Abstract
Antimicrobial susceptibilities of 233 Gram-positive and 180 Gram-negative strains to two novel bis-indoles were evaluated. Both compounds were potent inhibitors of Gram-positive bacteria, with MIC90 values of 0.004 to 0.5 μg/ml. One bis-indole, MBX 1162, exhibited potent activity against all Gram-negative strains, with MIC90 values of 0.12 to 4 μg/ml, even against high-level-resistant pathogens, and compared favorably to all comparator antibiotics. The bis-indole compounds show promise for the treatment of multidrug-resistant clinical pathogens.
Antibiotic resistance is reaching a crisis level because few options remain to treat certain pathogenic bacteria—mainly those causing hospital-acquired infection, but with the potential to occur in the community (8) and on the battlefield (2). Of special note are the following particularly problematic pathogens: multidrug-resistant (MDR) Acinetobacter baumannii, extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella species, Pseudomonas aeruginosa, vancomycin-resistant enterococci (VRE [Enterococcus faecium]), methicillin-resistant Staphylococcus aureus (MRSA), and coagulase-negative staphylococci such as methicillin-resistant Staphylococcus epidermidis (MRSE).
The continuing erosion of the efficacy of current antibiotics requires the discovery and development of new antibacterials that are not subject to existing mechanisms of target-based resistance. This can be accomplished by building derivatives of existing antibiotics which escape resistance mechanisms or by the development of entirely new chemical classes of antibiotics. The latter approach is preferred because preexisting target-based resistance mechanisms are unlikely to be present in the bacterial population. Here, we report MIC90 values versus several problematic bacterial pathogens for a recently described series of bis-indole compounds (6, 7). MBX 1066 (Fig. 1), along with MBX 1090, 1113 and 1128 (7) were identified in a screen of the NCI repository for compounds active against Bacillus anthracis. The indole groups of MBX 1066 and MBX 1090 face each other in a symmetrical fashion (“head-to-head”), while they are positioned in a tandem arrangement (“head-to-tail”) in MBX 1113 and 1128. The head-to-head compounds were found to be more potent than the head-to-tail compounds against Gram-negative species while being nearly equipotent against Gram-positive species. MBX 1066 exhibited low cytotoxicity against HeLa cells (50% cytotoxic concentration [CC50], 33 μg/ml) upon 3-day exposure, while the other compounds were slightly more cytotoxic (7). MBX 1066 displayed rapid bactericidal activity against both Gram-positive (Bacillus anthracis and B. subtilis) and Gram-negative (Yersinia pestis) bacteria (7) and demonstrated efficacy in murine models of Gram-positive (B. anthracis and S. aureus) and Gram-negative (Y. pestis) infections (7). MBX 1066 and related compounds bound serum proteins less than 25% (M. Butler, unpublished observation). Finally, experiments to isolate MBX 1066-resistant mutants by serial passage and spontaneous mutation selection were unsuccessful against both S. aureus and E. coli, although mutants resistant to a closely related compound, MBX 1090, were isolated (6, 7).
FIG. 1.
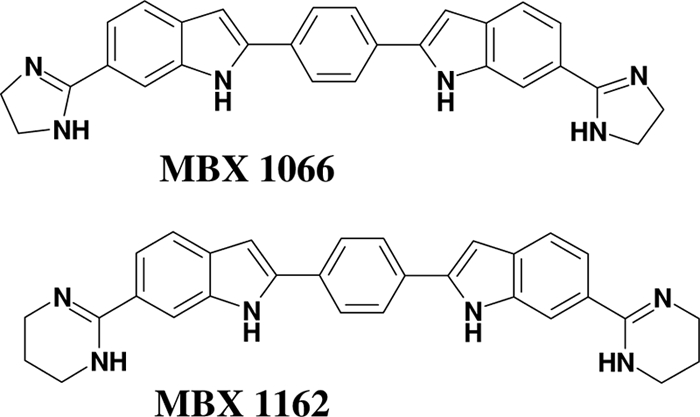
Structures of bis-indole compounds MBX 1066 and MBX 1162.
