Abstract
AcrAB-TolC is the major constitutively expressed efflux pump system that provides resistance to a variety of antimicrobial agents and dyes in Escherichia coli. However, no systematically optimized real-time dye efflux assay has been published for the measurement of its activity and for detection of possible competition between substrates. Here, we report on the development of such an assay using a lipophilic dye, Nile Red. Energy-depleted cells were loaded with the dye in the presence of low (10 μM or less) concentrations of the proton conductor carbonyl cyanide m-chlorophenylhydrazone (CCCP). The CCCP was then removed, and efflux was triggered by energization with glucose. Various known efflux pump inhibitors and antimicrobials were checked for the ability to slow down Nile Red efflux, presumably through competition. Besides the known inhibitors Phe-Arg-β-naphthylamide and 1-naphthyl-methylpiperazine, several tetracyclic compounds (doxorubicin, minocycline, chlortetracycline, doxycycline, and tetracycline) and tetraphenylphosphonium chloride were found to interfere with dye efflux. This inhibition could not be explained by the depletion of proton motive force. None of the other tested antimicrobials, including macrolides, fluoroquinolones, and β-lactams, had any impact on Nile Red efflux, even at concentrations of up to 1 mM.
The tripartite AcrAB-TolC efflux pump complex belongs to one of the seven resistance-nodulation-division (RND) family efflux pumps in Escherichia coli. Based on the substrate specificity with an exceptionally wide range and the high constitutional expression levels under physiological conditions, it acts as the major contributor to the intrinsic multidrug resistance in the bacterium (19).
It is also the best structurally characterized RND efflux pump complex, since the crystal structures of AcrB (15, 16, 27, 28), TolC (11), and AcrA (14, 30) have recently been obtained. Based on the asymmetric crystal structures of AcrB trimers, a model has been proposed in which the efflux pump undergoes a functional rotation through three distinct states (access/loose, binding/tight, and extrusion/expulsion) (15, 27). Moreover, in one crystallographic study of AcrB, the substrates doxorubicin and minocycline were localized within a phenylalanine-rich binding pocket close to residues Phe615 and Phe178 (15). It was thus suggested that the main extrusion route for substrates is via a series of channels in the periplasmic domain to the binding pocket and then out to the TolC funnel.
While the structural models are relatively advanced, the phenotypic characterization of AcrAB-TolC has mainly relied on comparisons between the MIC values of the wild-type and the AcrAB-TolC deletion strains (for example, see references 23 and 29). Although kinetic constants were obtained recently (12, 18), the substrates are limited to β-lactams, and a special strain is needed for the assay. Another approach has been to examine the ability of the AcrAB-TolC complex to compete against the intracellular accumulation of dyes (e.g., ethidium, and rhodamine 6G) added to the medium. However, accumulation assays can give only indirect evidence of AcrAB-TolC activity. Moreover, it is difficult to examine substrate competition with these methods. To circumvent these problems, efflux assays have been applied to proteoliposomes reconstituted with RND pump proteins (1, 33). However, this is a very cumbersome process and allows only a limited number of experiments.
Because of these difficulties, a few efflux assays have been attempted with whole cells that were preloaded with dyes. However, in the case of dyes like ethidium (10), doxorubicin, and rhodamine 6G (21), which appear to become concentrated mainly in the cytosol by binding to nucleic acids and/or proteins, the observed efflux is slow, presumably because it represents the combination of a dissociation step and perhaps more than one efflux step (31). These assays are thus likely to severely underestimate the true efflux rate.
