Abstract
Nanoparticles have emerged as key materials for biomedical applications because of their unique and tunable physical properties, multivalent targeting capability, and high cargo capacity1,2. Motivated by these properties and by current clinical needs, numerous diagnostic3–10 and therapeutic11–13 nanomaterials have recently emerged. Here we describe a novel nanoparticle targeting platform that uses a rapid, catalyst-free cycloaddition as the coupling mechanism. Antibodies against biomarkers of interest were modified with trans-cyclooctene and used as scaffolds to couple tetrazine-modified nanoparticles onto live cells. We show that the technique is fast, chemoselective, adaptable to metal nanomaterials, and scalable for biomedical use. This method also supports amplification of biomarker signals, making it superior to alternative targeting techniques including avidin/biotin.
It is often the case that affinity ligands and bioconjugation strategies must be optimized for each new preparation to maximize the binding properties of the targeted conjugates14,15. To streamline these efforts, there remains a critical need to develop advanced conjugation techniques that simplify operation, as well as extend detection limits by improving the efficiency of targeting and amplifying marker signals. Moreover, successful translation into clinical settings will require simple scale-up to successfully process tens or hundreds of samples. Here, we explore a modular and broadly applicable targeting platform based on bioorthogonal chemistry. We were particularly interested in (i) using a biocompatible chemistry with a fast reaction rate, (ii) using reaction partners with very small ‘footprints’ to maximize the number of covalent binding sites, and (iii) developing universal labelling approaches that build upon the vast array of available monoclonal antibodies. We reasoned that such a strategy would be valuable in further improving nanoparticle targeting.
We and others have recently described a covalent, bioorthogonal reaction between a 1,2,4,5-tetrazine (Tz) and a trans-cyclooctene (TCO) and used it for small molecule labelling16–18. The [4 + 2] cycloaddition is fast, chemoselective, does not require a catalyst, and proceeds in serum. We hypothesized that this chemistry could be adapted to targeting nanoparticle sensors in different configurations to improve binding efficiency and detection sensitivity. We have named this technique ‘bioorthogonal nanoparticle detection’ (BOND).
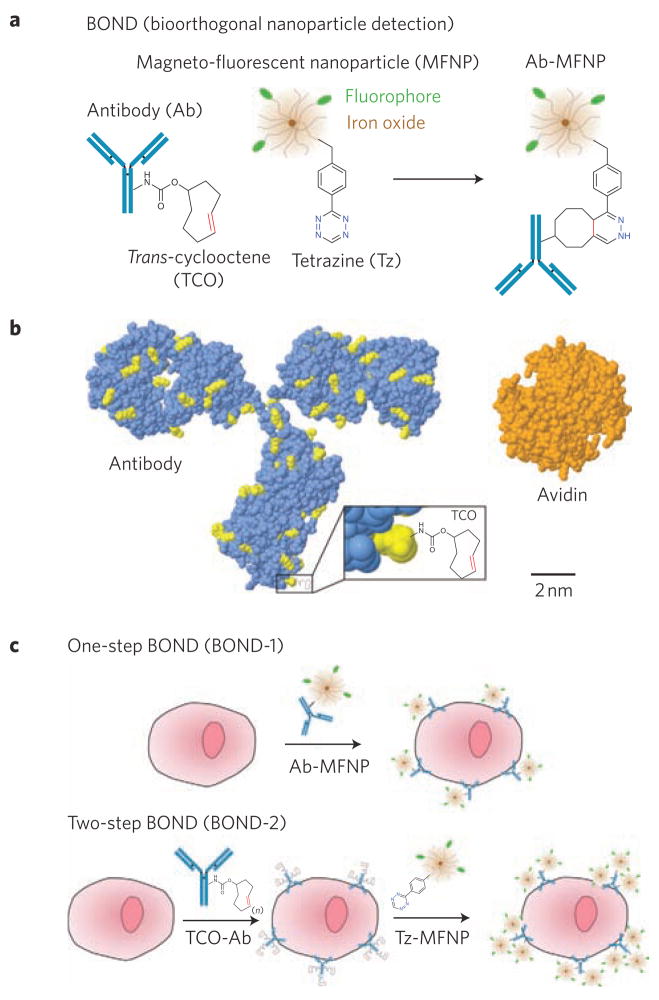
Figure 1 summarizes the chemistry, comparative molecular species dimensions, and experimental approaches of the different BOND strategies. We used magneto-fluorescent nanoparticles (MFNPs) to assess the performance of BOND using established fluorescence techniques and a novel miniaturized diagnostic magnetic resonance detector system developed for clinical point-of-care use19,20. To explore BOND in a biologically relevant system, we chose monoclonal antibodies as the scaffold for nanoparticle attachment due to their large size and the availability of numerous primary amine functional groups. For example, the monoclonal antibody trastuzumab, which is used clinically to treat breast cancers expressing HER221, has approximately 90 lysine residues that could be converted into nanoparticle reaction sites (Fig. 1b). We then comparatively tested BOND for targeting extracellular receptors on cancer cells using two assay types. In one setting, we directly coupled MFNPs to antibodies before cell exposure (BOND-1). In another setting, we used a two-step strategy (BOND-2) in which TCO-modified antibodies were used for primary target binding followed by covalent reaction with Tz–MFNP (Fig. 1c).