We have conducted a structure activity relationship (SAR) program around the head-to-head compounds, typified by MBX 1066. While the molecular target(s) of these compounds is not known, the fact that they share some structural features with compounds that bind in the minor groove of duplex DNA (1) suggests that these compounds may inhibit DNA synthesis by binding to DNA. In fact, we have shown that they are potent inhibitors of DNA synthesis (7). However, their efficacy in murine models of infection, together with favorable in vitro selectivity indices, indicates that these compounds discriminate to some degree between bacterial and mammalian targets (7).
Analogs of MBX 1066, particularly MBX 1162 (Fig. 1), exhibit improved Gram-negative activity while maintaining the Gram-positive potencies displayed by the parent compound. MBX 1162 is remarkably potent against antibiotic-resistant bacterial strains such as MDR A. baumannii, ESBL-producing Klebsiella pneumoniae, VRE, and MRSA, making it a promising new antibacterial agent. While MBX 1162 appeared somewhat more cytotoxic than MBX 1066 against HeLa cells (CC50, 4 μg/ml) upon 3-day exposure, it retained the favorable features of MBX 1066, including bactericidal activity against both Gram-positive and Gram-negative pathogens, low serum binding (M. Butler, unpublished), and absence of susceptibility to resistance development (T. Opperman, unpublished observation), and its enhanced antibacterial activity provided selectivity index values (CC50/MIC) comparable to those of MBX 1066 for Gram-negative species. In addition, MBX 1162 exhibited potent inhibition of DNA synthesis (T. Opperman and M. Butler, unpublished), suggesting its mechanism of action is similar to that of MBX 1066. Although we have observed an exceptionally broad antimicrobial profile for MBX 1066 and 1162 against single isolates of a variety of species, it is important to determine efficacy against larger groups of single species isolates, obtained from several clinical sources, looking specifically at populations of antibiotic-resistant clinical pathogens. To this end, we analyzed potencies against multiple strains of eight Gram-positive and eight Gram-negative species.
Two hundred thirty-three Gram-positive strains and 180 Gram-negative aerobic strains were tested by the broth microdilution method (4) against MBX 1066 and 1162 as well as four comparator antibiotics. The comparator antibiotics were selected to be the most appropriate for each family as well as for verifying particular resistances and were thus different for Gram-positive versus Gram-negative isolates. These included linezolid (ChemPacifica), daptomycin (Cubist), vancomycin (Sigma-Aldrich), and imipenem (United States Pharmacopeia) for the Gram-positive aerobic bacteria and imipenem, tigecycline (Wyeth), gentamicin (Sigma-Aldrich), and ciprofloxacin (United States Pharmacopeia) for the Gram-negative aerobic bacteria. In addition, 18 isolates of the Gram-positive anaerobe Clostridium difficile were analyzed (3) using clindamycin (Sigma-Aldrich), imipenem, and metronidazole (Sigma-Aldrich) as comparators. The growth medium used in these studies was the CLSI-recommended Mueller-Hinton broth II (MHB II), with the exception of the streptococci (MHB II plus 2% lysed horse blood), Haemophilus influenza (HTM medium), and C. difficile (supplemented brucella broth). The quality control reference strains, S. aureus ATCC 29213, E. faecalis ATCC 29212, Streptococcus pneumoniae ATCC 49619, E. coli ATCC 25922, P. aeruginosa ATCC 27853, and Bacteroides fragilis ATCC 25285, were tested in accordance with CLSI methodology, and the results were within published ranges (5). The locations of the sources of the clinical isolates are listed in Table 1.
TABLE 1.