Lipophilic membrane-partitioning dyes would therefore be better suited for the efflux assay. N,N,N-Trimethyl-4-(6-phenyl-1,3,5-hexatrien-1-yl)phenylammonium p-toluenesulfonate (TMA-DPH) efflux was examined in Pseudomonas aeruginosa but was found to be rather slow (22). N-Phenyl-1-naphtylamine (NPN) was used in another P. aeruginosa RND pump efflux study and was found to be extruded much faster (13). Similar results were obtained by another group using 2-(4-dimethylamino)styryl-N-ethylpyridinium iodide (DASPEI) in an acrAB-overexpressing E. coli strain (17). No attempt was made to optimize the assay conditions in these studies, and the last two studies carried out the preloading of dyes with relatively high concentrations (100 μM) of the proton conductor carbonyl cyanide m-chlorophenylhydrazone (CCCP) and short preloading times of only a few minutes, conditions that in our experience decrease efflux after the addition of an energy source, presumably due to the incomplete removal of CCCP. Moreover, no systematic substrate competition study has been carried out using direct efflux assays.
We therefore tried to optimize such an efflux essay by examining the roles of CCCP and the dye concentration, as well as cell growth and treatment conditions. We chose as the dye the lipophilic compound Nile Red, which fluoresces only weakly in aqueous solutions but becomes strongly fluorescent in nonpolar environments and has thus far mainly been used for staining intracellular lipids (5). After optimization of the experimental conditions, we also examined whether some routinely used antibiotics and previously published efflux pump inhibitors (EPIs) (2, 25) are capable of inhibiting real-time Nile Red efflux. (A report showing the utility of Nile Red for assays of different classes of efflux pumps in Candida albicans [6] appeared after most of our experimental work had been completed).
MATERIALS AND METHODS
Culture techniques and strains.
The wild-type E. coli K-12 strain AG100, its marR mutant AG102 overproducing AcrAB by severalfold, and its acrAB deletion mutant AG100A have been described (23). 3-AG100 is a multidrug-resistant mutant with acrAB overexpression obtained from strain AG100 after repeated exposure to a fluoroquinolone (9). 1-DC14 (acrAB::kan) has also been described (7). They were maintained at −80°C in 15% (vol/vol) glycerol for cryoprotection. They were grown at 37°C in LB broth (1% tryptone, 0.5% yeast extract, and 1% NaCl).
Chemicals.
Most chemicals, including antimicrobial agents, were obtained from Sigma. 1-Naphthyl-methylpiperazine (NMP) was from Ryan Scientific Inc. (Mt. Pleasant, SC). Linezolid was from WaterStone Technology, LLC (Carmel, IN). Nile Red was obtained from Invitrogen (Carlsbad, CA). The stock solutions of potential competitors were usually made at 20 mM, so that insignificant amounts of solvents were introduced into the reaction mixture. (In this article, we use the word “competitor” loosely to imply any compound that inhibits Nile Red efflux, presumably through competition.) Some pump substrates and competitors were dissolved in water (doxorubicin, tetraphenylphosphonium [TPP], deoxycholate, aminoglycosides, and β-lactams), sometimes after conversion to a hydrochloride salt (NMP, azithromycin, and doxycycline). Others were dissolved in 50% (vol/vol) ethanol (Phe-Arg-β-naphthylamide [PAβN], minocycline, tetracycline, chlortetracycline, chloramphenicol, and fluoroquinolones) or dimethyl sulfoxide (DMSO) in the case of all other lipophilic compounds. It was established that DMSO concentrations of up to 5% (vol/vol) or ethanol concentrations up to 2.5% (vol/vol) had no detrimental effect on Nile Red efflux assays when energization was carried out with glucose.
[methyl-3H]β-d-thiogalactopyranoside (TMG) was obtained from Moravek Biochemicals (Brea, CA) as 30-μg/ml (1-mCi/ml) stock solution in 33% ethanol (vol/vol).
Optimized Nile Red efflux (inhibition) assay.