Figure 1. Overview of BOND.
a, Schematic showing the conjugation chemistry between antibody and nanoparticle. The diagram is a schematic and not to scale. b, Comparative sizes (to scale) of a representative mouse IgG2a antibody (lysine residues available for TCO modification via amine-reactive chemistry are shown in yellow), a TCO modification and an avidin protein for comparison. Tetrazine is similar in size to TCO (~200 Da). Protein structures and sizes were obtained from the Protein Data Bank (antibody, 1IGT; avidin, 3FDC) c, Application of BOND for one-step (direct) and two-step targeting of nanoparticles to cells. Note that the antibody and tetrazine are present in multiple copies per nanoparticle (~2–3 antibodies, Ab; 84 tetrazine, Tz).
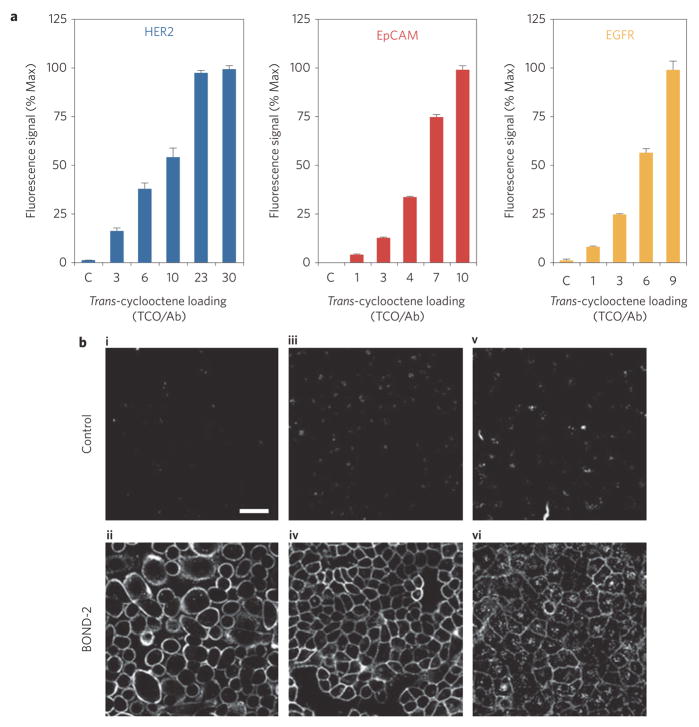
We first determined the extent to which TCO modification of antibodies promotes nanoparticle binding under the BOND-2 format. Figure 2 summarizes the results for three antibodies that were separately used to target HER2, EpCAM (CD326) and EGFR on cancer cell lines. For each antibody, TCO loading was modulated using various concentrations of an amine-reactive TCO. This resulted in a range of TCO valencies between approximately one and 30 per antibody (see Supplementary Methods and Figs S1–S3). Following sequential incubations with TCO–antibody and Tz–MFNP, we found that nanoparticle binding increased with successive TCO loading until saturating around at 20 TCO per antibody for HER2. TCO modifications had little effect on anti-HER2 affinity until loading levels reached 30 TCO per antibody (Supplementary Fig. S4). Furthermore, TCO modification did not affect the level of non-specific binding of MFNPs to control NIH/3T3 fibroblasts (Supplementary Fig. S4).
Figure 2. Effect of TCO loading on nanoparticle binding using BOND-2.
a, Fluorescence intensity measurements on live cells following sequential incubations with 10 μg ml−1 TCO-modified antibody and 10 nM Tz–MFNP using flow cytometry. Trastuzumab (anti-HER2), cetuximab (anti-EGFR) and anti-EpCAM antibodies were loaded with different numbers of TCO and measured by MALDI-TOF mass spectrometry (Supplementary Figs S1, S2). MFNP targeted HER2 on SK-BR-3 breast cancer cells, EpCAM on HCT 116 colon cancer cells, and EGFR on A549 lung cancer cells. b, Confocal microscopy images of similarly labelled live cells. Control: non-binding, TCO-modified control antibody (clone MOPC-21). HER2 (i,ii); EpCAM (iii,iv); EGFR (v, vi). Scale bar, 50 μm (i).
To further confirm the above results and to determine the spatial distribution of targeted nanoparticles, we performed confocal microscopy on live cancer cells (Fig. 2b). In these experiments, the cells were similarly incubated with TCO–antibody followed by Tz–MFNP. In all cases, a strong fluorescence signal was detected at the cell membranes; this was not observed for a TCO-modified control antibody. These data establish that the Tz/TCO cyclo-addition used for BOND-2 is sufficiently rapid and chemoselective to effectively target nanoparticles to live cells.