Activities of MBX 1066 and MBX 1162 and selected comparators against Gram-positive and Gram-negative isolates
| Organism and phenotype (no. of isolates tested)a | Agent | MIC (μg/ml)b |
||
|---|---|---|---|---|
| Range | 90% | 50% | ||
| Staphylococcus aureus | ||||
| All (39) | MBX 1066 | 0.004-0.5 | 0.25 | 0.12 |
| MBX 1162 | 0.008-0.5 | 0.5 | 0.12 | |
| Linezolid | 2-4 | 4 | 2 | |
| Vancomycin | 0.25-2 | 1 | 0.5 | |
| Imipenem | 0.008->8 | 4 | 2 | |
| Daptomycin | 0.12-1 | 0.5 | 0.25 | |
| MSSA (27) | MBX 1066 | 0.004-0.5 | 0.25 | 0.12 |
| MBX 1162 | 0.008-0.5 | 0.5 | 0.12 | |
| Linezolid | 2-4 | 4 | 2 | |
| Vancomycin | 0.5-2 | 1 | 0.5 | |
| Imipenem | 0.008-0.03 | 0.03 | 0.015 | |
| Daptomycin | 0.25-1 | 0.5 | 0.5 | |
| MRSA (12) | MBX 1066 | 0.06-0.12 | 0.12 | 0.06 |
| MBX 1162 | 0.03-0.12 | 0.12 | 0.06 | |
| Linezolid | 2-4 | 4 | 2 | |
| Vancomycin | 0.25-1 | 1 | 0.5 | |
| Imipenem | 0.12->8 | 8 | 1 | |
| Daptomycin | 0.12-0.5 | 0.25 | 0.25 | |
| Staphylococcus epidermidis | ||||
| All (39) | MBX 1066 | 0.004-0.06 | 0.03 | 0.015 |
| MBX 1162 | 0.008-0.06 | 0.06 | 0.015 | |
| Linezolid | 0.5-2 | 2 | 1 | |
| Vancomycin | 1-4 | 2 | 2 | |
| Imipenem | 0.015->8 | >8 | 0.015 | |
| Daptomycin | 0.5-1 | 1 | 0.5 | |
| MSSE (27) | MBX 1066 | 0.004-0.06 | 0.03 | 0.008 |
| MBX 1162 | 0.008-0.06 | 0.06 | 0.03 | |
| Linezolid | 0.5-2 | 2 | 1 | |
| Vancomycin | 1-4 | 2 | 1 | |
| Imipenem | 0.015-0.03 | 0.015 | 0.015 | |
| Daptomycin | 0.5-1 | 1 | 0.5 | |
| MRSE (12) | MBX 1066 | 0.004-0.03 | 0.03 | 0.015 |
| MBX 1162 | 0.008-0.06 | 0.06 | 0.015 | |
| Linezolid | 1-2 | 2 | 1 | |
| Vancomycin | 1-2 | 2 | 2 | |
| Imipenem | 0.5->8 | >8 | 8 | |
| Daptomycin | 0.5-1 | 1 | 0.5 | |
| Enterococcus faecalis | ||||
| All (39) | MBX 1066 | 0.004-0.12 | 0.06 | 0.03 |
| MBX 1162 | 0.004-0.25 | 0.06 | 0.03 | |
| Linezolid | 0.5-2 | 2 | 1 | |
| Vancomycin | 0.5->64 | >64 | 1 | |
| Imipenem | 0.25->8 | 2 | 2 | |
| Daptomycin | 0.03-4 | 2 | 1 | |
| VSE (27) | MBX 1066 | 0.004-0.12 | 0.06 | 0.06 |
| MBX 1162 | 0.004-0.25 | 0.06 | 0.06 | |
| Linezolid | 0.5-2 | 2 | 2 | |
| Vancomycin | 0.5-2 | 2 | 1 | |
| Imipenem | 0.25->8 | 4 | 1 | |
| Daptomycin | 0.03-4 | 2 | 1 | |
| VRE (12) | MBX 1066 | 0.015-0.06 | 0.06 | 0.03 |
| MBX 1162 | 0.008-0.03 | 0.03 | 0.015 | |
| Linezolid | 0.5-2 | 1 | 1 | |
| Vancomycin | >64 | >64 | >64 | |
| Imipenem | 0.5-2 | 2 | 2 | |
| Daptomycin | 0.25-2 | 2 | 0.5 | |
| Enterococcus faecium | ||||
| All (39) | MBX 1066 | 0.002-0.06 | 0.008 | 0.004 |
| MBX 1162 | 0.002-0.03 | 0.008 | 0.004 | |
| Linezolid | 1-4 | 4 | 2 | |