A 0.1-ml portion of an overnight culture (grown at 140 rpm on a shaker at 37°C) of the assayed E. coli strain was added to 20 ml LB broth in an Erlenmeyer flask and grown on a shaker (140 rpm; 37°C) for 14 to 16 h (the optical density at 660 nm [OD660] usually reached a plateau after 9 h). A 10-ml portion of the culture was centrifuged at 4,400 rpm for 10 min at room temperature. The pellet was resuspended in 20 mM potassium phosphate buffer (pH 7.0) containing 1 mM MgCl2 (PPB). After another centrifugation and resuspension step, the cells were adjusted to an OD660 of 1.0 in PPB. All subsequent operations were carried out at room temperature, except as noted. The cells were allowed to rest for about 15 min, and 2-ml aliquots were then transferred to Pyrex 15-ml conical centrifugation tubes (glass tubes were used because Nile Red becomes adsorbed to the walls of plastic tubes), and CCCP (5 mM stock solution in 50% DMSO) was added to a final concentration of 10 μM for the acrAB-overexpressing strain 3-AG100 or AG102 and 5 μM for strain AG100, AG100A, or 1-DC14.
After 15 min, a potential competitor was added to the desired final concentration if it was an inhibition experiment. After another 15 min, Nile Red (a stock solution of 5 mM in 10% dimethyl formamide-90% ethanol [vol/vol]) was added to a final concentration of 5 μM, and the cell suspension was incubated on a shaker (140 rpm; 37°C) for 3 h. The cells were left at room temperature for 60 min and then centrifuged for 5 min at 4,400 rpm. After the bulk of the supernatant was discarded, droplets of it clinging to the tube wall were thoroughly removed with absorbent tissues, and the cells were resuspended in 2 ml PPB in the presence of the competitor if any was used. (In the case of strongly fluorescent compounds, like tetracyclines and doxorubicin, the inhibitor was present only during the 3 h of incubation but not in the resuspension buffer to avoid the high background fluorescence in the external medium.) Immediately thereafter, 0.2 ml of this cell suspension was transferred to a quartz cuvette (Starna, Atascadero, CA) containing 1.8 ml of PPB, and the cuvette was placed in a QuantaMaster spectrofluorimeter (Photon Technology International). The slit width was 10 nm, excitation was at 552 nm, and emission was at 636 nm. The mixture was stirred with a magnetic stirrer. The fluorescence of the cell suspension was followed over 100 s, and Nile Red efflux was then triggered by rapid energization with 100 μl of 1 M glucose and monitored for another 200 s.
Uptake of TMG by the LacY transporter.
The accumulation of radioactively labeled TMG against a concentration gradient is a standard technique for estimation of the proton motive force, which energizes the active transport of TMG by the lactose permease LacY (8, 24, 26). To evaluate whether some compounds inhibit Nile Red efflux by diminishing the proton motive force, 100 μl of an overnight culture of 3-AG100 in LB broth was added to 20 ml LB broth in an Erlenmeyer flask and grown at 37°C (140 rpm) in the presence of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) until an OD660 of ∼0.8 was reached. The cell suspension was centrifuged at 4,400 rpm for 10 min, and the pellet was resuspended in 20 mM PPB. After another centrifugation and resuspension step, the cells were adjusted to an OD660 of 1.0 in PPB. A 100-μl portion of this cell suspension was then transferred to a microcentrifuge tube, and the respective competitor was added to a final concentration that had been shown to lead to ∼90% Nile Red efflux inhibition. In each experiment, negative (completely abolishing transport in the presence of 200 μM CCCP) and positive (no-competitor) controls were processed. After an exposure of 15 min, 3H-labeled TMG was added to a final concentration of 2 μM. After another 10 min, the cells were placed on 25-mm-diameter (pore size, 0.45 μm) Metricel GN-6 cellulose filters (Pall Corporation, NY) and washed with 5 ml of PPB using a vacuum filtration unit. Thereafter, the filters were dissolved in 5 ml of EcoLume scintillation cocktail (ICN Biomedicals, Costa Mesa, CA), and accumulation of radioactive TMG was quantified using a Beckman LS6500 liquid scintillation counter. At least three independent experiments were performed per tested competitor.
RESULTS
Development of an optimized Nile Red efflux protocol.
