome 1p36.2–36.3 (GenBank accession no. AL008733; shadowed boxes), directly demonstrating integration of the right T-DNA border into the host cell genomic DNA. There was no significant homology between the inserted pNeo sequence and its integration site within the HeLa-cell DNA (Fig. 3B), suggesting that T-DNA integration in mammalian cells, as in fungi (13, 14) and plants (34), is sequence-independent.
We could not compare the TAIL PCR data directly to the Southern analysis data, because the length of the amplified junction fragment (686 bp) was significantly smaller than the smallest restriction fragment detected in Southern blotting (≈2.7 kb, see Fig. 2, lane 6). Nevertheless, the nucleotide sequence of the junction fragment (Fig. 3B) was consistent with the Southern blot data, because it did not contain Eco0109I, HindIII, EcoRI, BamHI, PstI, or SalI restriction sites, which if present would have produced a different hybridization pattern on the Southern blot. Collectively, our Southern blot results and sequence analysis of the integration junction suggest that Agrobacterium can transport its T-DNA to HeLa cells and integrate it into their genome.
Effects of vir-Gene Mutations on T-DNA Transfer to HeLa Cells.
Most protein products of the vir genes play critical roles in the infection process and thus are indispensable for transformation of plant cells by Agrobacterium. If Agrobacterium transforms plant and human cells by the same molecular mechanism, HeLa-cell transformation should be blocked by mutations inactivating the major vir genes. Our observations that an Agrobacterium strain lacking its Ti plasmid, which supplies the vir genes that encode the T-DNA transfer machinery, was unable to transform all three human cell lines (Table 1) support this notion. We directly confirmed the involvement of vir genes in transformation of HeLa cells by using a series of Agrobacterium mutant strains, in which one of the five major vir loci, virA, virB, virG, virD, or virE, was inactivated by a Tn3-HoHo1 transposon insertion (18). Specifically, virA and virG were inactivated in pTi237 and pTi19 mutants, respectively, whereas virB, virD, and virE were inactivated in two independent mutants each, i.e., pTi243 and pTi30, pTi311 and pTi304, and pTi361 and pTi358, respectively (18, 36). In addition, Agrobacterium mutants ME42 and ME60 in the chromosomal virulence genes chvA and chvB, respectively (19, 20), also were tested.
These experiments demonstrated that all tested strains with mutations in the vir or chv genes lost their transforming ability, producing no geneticin-resistant cell lines under our experimental conditions. In control experiments, wild-type Agrobacterium infected HeLa cells, generating 62 geneticin-resistant transformants, corresponding to the transformation efficiency of 16 ± 3 stable transformants per 106 cells. Thus, Agrobacterium-mediated transformation of human cells may involve at least some of the specific virulence protein components of the T-DNA-transport machinery that are required for transformation of plant cells.
Discussion
Agrobacterium cells attached to both HeLa cells and plant protoplasts. Furthermore, Agrobacterium binding to, and genetic transformation of, HeLa cells depended on the presence of chvA and chvB genes, known to be required for bacterial attachment to plant hosts (19, 20). Generally, pathogenic bacteria have been proposed to employ similar mechanisms to attach themselves to the surfaces of plant and animal host cells (37). For example, plant vitronectin-like proteins likely function as host cell receptors used by Agrobacterium (37), and mammalian vitronectins also play a role in host colonization by several pathogenic bacteria, such as streptococci, Staphylococcus aureus, etc. (38–40). Thus, although binding in itself does not imply specific host-pathogen recognition, it is consistent with the ability of Agrobacterium to stably transform human cells. Such genetic transformation was demonstrated directly by production of transgenic mammalian cell lines resistant to neomycin after coincubation with Agrobacterium.