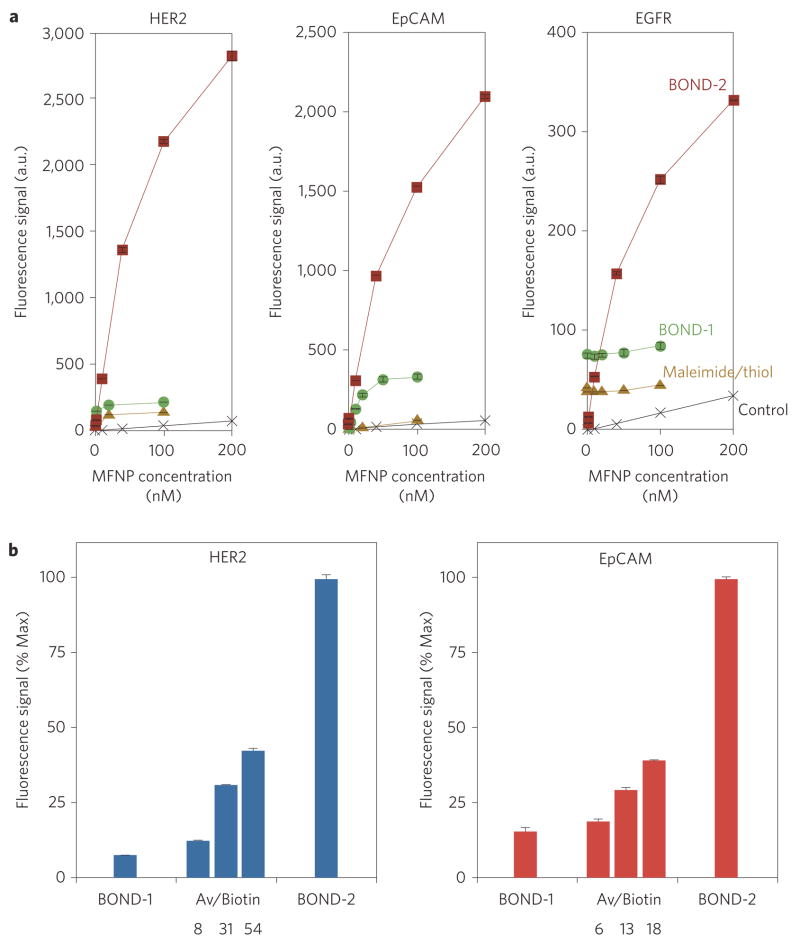
We next determined the comparative performance of BOND-2 relative to direct labelling with MFNP immuno-conjugates produced via maleimide/thiol chemistry and BOND-1 (Fig. 3a). For BOND-2, the optimized preparations determined in the above experiments were used. We found that BOND-2 consistently yielded higher nanoparticle binding to cells compared to either of the immuno-conjugates, higher by more than a factor of 15 for HER2 and approaching 10 for the other cases. The BOND-1 and maleimide/thiol immuno-conjugates bound to a similar level, but tended to vary across the different markers depending on antibody affinity (5 nM for cetuximab, 9 nM for trastuzumab22,23). These observations suggest that TCO-decorated antibodies can serve as scaffolds for subsequent nanoparticle attachment. This strategy effectively amplifies the achievable signal, the extent of which increases with the number of available TCO reaction sites.
Figure 3. Comparison of different nanoparticle targeting strategies.
SK-BR-3, HCT 116 and A549 cells were labelled with different concentrations of MFNP using the two-step BOND-2 or direct MFNP immuno-conjugates, and the fluorescence signal was measured using flow cytometry. MFNP immuno-conjugates were prepared either via maleimide/thiol or TCO/Tz (BOND-1) chemistries. Control samples were incubated with Tz–MFNP only. BOND-2 resulted in significantly higher nanoparticle binding, exceeding the direct immuno-conjugates by a factor of 15 for HER2. b, Fluorescence intensity of SK-BR-3 and HCT 116 cells labelled with 10 μg ml−1 biotin-modified antibody and 100 nM avidin–MFNP was measured using flow cytometry. Biotinylated anti-HER2 and anti-EpCAM antibodies were prepared analogously to the TCO modifications, and biotin levels were determined by MALDI-TOF mass spectrometry (Supplementary Figs S1, S2). Nanoparticle binding increased with biotin loading but remained lower than BOND-2 in both cases. Values for BOND-1 and BOND-2 were taken from a (100 nM MFNP).