Preliminary Nile Red efflux assays in the acrAB-overexpressing strain 3-AG100 showed that CCCP concentrations of 100 μM or greater, which are usually used, according to the literature, to block efflux and to allow preloading of dyes (13, 17, 22), significantly impaired Nile Red efflux even after repeated washing of the cells (data not shown). However, at much lower CCCP concentrations of 10 to 20 μM, which still led to the Nile Red loading level corresponding to 90 to 100% of the level seen with 100 μM CCCP, there was substantial preenergization efflux of Nile Red after resuspension of the cells in CCCP-free buffer, suggesting incomplete de-energization of the cells. Since in these experiments the cells were grown to mid-exponential phase in LB broth, we tried to alter the growth conditions so that the preenergization efflux of Nile Red could be avoided. We first tried M9 or M63 minimal medium supplemented with 0.4% glucose, but there was no decrease in preenergization efflux. However, when cells were grown in LB broth overnight for 14 to 16 h (well into the stationary phase; see Materials and Methods), preenergization efflux was dramatically reduced.
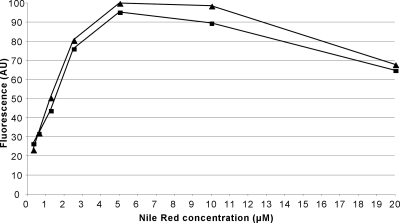
Additional experiments were carried out to determine the optimal concentrations and lengths of CCCP/Nile Red incubation. We found that incubation periods longer than 3 h did not improve the Nile Red loading further. Nile Red concentrations of 5 μM gave the best overall fluorescence yield, with increasing concentrations leading to decreased fluorescence (Fig. 1), possibly due to self-quenching within the cell membrane. CCCP concentrations as low as 5 to 10 μM in strain 3-AG100 and 2.5 to 5 μM in strain AG100 led to sufficient dye loading over the 3 h of incubation without compromising the substrate efflux after washing. CCCP concentrations of 40 μM or higher were found to slow down efflux considerably, even after washing (data not shown). We also tried energization with ammonium formate instead of glucose. However, we later dropped it in favor of glucose, since it was found that the formate metabolism was very sensitive to numerous compounds, including solvents (as low as 1% ethanol) and certain antibiotics (especially chloramphenicol and gentamicin).
FIG. 1.
Nile Red loading (determined as relative fluorescence units) in strain 3-AG100 before the energization of efflux, depicted as a function of the Nile Red concentration. Two independent experiments are shown. Fluorescence intensity is shown in arbitrary units (AU).
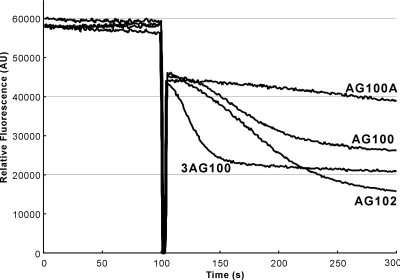
After the addition of glucose, which presumably produced the proton motive force across the cytoplasmic membrane, Nile Red was rapidly transported out from the cells of acrAB-overexpressing strains (only 20 s for 50% Nile Red efflux in strain 3-AG100 and about 70 s in AG102 [Fig. 2]), but it exited only very slowly from strain 1-DC14 or AG100A with acrB deleted (more than 200 s for 50% Nile Red efflux). The efflux occurred at a modest rate in strain AG100, which expresses acrAB at the wild-type level (Fig. 2).
FIG. 2.
Nile Red efflux after addition of 50 mM glucose at 100 s to de-energized cells displaying various levels of acrAB expression, from none (AG100A) to wild-type level constitutive expression (AG100) and overexpression (a modest level [AG102] and a high level [3-AG100]).
Competition with Nile Red efflux.