| Vancomycin | 0.5->64 | >64 | 1 | |
| Imipenem | 1->8 | >8 | >8 | |
| Daptomycin | 1-8 | 4 | 4 | |
| VSE (27) | MBX 1066 | 0.002-0.06 | 0.015 | 0.004 |
| MBX 1162 | 0.002-0.03 | 0.015 | 0.004 | |
| Linezolid | 2-4 | 4 | 2 | |
| Vancomycin | 0.5-4 | 1 | 0.5 | |
| Imipenem | 1->8 | >8 | >8 | |
| Daptomycin | 1-8 | 4 | 4 | |
| VRE (12) | MBX 1066 | 0.002-0.008 | 0.004 | 0.004 |
| MBX 1162 | 0.004-0.008 | 0.004 | 0.004 | |
| Linezolid | 1-2 | 2 | 2 | |
| Vancomycin | 64->64 | >64 | >64 | |
| Imipenem | 8 | 8 | 8 | |
| Daptomycin | 1-4 | 4 | 2 | |
| Streptococcus pneumoniae | ||||
| All (53) | MBX 1066 | 0.008-2 | 0.03 | 0.03 |
| MBX 1162 | 0.015-0.06 | 0.03 | 0.03 | |
| Linezolid | 0.5-2 | 1 | 1 | |
| Vancomycin | 0.12-0.5 | 0.25 | 0.25 | |
| Imipenem | <0.08-1 | 0.03 | 0.015 | |
| Daptomycin | <0.03-1 | 0.25 | 0.06 | |
| PSSP (27) | MBX 1066 | 0.008-0.12 | 0.03 | 0.015 |
| MBX 1162 | 0.015-0.03 | 0.03 | 0.03 | |
| Linezolid | 0.5-2 | 2 | 1 | |
| Vancomycin | 0.12-0.25 | 0.25 | 0.25 | |
| Imipenem | <0.008-0.03 | <0.008 | <0.008 | |
| Daptomycin | <0.03-0.5 | 0.25 | 0.06 | |
| PISPc (14) | MBX 1066 | 0.008-2 | 0.12 | 0.015 |
| MBX 1162 | 0.015-0.06 | 0.03 | 0.03 | |
| Linezolid | 0.5-2 | 1 | 1 | |
| Vancomycin | 0.25-0.5 | 0.25 | 0.25 | |
| Imipenem | <0.008-0.25 | 0.25 | 0.03 | |
| Daptomycin | <0.03-1 | 0.25 | 0.06 | |
| PRSP (12) | MBX 1066 | 0.03-0.06 | 0.06 | 0.03 |
| MBX 1162 | 0.015-0.06 | 0.06 | 0.03 | |
| Linezolid | 0.5-1 | 1 | 1 | |
| Vancomycin | 0.25-0.5 | 0.25 | 0.25 | |
| Imipenem | 0.12-1 | 1 | 0.25 | |
| Daptomycin | <0.03-0.12 | 0.12 | 0.06 | |
| Streptococcus agalactiae | ||||
| All (12) | MBX 1066 | 0.03-0.12 | 0.06 | 0.06 |
| MBX 1162 | 0.06-0.12 | 0.06 | 0.06 | |
| Linezolid | 1-2 | 2 | 2 | |
| Vancomycin | 0.5-1 | 0.5 | 0.5 | |
| Imipenem | 0.06-8 | 0.06 | 0.06 | |
| Daptomycin | 0.12-2 | 1 | 0.5 | |
| Streptococcus pyogenes | ||||
| All (12) | MBX 1066 | 0.03 | 0.03 | 0.03 |
| MBX 1162 | 0.03 | 0.03 | 0.03 | |
| Linezolid | 1-2 | 2 | 1 | |
| Vancomycin | 0.5 | 1 | 1 | |
| Imipenem | 0.06 | 0.06 | 0.06 | |
| Daptomycin | 0.03-2 | 2 | 0.5 | |
| Clostridium difficile | ||||
| (anaerobic bacteria) | ||||
| All (18) | MBX 1066 | 0.03-0.25 | 0.12 | 0.06 |
| MBX 1162 | 0.03-0.12 | 0.12 | 0.06 | |
| Clindamycin | 0.25->8 | >8 | 4 | |
| Imipenem | 0.5->8 | 8 | 4 | |
| Metronidazole | 0.06->8 | 0.5 | 0.12 | |
| Escherichia coli | ||||
| All (27) | MBX 1066 | 0.03-2 | 0.5 | 0.12 |
| MBX 1162 | 0.06-0.25 | 0.25 | 0.12 | |
| Imipenem | 0.06-0.5 | 0.25 | 0.25 | |
| Tigecycline | 0.12-0.25 | 0.25 | 0.12 | |