To determine whether the amplification was unique to the bioorthogonal chemistry applied or simply a consequence of using a two-step labelling strategy, we tested avidin/biotin as the secondary coupling mechanism. We used avidin/biotin for this purpose because it is the gold standard for biological, non-covalent binding interactions24. However, it is known that the large molecular size of avidin (~6 nm, ~67 kDa) and its potential for eliciting immune responses in vivo limit its use for many clinical applications25. We biotinylated antibodies using similar procedures to achieve a range of loadings (see Supplementary Methods and Figs S1–S3). Although nanoparticle binding using avidin/biotin exceeded the direct conjugates and could be improved by increasing biotin valency, the overall signal remained considerably lower compared to BOND-2, despite higher biotin loadings (Fig. 3b). We believe that this finding can be attributed to the large size of avidin, which could potentially mask adjacent biotin sites. Second, biotin must associate within a deep cleft inside the avidin protein, which could physically or spatially constrain certain binding configurations. Conversely, Tz is a small molecule and can interact with TCO on the surface of the antibody without physical limitation or encroachment on neighbouring TCO sites. Finally, the Tz valency (84) was considerably higher than the avidin valency (8) on the nanoparticle due to its smaller size, increasing binding probability. The above arguments are supported by the fact that nanoparticle binding saturated at a lower TCO valency (~20) in comparison to biotin (>30). Thus, relatively higher quantities and more diversely spaced reaction sites are required to further increase nanoparticle binding for avidin/biotin. The net result is that the small-molecule bioorthogonal chemistry allows nanoparticles to pack more densely onto the antibody scaffolds, yielding greater signal amplification. It should be noted that the MFNP, although larger than avidin (~28 nm versus ~6 nm hydrodynamic diameter), is not limiting because the bulk of the size can be attributed to a dextran matrix, which can promote binding at a longer range through the presentation of extended reactive linkages. Table 1 summarizes the results from the various labelling techniques used in this study.
Table 1.
Comparison of nanoparticle targeting strategies
| BOND-2 | Avidin/biotin | Direct conjugation | |
|---|---|---|---|
| Antibody modification | Trans-cyclooctene | Biotin | Various (thiol, amine, cycloaddition) |
| Nanoparticle–antibody linkage | Covalent | Non-covalent | Covalent |
| Antibody valency potential | ~30–50 | ~30–50 | NA |
| Nanoparticle modification | Tetrazine | Avidin | Various (thiol, amine, cycloaddition) |
| Nanoparticle valency (NV) | 84 | 8 | 2–3 |
| Kinetic reaction rate (monovalent, kon, M−1s−1)★ | 6 × 103 | 1 × 107 | 105–106 |
| Net nanoparticle reaction potential (NV · kon, M−1 s−1) | 5 × 105 | 8 × 107 | ~105–106 |
| Relative labelling efficacy† | 15 | 5 | 1 |
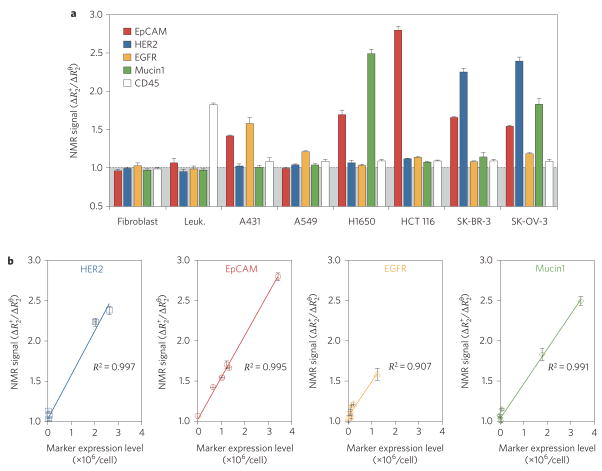
The ability to rapidly profile cancer cells in peripheral blood26,27 or fine needle aspirates20 has important clinical applications for early cancer detection and in devising treatment decisions28. We therefore adapted BOND-2 to molecular profiling of small populations of cancer cells by diagnostic magnetic resonance (Fig. 4). MFNPs were targeted to tumour cells using BOND-2, and the transverse relaxation rate (R2) was measured for ~1,000 cells using a miniaturized diagnostic magnetic resonance device20. At these scant sample sizes, which are in line with clinical specimens, fluorescent signal detection was difficult. However, parallel magnetic resonance measurements could be performed rapidly and at good signal-to-noise levels (Fig. 4a). As expected, markers signals were near normal levels for benign fibroblasts and leukocytes (with the exception of CD45, naturally expressed in the latter). Tumour cells showed considerable heterogeneity in the expression of the different markers, a finding that correlated well with the actual expression levels that were independently determined by flow cytometry using larger sample sizes (Fig. 4b). (Marker expression levels are listed in Supplementary Table S1.) The sensitivity of magnetic detection, including both diagnostic magnetic resonance and magnetic resonance imaging, using BOND-2 and the other targeting techniques are presented in Supplementary Fig. S5. Collectively, these data demonstrate the feasibility of profiling scant cell populations using the efficient and modular nanoparticle targeting strategy of BOND-2.
Figure 4. Profiling cancer cells using diagnostic magnetic resonance.