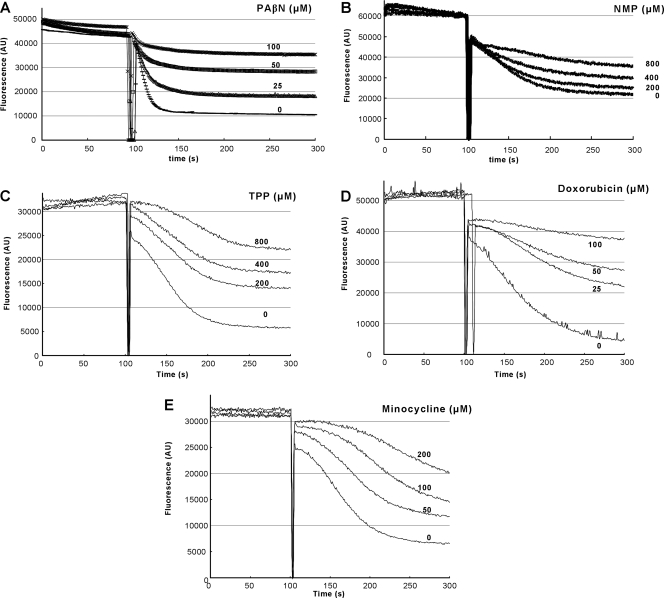
Various antimicrobials, most of which have been previously shown to be substrates of the AcrAB-TolC pump complex (20, 29), were examined to see if they inhibited, presumably through competition, the energized efflux of Nile Red. We also included in this series of experiments the known EPIs PAβN and NMP, as well as negative controls, such as aminoglycosides. All compounds (with the exception of doxorubicin and PAβN, which were powerful inhibitors) were tested at 200 μM and 1,000 μM, and representative experiments demonstrating positive competition are shown in Fig. 3. Additionally, we estimated the time that was needed by the cells to extrude half of the preloaded Nile Red into the external medium as a (semi)quantitative indicator of the extent of inhibition; this is called tefflux50% in Table 1.
FIG. 3.
Dose-dependent inhibition of Nile Red efflux by presumed competitors. Efflux was triggered at 100 s by the addition of 50 mM glucose. The intensity of fluorescence emission from Nile Red is shown in arbitrary units (AU). (A) Effect of PAβN in strain 3-AG100. (B) Effect of NMP in strain AG102. (C) Effect of TPP in strain AG102. (D) Effect of doxorubicin in strain AG102. As indicated in the text, doxorubicin was present only in the preincubation buffer and was removed from the assay mixture. (E) Effect of minocycline in strain AG102. Minocycline was present only in the preincubation buffer and was removed from the assay mixture.
TABLE 1.
Inhibition of Nile Red efflux in strains 3-AG100 and AG102a
| Compound |
tefflux50% (s) at: |
|
|---|---|---|
| 200 μM | 1,000 μM | |
| PAβN | >200 | >200 |
| Doxorubicin | >200 (>200) | >200 (>200) |
| Minocycline | >200 (>200) | >200 (>200) |
| Chortetracycline | >200 (200) | >200 (>200) |
| Doxycycline | 30 (170) | >200 (>200) |
| Tetracycline | 25 (90) | >200 (>200) |
| NMP | 25 (100) | >200 (>200) |
| TPP | 25 (105) | >200 (>200) |
| Chloramphenicol | 20 | 20 |
| Kanamycin | ND | 20 |
| Gentamicin | ND | 20 |
| Streptomycin | ND | 20 |
| Spectinomycin | ND | 20 |
| Rifampin | ND | 20 |
| Azithromycin | ND | 20 |
| Erythromycin | ND | 20 |
| Clarithromycin | ND | 20 |
| Ciprofloxacin | ND | 20 |
| Norfloxacin | ND | 20 |
| Nalidixic acid | ND | 20 |
| Fusidic acid | ND | 20 |
| Deoxycholate | ND | 20 |
| Ampicillin | ND | 20 |
| Nafcillin | ND | 20 |
| Linezolid | ND | 20 |
| Novobiocin | ND | 20 |
| 1,10-Phenanthroline | ND | 20 |
| No competitor | 20 (70) | 20 (70) |
AG102 is shown in parentheses. The extent of inhibition is expressed as the mean time for 50% efflux after energization with 50 mM glucose by various compounds at 200 μM and 1,000 μM. The time was rounded to 5 s. At least three independent experiments were performed for each inhibitor.