| Gentamicin | 0.5->8 | >8 | 1 | |
| Ciprofloxacin | 0.015->2 | >2 | 0.03 | |
| Klebsiella pneumoniae | ||||
| All (39) | MBX 1066 | 0.25->16 | 8 | 2 |
| MBX 1162 | 0.06-1 | 0.5 | 0.25 | |
| Imipenem | 0.06-32 | 1 | 0.12 | |
| Tigecycline | 0.25-8 | 2 | 0.5 | |
| Gentamicin | 0.12->32 | >32 | 0.5 | |
| Ciprofloxacin | 0.06->8 | >8 | 0.25 | |
| ESBL (12) | MBX 1066 | 0.5->16 | >16 | 1 |
| MBX 1162 | 0.06-0.5 | 0.5 | 0.12 | |
| Imipenem | 0.12-2 | 1 | 0.25 | |
| Tigecycline | 0.25-8 | 2 | 0.5 | |
| Gentamicin | 0.25->32 | >32 | 0.5 | |
| Ciprofloxacin | 0.06->8 | >8 | >8 | |
| Serratia marcescens | ||||
| All (12) | MBX 1066 | 0.06-2 | 2 | 1 |
| MBX 1162 | 0.12-0.5 | 0.25 | 0.12 | |
| Imipenem | 2->8 | >8 | 4 | |
| Tigecycline | 0.5-2 | 1 | 1 | |
| Gentamicin | 0.25-2 | 2 | 0.5 | |
| Ciprofloxacin | 0.06->2 | 1 | 0.25 | |
| Proteus mirabilis | ||||
| All (12) | MBX 1066 | 8->16 | >16 | >16 |
| MBX 1162 | 0.12-2 | 2 | 1 | |
| Imipenem | 2-8 | 8 | 4 | |
| Tigecycline | 1-4 | 4 | 4 | |
| Gentamicin | 0.5-16 | 8 | 1 | |
| Ciprofloxacin | 0.015->8 | >8 | 0.06 | |
| Acinetobacter baumannii | ||||
| All (40) | MBX 1066 | 0.06->16 | >16 | 8 |
| MBX 1162 | 0.12-4 | 4 | 0.5 | |
| Imipenem | 0.06->32 | >32 | 0.5 | |
| Tigecycline | 0.06->32 | 4 | 0.5 | |
| Gentamicin | 0.25->32 | >32 | 2 | |
| Ciprofloxacin | 0.015->8 | >8 | 0.5 | |
| MDR (13) | MBX 1066 | 1->16 | >16 | >16 |
| MBX 1162 | 0.12-4 | 4 | 2 | |
| Imipenem | 0.06->32 | >32 | 4 | |
| Tigecycline | 0.25->32 | 4 | 2 | |
| Gentamicin | 0.5->32 | >32 | >32 | |
| Ciprofloxacin | 0.12->8 | >8 | >8 | |
| Pseudomonas aeruginosa | ||||
| All (27) | MBX 1066 | 0.06->16 | >16 | >16 |
| MBX 1162 | 0.03->16 | 1 | 0.25 | |
| Imipenem | 0.5->8 | >8 | 1 | |
| Gentamicin | 0.25->8 | >8 | 2 | |
| Ciprofloxacin | 0.12->2 | >2 | 0.25 | |
| Burkholderia cepacia | ||||
| All (11) | MBX 1066 | ≤0.015-4 | 0.06 | ≤0.015 |
| MBX 1162 | 0.03-0.25 | 0.12 | 0.06 | |
| Imipenem | 4->8 | >8 | 4 | |
| Tigecycline | 1-4 | 4 | 2 | |
| Gentamicin | >8 | >8 | >8 | |
| Ciprofloxacin | 0.5-2 | 2 | 2 | |
| Haemophilus influenzae | ||||
| All (12) | MBX 1066 | 1->16 | >16 | 4 |
| MBX 1162 | 0.5-4 | 4 | 1 | |
| Levofloxacin | 0.008-1 | 0.06 | 0.015 | |
| Cefotaxime | 0.03->4 | >4 | 1 | |
| Amoxicillin/ | 0.5/0.25-16/8 | 8/4 | 1/0.5 | |
| clavulanate | ||||
Bacterial sources: Clarian Health Partners, Indianapolis, IN; GR Micro, London, United Kingdom; University of California Los Angeles Medical Center, Los Angeles, CA; Mount Sinai Hospital, New York, NY, Pfizer Ann Arbor, Ann Arbor, MI; American Type Culture Collection, Manassas, VA.