Magnetic profiling of cell samples (human tumour cell lines: A431, A549, NCI-H1650, HCT 116, SK-BR-3 and SK-OV-3; control: NIH/3T3 fibroblasts, peripheral blood leukocytes) for a panel of cancer markers in scant samples (~1,000 cells) using a recently developed miniaturized diagnostic magnetic resonance device. Cells were labelled with TCO–antibodies followed by Tz–MFNP before measurement of transverse relaxation time (R2). a, Marker expression levels, determined based on the ratio of the positive marker ( ) and control ( ) signals (see Supplementary Methods), were heterogeneous for tumour cells but normal for the control fibroblasts and leukocytes, with the exception of the leukocyte marker CD45. b, The tumour signals showed excellent correlation with measured marker expression levels, as determined independently by flow cytometry (values listed in Supplementary Table S1).
The bioorthogonal [4 + 2] cycloaddition chemistry between TCO and Tz results in higher nanoparticle binding to mammalian cells compared to other standard techniques. This was achieved because the high valencies and small sizes of the reactants promoted attachment of multiple nanoparticles to each antibody scaffold, amplifying the signal per marker. In contrast, direct nanoparticle immuno-conjugates are limited to at most one nanoparticle per marker, and potentially less than one due to multivalent binding. Because the TCO–antibody is applied in excess before nanoparticle exposure and contains numerous TCO moieties, most marker sites should be occupied by separate antibody scaffolds (Supplementary Fig. S4), and crosslinking of neighbouring antibodies by a nanoparticle consumes an additional TCO rather than an entire marker. We therefore speculate that a portion of the amplification observed for BOND-2 (and avidin/biotin) can be attributed to more efficient antigen recognition. BOND-2 also outperformed a similar two-step targeting strategy using avidin/biotin, most likely as a result of biotin masking or steric constraints that are imposed by the large footprint of avidin. Consequently, the covalent Tz/TCO reaction permits more nanoparticles to bind per antibody scaffold. The increased detection sensitivity resulting from amplification and the modular nature of the BOND-2 technique make it ideally suited for clinically oriented molecular profiling applications. We have demonstrated such an application here with magnetic detection of tumour cells using a miniaturized magnetic resonance detector that was designed for point-of-care clinical use.
We expect that the described BOND platform will have widespread use in diverse nanoparticle targeting applications, including alternative bioorthogonal small-molecule chemistries, affinity molecule scaffolds (proteins, peptides, aptamers, natural products, engineered hybrids) and nanoparticle sensors such as quantum dots, carbon nanotubes, gold nanoshells and polymer matrices/vesicles.
Methods
Preparation of MFNPs
We used crosslinked iron oxide (CLIO) as our model nanoparticle. The synthesis of CLIO bearing 89 primary amine functional groups is described in the Supplementary Methods. Amine-terminated MFNP (amino-MFNP) was created by treating the CLIO with a limiting quantity of an amine-reactive cyanine dye (VivoTag 680, VT680, VisEn Biomedical). This reaction was performed in phosphate buffered saline (PBS) at pH 8.0 for 1 h, followed by purification using gel filtration (Sephadex G-50, GE Healthcare). Approximately 4.7 VT680 molecules were conjugated per MFNP based on absorbance measurements, leaving ~84 available primary amine sites.
Tetrazine modification of MFNPs
Amino–MFNP was modified with 2,5-dioxopyrrolidin-1-yl 5-(4-(1,2,4,5-tetrazin-3-yl)benzylamino)-5-oxopentanoate (Tz-NHS, see Supplementary Methods) to create tetrazine–MNFP (Tz–MFNP). The reaction was performed using 500 equiv. of Tz–NHS relative to amino–MFNP, and proceeded in PBS containing 5% dimethylformamide (DMF) for 3 h at room temperature. Excess Tz–NHS was removed using Sephadex G-50. No primary amine groups were detectable at this point using the N-succinimidyl-3-(2-pyridyldithio)propionate (SPDP) method (see Supplementary Methods), and thus the tetrazine valency was assumed to be 84.
Antibody modifications
Monoclonal antibodies were modified with (E)-cyclooct-4-enyl 2,5-dioxopyrrolidin-1-yl carbonate (TCO-NHS) that was synthesized as previously reported by our group16. For each case, 0.5 mg of antibody was buffer exchanged into PBS (pH 8.0) using 2 ml Zeba desalting columns (Thermo Fisher). TCO–NHS was then reacted in 10% DMF for 3 h at room temperature. Anti-HER2 antibody (trastuzumab, Genentech) was reacted with 30, 100, 300, 1,000 and 3,000 equiv. of TCO–NHS. Equivalents were 10, 30, 100, 300 and 1,000 for the anti-EpCAM antibody (clone 158206, R&D Systems), and 10, 100, 300 and 1,000 for the anti-EGFR antibody (cetuximab, Imclone Systems). Control mouse IgG1 (clone MOPC-21, BioLegend), anti-Mucin1 (clone M01102909, Fitzgerald Industries) and anti-CD45 (clone HI30, BioLegend) antibodies were reacted with 1,000 equiv. of TCO–NHS exclusively. Samples were purified using Zeba columns, and concentration was determined by absorbance measurement. Biotinylation was performed for the anti-HER2 and anti-EpCAM antibodies using EZ-Link NHS–biotin (Thermo Fisher) at 10, 100 and 300 equiv. in a similar fashion. TCO and biotin valencies were determined based on changes in molecular weight using MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) mass spectrometry (see Supplementary Methods). Functional TCO and biotin loadings were also measured using a Tz-VT680 probe and the HABA assay, respectively (see Supplementary Methods).