Sequence analysis of the pNeo–HeLa DNA junction showed that the integration event occurred at the right T-DNA border, suggesting bona fide T-DNA transfer and lending support to the notion that Agrobacterium transformed HeLa cells by a mechanism similar to what it uses for transformation of plants cells. Indeed, Agrobacterium-mediated HeLa-cell transformation required the activity of vir genes necessary for plant transformation (18). Specifically, mutants in the virA, virB, virG, virD, and virE genes, which are avirulent on plant hosts (18), also failed to transform HeLa cells. During Agrobacterium infection of plant cells, these vir genes play critical roles. Specifically, VirA and VirG sense plant phenolics and promote transcriptional activation of the vir genes (reviewed in ref. 41), VirB proteins are required for T-DNA export from Agrobacterium into the host cell (reviewed in ref. 42), and VirD and VirE proteins generate and associate with a mobile single-stranded copy of the T-DNA and mediate its import into the host cell nucleus and possibly its integration into the host genome (reviewed in ref. 7). Thus, Agrobacterium likely uses its plant-transforming protein machinery for transformation of HeLa cells. This assumption implies that such basic processes as nuclear import and DNA integration are similar enough between plant and animal cells to be taken advantage of by Agrobacterium. Indeed, the VirD2 protein localizes to the cell nucleus both in plant (43–47) and animal systems (48, 49), and its cellular receptor, AtKAPα, also is conserved between these organisms (50). Also, although its nuclear import in plant and animals remains controversial (48, 49), VirE2 together with VirD2 facilitate nuclear import of DNA into mammalian cell nuclei in vitro (48). For example, in mammalian cells, VirE2 may act to shape the T-DNA into a transferable form and/or protect it from cellular nucleases (48, 51, 52).
An interesting aspect of Agrobacterium-mediated transformation of mammalian cells is that it occurred at 37°C. Previously, exposure to high temperatures has been shown to reversibly inhibit VirA, which is involved in perceiving the vir-inducing plant signals (53), and/or other components of the T-DNA transfer machinery (54). Our transformation protocol includes pregrowth of Agrobacterium at 28°C in the presence of the vir-inducer AS. Because at 28°C vir genes are well expressed (18), the preinduced Agrobacterium cells may be competent already for transformation, allowing the T-DNA transfer even at higher temperatures.
Our results also suggest that uninduced Agrobacterium can still transform HeLa cells, although with significantly lower efficiency. This transforming activity may be due to various reasons such as vir-gene induction by the components of the HeLa-cell growth medium, e.g., phenol red pH indicator or FBS, or even HeLa-cell exudates. Furthermore, a secondary vir-gene-independent mechanism such as taking up of Agrobacterium by endocytosis may contribute also to the transformation process. However, because the T-DNA integration occurred at the predicted specifically nicked border sequence, the VirD2 and VirD1 proteins, which potentiate this nick (reviewed in ref. 7), most likely were involved. This notion is strengthened further by our observations that vir-gene mutants of Agrobacterium or Agrobacterium carrying the T-DNA-containing pNeo plasmid but not the vir-genes-containing Ti plasmid were unable to transform HeLa cells. Clearly, additional experiments will be required to elucidate the exact mechanism by which Agrobacterium transforms mammalian cells. Presently, however, there is little doubt that Agrobacterium T-DNA can be transferred to and integrated into the mammalian cell genome.
That Agrobacterium carrying a neomycin resistance gene within its T-DNA generated stable antibiotic-resistant lines of human cells expands its potential host range from plants, yeast, and filamentous fungi to mammalian cells. Here, transformation of human cells has been observed in laboratory conditions; whether it may be relevant biologically in nature remains unknown. Interestingly, Agrobacterium or Agrobacterium-related species have been suggested to be involved in several human diseases (55, 56).
Acknowledgments
We thank Dr. Stanton Gelvin for advice and generosity in providing numerous Agrobacterium strains, Dr. Nir Ohad for help with microscopy of petunia protoplasts, and Dr. Joseph Roitelman for help with HeLa-cell cultures. We also thank the anonymous reviewers and the member-editor for constructive criticisms and suggestions that improved this manuscript. This work was supported by grants from the National Institutes of Health, National Science Foundation Functional Genomic Initiative, U.S. Department of Agriculture, and U.S.–Israel Binational Science Foundation (to V.C.); by a grant from U.S.–Israel Binational Research and Development Fund (BARD; to V.C. and Y.G.); by a scholarship from the Council for Tobacco Research (to C.D.); and by BARD postdoctoral fellowships (to T.K. and T.T.). V.C. was supported also in part by a Research Fellow Award from BARD.