90% and 50%, MIC90 and MIC50, respectively.
PISP, penicillin-intermediate S. pneumoniae.
Against Gram-positive species, MBX 1066 and 1162 displayed greater potencies than all comparator antibiotics (linezolid, vancomycin, imipenem, and daptomycin) against the antibiotic-resistant isolates (MRSA, MRSE, VRE [E. faecalis and E. faecium], penicillin-resistant S. pneumoniae [PRSP]) and all Enterococcus isolates, as well as the anaerobic C. difficile isolates (versus clindamycin, imipenem, and metronidazole) (Table 1). In addition, they demonstrated significant potency against the antibiotic-sensitive isolates of methicillin-susceptible S. aureus (MSSA), methicillin-susceptible S. epidermidis (MSSE), and penicillin-susceptible S. pneumoniae (PSSP), but their MIC90 values were slightly poorer than those of at least one comparator. Finally, they displayed equivalent efficacy to imipenem against S. agalactiae, while demonstrating greater potencies than the other three antibiotics. The overall MIC ranges for MBX 1066 and 1162 against Gram-positive isolates were 0.002 to 2 and 0.002 to 0.5 μg/ml, respectively, indicating that MBX 1162 is slightly more potent.
Against Gram-negative species, MBX 1162 was clearly more potent than MBX 1066. It was also more potent than all comparator antibiotics (imipenem, tigecycline, gentamicin, and ciprofloxacin), by its MIC90 and/or MIC range, in most cases, except that it exhibited lower potency than levofloxacin against H. influenzae isolates and equivalent potency to imipenem and tigecycline against E. coli isolates (Table 1). Of special interest, MBX 1162 was most potent against ESBL-producing K. pneumoniae strains and against all isolates of A. baumannii, including the MDR isolates (Table 1).
The critical need for new antibiotics, especially those that are effective against antibiotic-resistant and antibiotic-sensitive isolates of clinical pathogens, makes the results presented here highly significant. The bis-indole compounds are currently being pursued as topical agents for the treatment of wounds/skin infections and oral/parenteral agents for the treatment of systemic infections caused by antibiotic-resistant strains of A. baumannii, P. aeruginosa, K. pneumoniae, E. coli, Serratia marcescens, P. mirabilis, S. aureus, and Enterococcus species.
Acknowledgments
We thank Beverly H. Murray and Nanette L. Nichols of Micromyx for their contributions to this research.
This project was supported by DOD contract no. HDTRA-06-C-0042.
Footnotes
Published ahead of print on 12 July 2010.
REFERENCES
- 1.Brosh, R. M., Jr., J. K. Karow, E. J. White, N. D. Shaw, I. D. Hickson, and V. A. Bohr. 2000. Potent inhibition of Werner and Bloom helicases by DNA minor groove binding drugs. Nucleic Acids Res. 28:2420-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calhoun, J. H., C. K. Murray, and M. M. Manring. 2008. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin. Orthop. Relat. Res. 466:1356-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CLSI. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria: approved standard M11-A7. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed]
- 4.CLSI. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, M7-A7: approved standard, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.CLSI. 2006. Performance standards for antimicrobial susceptibility testing, M100-S16: 16th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Opperman, T. J., J. D. Williams, C. Houseweart, R. G. Panchal, S. Bavari, N. P. Peet, D. T. Moir, and T. L. Bowlin. 2010. Efflux-mediated bis-indole resistance in Staphylococcus aureus reveals differential substrate specificities for MepA and MepR. Bioorg. Med. Chem. 18:2123-2130. [DOI] [PubMed] [Google Scholar]
- 7.Panchal, R. G., R. L. Ulrich, D. Lane, M. M. Butler, C. Houseweart, T. Opperman, J. D. Williams, N. P. Peet, D. T. Moir, T. Nguyen, R. Gussio, T. Bowlin, and S. Bavari. 2009. Novel broad-spectrum bis-(imidazolinylindole) derivatives with potent antibacterial activities against antibiotic-resistant strains. Antimicrob. Agents Chemother. 53:4283-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh, F. M., and S. G. Amyes. 2004. Microbiology and drug resistance mechanisms of fully resistant pathogens. Curr. Opin. Microbiol. 7:439-444. [DOI] [PubMed] [Google Scholar]