Cell labelling and detection
The human cancer cell lines SK-BR-3, HCT 116, A549, NCI-H1650, A431 and SK-OV-3 and the mouse embryonic fibroblast cell line NIH/3T3 were all obtained from ATCC and maintained in DMEM media supplemented with 10% fetal bovine serum (FBS) and 5% penicillin/streptomycin. Before experiments, cells were grown to ~90% confluency, released using 0.05% Tryspin/0.53 mM EDTA, and prepared by washing twice with PBS containing 2% FBS (PBS + ). Cells (250,000/sample) were then labelled with TCO- or biotin- modified monoclonal antibody (10 μg ml−1) in 0.1 ml PBS+ for 10 min at room temperature. Following centrifugation and aspiration of the antibody solution, cells were directly resuspended with Tz–MFNP (0.2–200 nM) or avidin–MFNP (100 nM), incubated for 30 min at room temperature, and washed twice by centrifugation with ice-cold PBS +. Antibodies were omitted for control samples. For direct antibody–MFNP conjugates, only the MFNP binding period (0.2–100 nM) was used. Avidin– and antibody–MFNP conjugates were prepared as described in the Supplementary Methods. MFNP molar concentration was determined based on an estimated 447 kDa molecular weight for CLIO (8,000 Fe atoms per core crystal, 55.85 Da each29). At the conclusion of the labelling procedures, VT680 fluorescence was assessed using an LSRII flow cytometer (Becton Dickinson) and mean fluorescence intensity was determined using FlowJo software. All measurements were performed in triplicate, and the data are presented as the mean ± standard error. For confocal microscopy studies, cells were grown on glass slides with removable chamber wells (Lab-Tek; Thermo Fisher). Cell labelling was performed as described above, and VT680 fluorescence was imaged using a multichannel upright laser-scanning confocal microscope (FV1000; Olympus) with a ×40 water immersion objective lens. Images were acquired with Fluoview software (version 4.3; Olympus) and analysed using ImageJ software (version 1.41; Bethesda MD). For magnetic resonance detection, human tumour cells lines, control NIH/3T3 fibroblasts and fresh human peripheral blood leukocytes were labelled with a TCO–antibody (control; anti-HER2, EpCAM, EGFR, Mucin1 or CD45) and Tz–MFNP as described above. Magnetic resonance measurements were performed using a previously described miniaturized nuclear magnetic resonance device (see Supplementary Methods)20.
Supplementary Material
Figure S1: Mass spectrometry to quantitate antibody modifications with TCO or biotin for anti-HER2 antibody. TCO and biotin loadings on the anti-HER2 antibody were determined by MALDI-TOF mass spectrometry based on changes in molecular weight. a, Representative molecular weight/ioniziation (M/z) histograms for the unmodified anti-HER2 antibody, and following reaction with 1000-fold excess of TCO-NHS. The singly ionised peak (z=1) has an average molecular weight of ~148,000 dalton for the unmodified antibody, with a doubly ionised peak (z=2) also detectable at ~75,000 dalton. The approximately 3500 dalton change in molecular weight with addition of the TCO corresponds to ~23 TCO/antibody based on 152.2 dalton mass added. b, Merged data for TCO and biotin modifications to the anti-HER2 antibody. Biotin valencies were determined based on 226.1 dalton mass added. Average molecular weight and data merges were performed using MatLab.
Figure S2: Mass spectrometry to quantitate antibody modifications with TCO or biotin for other antibodies. A similar analysis to Supplementary Fig. S1 was perfomed for the a, TCO and biotin modifications of the anti-EpCAM antibody and b, TCO modification of the anti-EGFR, control, anti-CD45, and anti-Mucin1 antibodies. The anti-EpCAM antibody and -EGFR antibody (at high TCO loadings) did not ionise well, resulting in histograms with significantly more noise.
Figure S3: Correlation of TCO and biotin valencies between the MALDI-MS measurements and functional assays. a, TCO valencies correlated well between mass measurements and functional assays performed using at Tz-fluorophore (VT680) for each of the antibodies tested. However, the functional values were considerably lower than the mass values, presumably because certain TCO sites could not be accessed or due to physical interaction between the large, highly-charged VT680 molecules. b, Similar results were obtained for biotin valencies, this time where the functional metric was avidin binding using the HABA assay. TCO and biotin valencies used throughout the text reflect the MALDI-MS results so that the TCO and biotin species can be compared directly.