Abbreviations
- Ti
tumor inducing
- T-DNA
transferred DNA
- AS
acetosyringone
- TAIL
thermal asymmetric interlaced
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041327598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041327598
References
- 1.Gaudin V, Vrain T, Jouanin L. Plant Physiol Biochem (Paris) 1994;32:11–29. [Google Scholar]
- 2.Bevan M W. Nucleic Acids Res. 1984;12:1811–1821. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage P, Walden R, Draper J. In: Plant Genetic Transformation and Gene Expression, A Laboratory Manual. Draper J, Scott R, Armitage P, Walden R, editors. Oxford: Blackwell Scientific; 1988. pp. 1–67. [Google Scholar]
- 4.Hooykaas P J J, Beijersbergen A G M. Annu Rev Phytopathol. 1994;32:157–179. [Google Scholar]
- 5.Zambryski P. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:465–490. [Google Scholar]
- 6.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzfira T, Citovsky V. Mol Plant Pathol. 2000;1:201–212. doi: 10.1046/j.1364-3703.2000.00026.x. [DOI] [PubMed] [Google Scholar]
- 8.Matthysse A G, Yarnall H, Boles S B, McMahan S. Biochim Biophys Acta. 2000;1490:208–212. doi: 10.1016/s0167-4781(99)00250-x. [DOI] [PubMed] [Google Scholar]
- 9.Matthysse A G. Crit Rev Microbiol. 1986;13:281–307. doi: 10.3109/10408418609108740. [DOI] [PubMed] [Google Scholar]
- 10.Stachel S E, Messens E, Van Montagu M, Zambryski P. Nature (London) 1985;318:624–629. [Google Scholar]
- 11.Piers K L, Heath J D, Liang X, Stephens K M, Nester E W. Proc Natl Acad Sci USA. 1996;93:1613–1618. doi: 10.1073/pnas.93.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P J J. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bundock P, Hooykaas P J. Proc Natl Acad Sci USA. 1996;93:15272–15275. doi: 10.1073/pnas.93.26.15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Groot M J, Bundock P, Hooykaas P J, Beijersbergen A G. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 15.Hajdukiewicz P, Svab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 16.Zambryski P, Joos H, Genetello C, Leemans J, Van Montagu M, Schell J. EMBO J. 1983;2:2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garfinkel D J, Nester E W. J Bacteriol. 1980;144:732–743. doi: 10.1128/jb.144.2.732-743.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stachel S E, Nester E W. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douglas C J, Halperin W, Nester E W. J Bacteriol. 1982;152:1265–1275. doi: 10.1128/jb.152.3.1265-1275.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douglas C J, Staneloni R J, Rubin R A, Nester E W. J Bacteriol. 1985;161:850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark E M, Izhar S, Hanson M R. Mol Gen Genet. 1985;199:440–445. [Google Scholar]
- 22.Ballas N, Zakai N, Loyter A. Exp Cell Res. 1987;170:228–234. doi: 10.1016/0014-4827(87)90132-7. [DOI] [PubMed] [Google Scholar]
- 23.Lichtenstein C, Draper J. In: DNA Cloning: A Practical Approach. Glover D M, editor. Vol. 2. Oxford: IRL; 1986. pp. 67–119. [Google Scholar]
- 24.Tzfira T, Jensen C S, Wangxia W, Zuker A, Altman A, Vainstein A. Plant Mol Biol Rep. 1997;15:219–235. [Google Scholar]
- 25.Clough S J, Bent A F. Plant J. 1998;16:753–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 26.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1987. [Google Scholar]
- 27.Liu Y G, Mitsukawa N, Oosumi T, Whittier R F. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 28.Matthysse A G, Wyman P M, Holmes K V. Infect Immun. 1978;22:516–522. doi: 10.1128/iai.22.2.516-522.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthysse A G, Yarnall H A, Young N. J Bacteriol. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felgner P L, Gadek T R, Holm M, Roman R, Chan H W, Wenz M, Northrop J P, Ringold G M, Danielsen M. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foecking M K, Hofstetter H. Gene. 1986;45:101–105. doi: 10.1016/0378-1119(86)90137-x. [DOI] [PubMed] [Google Scholar]