The known EPIs PAβN and NMP in fact inhibited Nile Red efflux in a dose-dependent manner. PAβN almost completely inhibited Nile Red efflux at 100 μM (Fig. 3A), whereas at least 800 μM NMP was needed to achieve comparable inhibition (Fig. 3B). Surprisingly, TPP was found to be as potent as NMP in its ability to block Nile Red efflux (Fig. 3C). Also all of the tested tetracyclic agents (tetracycline, chlortetracycline, doxycycline, and doxorubicin) were found to retard Nile Red efflux (Table 1), with doxorubicin (Fig. 3D), minocycline (Fig. 3E), and chlortetracycline (not shown) being the most potent compounds. Interestingly, all of the above-mentioned inhibitors were also found to be active when present only in the incubation buffer and not in the resuspension buffer.
Effects of inhibitors on the proton motive force.
Because the AcrB-mediated efflux of Nile Red is energized by the proton motive force, there was a possibility that the inhibition of efflux occurred indirectly through the dissipation of proton motive force by the inhibitors. We therefore examined the extent of uphill accumulation of [3H]TMG, catalyzed by a similarly proton motive force-coupled LacY transporter, as an indicator of membrane energization (8). As seen in Table 2, inhibitors often reduced the accumulation of TMG, but the remaining level of proton motive force, as judged from the extent of accumulation, was still usually higher than 44%. Tetracycline and chlortetracycline dissipated the proton motive force somewhat further, yet its levels (27 and 14%) were much higher than that observed in the presence of CCCP (nearly complete dissipation to 4.5%) (Table 1). In this assay, we included 800 μM 1,10-phenanthroline, which does not inhibit Nile Red efflux (Table 1). It decreased [3H]TMG accumulation to 34% (not shown). This result shows that such a large reduction in the proton motive force cannot by itself decrease Nile Red efflux visibly and reinforces our conclusion that the inhibition was not the indirect effect of energy dissipation.
TABLE 2.
[3H]TMG accumulation in strain 3-AG100 in the presence of various compounds at concentrations that inhibited Nile Red efflux almost completely (∼90%)
| Compound | Radioactivity (cpm)a | SDa | % Accumulation |
|---|---|---|---|
| None | 84,700 | 28,800 | 100 |
| TPP (800 μM) | 82,000 | 20,300 | 97 |
| Doxorubicin (100 μM) | 65,700 | 14,600 | 78 |
| PAβN (100 μM) | 50,900 | 14,100 | 60 |
| NMP (800 μM) | 44,700 | 22,800 | 53 |
| Minocycline (200 μM) | 38,500 | 5,700 | 45 |
| Doxycycline (800 μM) | 37,500 | 5,300 | 44 |
| Tetracycline (800 μM) | 23,100 | 8,700 | 27 |
| Chlortetracycline (200 μM) | 12,200 | 6,400 | 14 |
| CCCP (200 μM) | 3,800 | 1,500 | 4.5 |
At least three independent experiments were performed per inhibitor.
DISCUSSION
We developed an optimized real-time Nile Red efflux assay for studies of substrate efflux and competition in whole E. coli cells. Nile Red was an excellent substrate of the AcrAB-TolC pump complex and was extruded from preloaded cells rapidly if strains with elevated pump protein expression were used. It is also nontoxic for E. coli and gives a very good signal-to-noise ratio, since its fluorescence yield is high when it is associated with cell membrane phospholipids but it is almost nonfluorescent when present in the external medium. Moreover, due to its long maximum emission wavelength (636 nm), problems with interference when measuring efflux in the presence of visibly absorbing or fluorescent inhibitors are not as pronounced as with other dyes.
The only drawback is that loading of the cells at an external concentration of more than 5 μM leads to decreased Nile Red fluorescence (Fig. 1), which is most likely due to increased self-quenching. On the other hand, use of very low concentrations of Nile Red was not practical, because a significant fraction of the added dye was adsorbed to the walls of the tubes and cuvettes. In principle, the course of efflux of a dye should follow the integrated form of the Michaelis-Menten equation, and thus, the curve fitting was expected to give us the kinetic constants of Nile Red efflux. However, in practice, it was impossible to carry out this analysis because of self-quenching and perhaps because the membrane could not be energized instantaneously upon the addition of glucose.