Figure S4: Effect of TCO loading on antibody affinity and background nanoparticle adhesion. a, Modification of the anti-HER2 antibody with TCO had negligible effect on binding affinity at low loadings, consistently exhibiting a KD = ~1–2 nM up to approximately 10 TCO/Ab. Affinity decreased by approximately 3-fold at 23 TCO/Ab (KD = ~4 nM) and 10-fold at 30 TCO/Ab (KD = ~10 nM). These results also indicate that antibody binding is saturated at the concentration employed for cell labelling studies (10 μg/mL, or ~67 nM). b, Modification of the anti-HER2 antibody with TCO did not affect the level of MFNP binding to control NIH/3T3 fibroblast cells under the BOND-2 scheme. This result demonstrates that the TCO does not make the antibody more “sticky” to cells, even at high TCO loadings, and further confirms the specificity of the BOND-2 technique. Fluorescence values are comparable to controls for HER2-positive SK-BR-3 cells (see Fig. 2).
Figure S5: Diagnostic magnetic resonance detection of the different nanoparticle targeting strategies. (a) Magnetic resonance detection sensitivity for cancer cells based on HER2. BOND-2 outperformed the other approaches, with a detection threshold of approximately 200 cells in comparison to > 1000 cells for direct immuno-conjugates. (b) Detection thresholds for each technique, which was determined based on the number of cells required to register a 2.5% change in transverse relaxation time (T2). Note that using more magnetic nanoparticles can significantly improve detection thresholds to near single cell levels6.
Figure S6: Magnetic resonance imaging of the different nanoparticle targeting strategies. Magnetic resonance imaging of targeted cells. Cancer (SK-BR-3) and normal (NIH/3T3) cells were labelled using the different targeting strategies and imaged by a magnetic resonance scanner (4.7 T; Bruker Pharmascan). Consistent with fluorescence and diagnostic magnetic resonance measurements, BOND-2 yielded the most pronounced T2 changes due to higher nanoparticle binding. Prior to imaging, labelled cells (106) were lysed and embedded in agar.
Supplementary Table S1: Biomarker expression levels for the cell lines used for magnetic resonance profiling (Fig. 4). The number of biomarkers per cell was determined by flow cytometry using fluorescent antibody staining and calibration beads (see Supplementary Methods).
Acknowledgments
The authors gratefully acknowledge N. Sergeyev for synthesizing CLIO and C. Wang for assistance with MALDI-TOF measurements. We especially thank G. Thurber, M. Pittet, F. Swirski and M. Nahrendorf for their many helpful suggestions. We also thank Y. Fisher-Jeffes for reviewing the manuscript. This work was funded in part by NCI grant P50CA86355 and RO1 EB004626.
Footnotes
Author contributions
J.B.H. designed and performed the experiments, analysed the data and wrote the manuscript. N.K.D. and S.A.H. developed and synthesized the bioorthogonal chemistries. H.L. performed the magnetic resonance measurements. R.W. provided overall guidance, designed experiments, reviewed the data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Additional information
The authors declare no competing financial interests. Supplementary information accompanies this paper at www.nature.com/naturenanotechnology. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nature Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 2.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giljohann DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi HS, et al. Design considerations for tumour-targeted nanoparticles. Nature Nanotech. 2010;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Y, Gao X. Plasmonic fluorescent quantum dots. Nature Nanotech. 2009;4:571–576. doi: 10.1038/nnano.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, et al. Protein microarrays with carbon nanotubes as multicolor Raman labels. Nature Biotechnol. 2008;26:1285–1292. doi: 10.1038/nbt.1501. [DOI] [PubMed] [Google Scholar]
- 7.Qian X, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nature Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, et al. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nature Med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nature Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 10.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 11.Peer D, et al. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotech. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 12.Cho K, Wang X, Nie S, Chen ZG, Shin DM. Therapeutic nanoparticles for drug delivery in cancer. Clin Cancer Res. 2008;14:1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 13.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nature Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw SY, et al. Perturbational profiling of nanomaterial biologic activity. Proc Natl Acad Sci USA. 2008;105:7387–7392. doi: 10.1073/pnas.0802878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing Y, et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nature Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 16.Devaraj NK, Upadhyay R, Haun JB, Hilderbrand SA, Weissleder R. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/transcyclooctene cycloaddition. Angew Chem Int Ed. 2009;48:7013–7016. doi: 10.1002/anie.200903233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devaraj NK, Weissleder R, Hilderbrand SA. Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjug Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels–Alder reactivity. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nature Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. Rapid detection and profiling of cancer cells in fine-needle aspirates. Proc Natl Acad Sci USA. 2009;106:12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudis CA. Trastuzumab—mechanism of action and use in clinical practice. N Engl J Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 22.Hong KW, et al. A novel anti-EGFR monoclonal antibody inhibiting tumor cell growth by recognizing different epitopes from cetuximab. J Biotechnol. 2010;145:84–91. doi: 10.1016/j.jbiotec.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Troise F, Cafaro V, Giancola C, D’Alessio G, De Lorenzo C. Differential binding of human immunoagents and Herceptin to the ErbB2 receptor. FEBS J. 2008;275:4967–4979. doi: 10.1111/j.1742-4658.2008.06625.x. [DOI] [PubMed] [Google Scholar]