- 32.Binns A N, Thomashow M F. Annu Rev Microbiol. 1988;42:575–606. [Google Scholar]
- 33.Zambryski P. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 309–333. [Google Scholar]
- 34.Tinland B. Trends Plant Sci. 1996;1:178–184. [Google Scholar]
- 35.Wang K, Stachel S E, Timmerman B, Van Montagu M, Zambryski P. Science. 1987;235:587–591. doi: 10.1126/science.235.4788.587. [DOI] [PubMed] [Google Scholar]
- 36.Stachel S E, An G, Flores C, Nester E W. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner V T, Matthysse A G. J Bacteriol. 1992;174:5999–6003. doi: 10.1128/jb.174.18.5999-6003.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsson M, Wadstrom T. FEMS Microbiol Immunol. 1990;65:55–62. doi: 10.1111/j.1574-6968.1990.tb03479.x. [DOI] [PubMed] [Google Scholar]
- 39.Valentin-Weigand P, Grulich-Henn J, Chhatwal G S, Muller-Berghaus G, Blobel H, Preissner K T. Infect Immun. 1988;56:2851–2855. doi: 10.1128/iai.56.11.2851-2855.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chhatwal G S, Preissner K T, Muller-Berghaus G, Blobel H. Infect Immun. 1987;55:1878–1883. doi: 10.1128/iai.55.8.1878-1883.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winans S C, Mantis N J, Chen C Y, Chang C H, Han D C. Res Microbiol. 1994;145:461–473. doi: 10.1016/0923-2508(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 42.Christie P J. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tinland B, Koukolikova-Nicola Z, Hall M N, Hohn B. Proc Natl Acad Sci USA. 1992;89:7442–7446. doi: 10.1073/pnas.89.16.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Citovsky V, Warnick D, Zambryski P. Proc Natl Acad Sci USA. 1994;91:3210–3214. doi: 10.1073/pnas.91.8.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrera-Estrella A, Van Montagu M, Wang K. Proc Natl Acad Sci USA. 1990;87:9534–9537. doi: 10.1073/pnas.87.24.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossi L, Hohn B, Tinland B. Mol Gen Genet. 1993;239:345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- 47.Howard E, Zupan J, Citovsky V, Zambryski P. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 48.Ziemienowicz A, Gorlich D, Lanka E, Hohn B, Rossi L. Proc Natl Acad Sci USA. 1999;96:3729–3733. doi: 10.1073/pnas.96.7.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guralnick B, Thomsen G, Citovsky V. Plant Cell. 1996;8:363–373. doi: 10.1105/tpc.8.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ballas N, Citovsky V. Proc Natl Acad Sci USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Citovsky V, Guralnick B, Simon M N, Wall J S. J Mol Biol. 1997;272:718–727. doi: 10.1006/jmbi.1997.1230. [DOI] [PubMed] [Google Scholar]
- 52.Citovsky V, Wong M L, Zambryski P. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin S, Song Y N, Deng W Y, Gordon M P, Nester E W. J Bacteriol. 1993;175:6830–6835. doi: 10.1128/jb.175.21.6830-6835.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fullner K J, Nester E W. J Bacteriol. 1996;178:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauter C. Lancet. 1995;346:1433. doi: 10.1016/s0140-6736(95)92451-5. [DOI] [PubMed] [Google Scholar]
- 56.Chalandon Y, Roscoe D L, Nantel S H. Bone Marrow Transplant. 2000;26:101–104. doi: 10.1038/sj.bmt.1702470. [DOI] [PubMed] [Google Scholar]
- 57.Gielen J, Terryn N, Villarroel R, Van Montagu M. J Exp Bot. 1999;50:1421–1422. [Google Scholar]
- 58.Uchimya H, Murashige T. Plant Physiol. 1974;54:936–944. doi: 10.1104/pp.54.6.936. [DOI] [PMC free article] [PubMed] [Google Scholar]