One of the important outcomes of this study was that several antibiotics were shown to compete against Nile Red efflux. With the previously reported EPI PAβN, NPN efflux was already shown to be inhibited (13). With the other EPI, NMP, inhibition of efflux was shown indirectly by the increased intracellular accumulation of levofloxacin and ethidium (2). However, direct competition between dye efflux and common antimicrobials has not been demonstrated to our knowledge. In fact attempts in our laboratories to show competition between antimicrobials through the determination of MIC values were not successful (unpublished results). Here, we have shown clearly that several antibiotics, as well as the known EPIs PAβN and NMP, act as powerful, dose-dependent inhibitors of Nile Red efflux, presumably as competitors of Nile Red (Fig. 3 and Table 1). This apparent inhibition cannot be explained solely by the depletion of proton motive force by the competitors (Table 1).
The newly discovered inhibitors of Nile Red efflux include various tetracyclines (tetracycline, doxycycline, minocycline, and chlortetracycline), as well as an anthracycline (doxorubicin) and TPP (Table 1). Curiously, most of the newly discovered inhibitors are tetracyclic compounds, although the known EPIs are not. Two of the tetracyclic inhibitors, which were detected in a recent AcrB cocrystalization study within a phenylalanine-rich substrate binding pocket (15), were found to be the most powerful inhibitors, doxorubicin and minocycline, exerting very strong inhibition at concentrations as low as 100 to 200 μM (Fig. 3D and E), which is similar in potency to the well-characterized EPI PAβN (Fig. 3A).
Some inhibitors, with the exception of doxorubicin, were found to be active at concentrations above their own MICs. However, this does not mean that the observed efflux inhibition is due to their antimicrobial activity. Tetracyclines are bacteriostatic agents that inhibit protein synthesis but are not likely to alter other aspects of bacterial metabolism in the short term. Moreover, the translational inhibitors linezolid and the macrolides had absolutely no impact on Nile Red efflux at concentrations of up to 1 mM (in the cases of linezolid and erythromycin even up to 2 mM).
One surprising result of this study was that the apparent competition or inhibition was limited to the group of agents described above. Thus, other substrates of the AcrAB-TolC system tested, such as chloramphenicol, rifampin, macrolides, fluoroquinolones, nalidixic acid, linezolid, novobiocin, nafcillin, and deoxycholate, showed no evidence of competition (Table 1), even though we were using very high concentrations, up to 1 mM. A recent study (4) has also failed to show efflux competition between various substrates, including steroids. This lack of observable competition may be due to the fact that the capacity of this transport system is very high, so that competition cannot be detected, as hypothesized in reference 4. We have recently succeeded in measuring the efflux rates of β-lactam substrates by the AcrAB-TolC complex (12, 18): some substrates (e.g., cephaloridine and oxacillin) have high Vmax values corresponding to turnover numbers of 500 to 1,000, although others (e.g., nitrocefin) have much lower turnover numbers, around 10.