- 24.Green NM. Avidin and streptavidin. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 25.Chinol M, et al. Biochemical modifications of avidin improve pharmacokinetics and biodistribution, and reduce immunogenicity. Br J Cancer. 1998;78:189–197. doi: 10.1038/bjc.1998.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nature Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds F, O’Loughlin T, Weissleder R, Josephson L. Method of determining nanoparticle core weight. Anal Chem. 2005;77:814–817. doi: 10.1021/ac049307x. [DOI] [PubMed] [Google Scholar]
- 30.Piran U, Riordan WJ. Dissociation rate constant of the biotin–streptavidin complex. J Immunol Methods. 1990;133:141–143. doi: 10.1016/0022-1759(90)90328-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Mass spectrometry to quantitate antibody modifications with TCO or biotin for anti-HER2 antibody. TCO and biotin loadings on the anti-HER2 antibody were determined by MALDI-TOF mass spectrometry based on changes in molecular weight. a, Representative molecular weight/ioniziation (M/z) histograms for the unmodified anti-HER2 antibody, and following reaction with 1000-fold excess of TCO-NHS. The singly ionised peak (z=1) has an average molecular weight of ~148,000 dalton for the unmodified antibody, with a doubly ionised peak (z=2) also detectable at ~75,000 dalton. The approximately 3500 dalton change in molecular weight with addition of the TCO corresponds to ~23 TCO/antibody based on 152.2 dalton mass added. b, Merged data for TCO and biotin modifications to the anti-HER2 antibody. Biotin valencies were determined based on 226.1 dalton mass added. Average molecular weight and data merges were performed using MatLab.
Figure S2: Mass spectrometry to quantitate antibody modifications with TCO or biotin for other antibodies. A similar analysis to Supplementary Fig. S1 was perfomed for the a, TCO and biotin modifications of the anti-EpCAM antibody and b, TCO modification of the anti-EGFR, control, anti-CD45, and anti-Mucin1 antibodies. The anti-EpCAM antibody and -EGFR antibody (at high TCO loadings) did not ionise well, resulting in histograms with significantly more noise.
Figure S3: Correlation of TCO and biotin valencies between the MALDI-MS measurements and functional assays. a, TCO valencies correlated well between mass measurements and functional assays performed using at Tz-fluorophore (VT680) for each of the antibodies tested. However, the functional values were considerably lower than the mass values, presumably because certain TCO sites could not be accessed or due to physical interaction between the large, highly-charged VT680 molecules. b, Similar results were obtained for biotin valencies, this time where the functional metric was avidin binding using the HABA assay. TCO and biotin valencies used throughout the text reflect the MALDI-MS results so that the TCO and biotin species can be compared directly.
Figure S4: Effect of TCO loading on antibody affinity and background nanoparticle adhesion. a, Modification of the anti-HER2 antibody with TCO had negligible effect on binding affinity at low loadings, consistently exhibiting a KD = ~1–2 nM up to approximately 10 TCO/Ab. Affinity decreased by approximately 3-fold at 23 TCO/Ab (KD = ~4 nM) and 10-fold at 30 TCO/Ab (KD = ~10 nM). These results also indicate that antibody binding is saturated at the concentration employed for cell labelling studies (10 μg/mL, or ~67 nM). b, Modification of the anti-HER2 antibody with TCO did not affect the level of MFNP binding to control NIH/3T3 fibroblast cells under the BOND-2 scheme. This result demonstrates that the TCO does not make the antibody more “sticky” to cells, even at high TCO loadings, and further confirms the specificity of the BOND-2 technique. Fluorescence values are comparable to controls for HER2-positive SK-BR-3 cells (see Fig. 2).
Figure S5: Diagnostic magnetic resonance detection of the different nanoparticle targeting strategies. (a) Magnetic resonance detection sensitivity for cancer cells based on HER2. BOND-2 outperformed the other approaches, with a detection threshold of approximately 200 cells in comparison to > 1000 cells for direct immuno-conjugates. (b) Detection thresholds for each technique, which was determined based on the number of cells required to register a 2.5% change in transverse relaxation time (T2). Note that using more magnetic nanoparticles can significantly improve detection thresholds to near single cell levels6.
Figure S6: Magnetic resonance imaging of the different nanoparticle targeting strategies. Magnetic resonance imaging of targeted cells. Cancer (SK-BR-3) and normal (NIH/3T3) cells were labelled using the different targeting strategies and imaged by a magnetic resonance scanner (4.7 T; Bruker Pharmascan). Consistent with fluorescence and diagnostic magnetic resonance measurements, BOND-2 yielded the most pronounced T2 changes due to higher nanoparticle binding. Prior to imaging, labelled cells (106) were lysed and embedded in agar.
Supplementary Table S1: Biomarker expression levels for the cell lines used for magnetic resonance profiling (Fig. 4). The number of biomarkers per cell was determined by flow cytometry using fluorescent antibody staining and calibration beads (see Supplementary Methods).