An alternative interpretation of these widely different competition data is that the binding sites within the relatively big substrate binding cavity may be different for different compounds. We carried out computer docking studies (32) of the binding of various substrates to the AcrB binding cavity identified by Murakami et al. (15). We found that most substrates bind to the cavity in one of the two ways. Some substrates were predicted to bind to the narrow groove at the end of binding site far away from the plane of the membrane and were called “groove binders.” In contrast, some others appeared to bind to the wider area closer to the plane of the membrane and were called “cave-binders.” The limited range of experiments using the efflux of nitrocefin and the labeling of Phe615Cys by fluorescein-maleimide indeed suggested that the groove binders compete against each other but that there was little competition between a groove binder and a cave binder. Nile Red, used in the present experiments, appears to bind predominantly to the groove area (see Fig. S2C in reference 32). Although its “lowest-energy” binding mode was to the cave, the differences in the calculated binding energies were so small that we cannot give much weight to this one binding pose. In this light, it becomes understandable that Nile Red efflux was inhibited, presumably through competition, by known groove binders, such as tetracycline, minocycline, chlortetracycline, doxycycline, and doxorubicin (Table 1). Similarly, a cave binder (chloramphenicol) did not interfere with Nile Red efflux. However, the correspondence is not perfect, as groove binders, such as novobiocin, erythromycin, and nafcillin, did not show inhibition of Nile Red efflux, a result that could have resulted from differences in affinities to the binding site among various ligands. Among known EPIs, PAβN is predicted to behave most of the time as a groove binder (32) and indeed was a strong inhibitor. NMP, which appeared to be a typical cave binder (32), surprisingly showed strong inhibition, albeit at high concentrations (Table 1); this might indicate that EPI acts not only by competition, but also by some other unique mechanisms. Our results are also consistent, to some extent, with the results of site-directed mutagenesis of Phe residues surrounding the substrate binding site (3), as discussed previously (32). However, site-directed mutagenesis has revealed more specific interactions, such as the effect of Phe615 on macrolide efflux (1), and these data indicate that we must combine different approaches to come to a more concrete understanding of the substrate binding and competition among substrates.
Acknowledgments
This study was supported in part by a grant from the U.S. Public Health Service (AI-09644).
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Aires, J. R., and H. Nikaido. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 187:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohnert, J. A., and W. V. Kern. 2005. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 49:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohnert, J. A., S. Schuster, M. A. Seeger, E. Fahnrich, K. M. Pos, and W. V. Kern. 2008. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J. Bacteriol. 190:8225-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elkins, C. A., and L. B. Mullis. 2007. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob. Agents Chemother. 51:923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenspan, P., and S. D. Fowler. 1985. Spectrofluorometric studies of the lipid probe, Nile Red. J. Lipid Res. 26:781-789. [PubMed] [Google Scholar]
- 6.Ivnitski-Steele, I., A. R. Holmes, E. Lamping, B. C. Monk, R. D. Cannon, and L. A. Sklar. 2009. Identification of Nile Red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal. Biochem. 394:87-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashket, E. R. 1985. The proton motive force in bacteria: a critical assessment of methods. Annu. Rev. Microbiol. 39:219-242. [DOI] [PubMed] [Google Scholar]
- 9.Kern, W. V., M. Oethinger, A. S. Jellen-Ritter, and S. B. Levy. 2000. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 44:814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klyachko, K. A., S. Schuldiner, and A. A. Neyfakh. 1997. Mutations affecting substrate specificity of the Bacillus subtilis multidrug transporter Bmr. J. Bacteriol. 179:2189-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 12.Lim, S. P., and H. Nikaido. 2010. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter acrab-tolc of Escherichia coli. Antimicrob. Agents Chemother. 54:1800-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikolosko, J., K. Bobyk, H. I. Zgurskaya, and P. Ghosh. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173-179. [DOI] [PubMed] [Google Scholar]
- 16.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 17.Murakami, S., N. Tamura, A. Saito, T. Hirata, and A. Yamaguchi. 2004. Extramembrane central pore of multidrug exporter AcrB in Escherichia coli plays an important role in drug transport. J. Biol. Chem. 279:3743-3748. [DOI] [PubMed] [Google Scholar]
- 18.Nagano, K., and H. Nikaido. 2009. Kinetic behavior of the major multidrug efflux pump acrb of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:5854-5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 20.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ocaktan, A., H. Yoneyama, and T. Nakae. 1997. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa. J. Biol. Chem. 272:21964-21969. [DOI] [PubMed] [Google Scholar]
- 23.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purdy, D. R., and A. L. Koch. 1976. Energy cost of galactoside transport to Escherichia coli. J. Bacteriol. 127:1188-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renau, T. E., R. Leger, E. M. Flamme, J. Sangalang, M. W. She, R. Yen, C. L. Gannon, D. Griffith, S. Chamberland, O. Lomovskaya, S. J. Hecker, V. J. Lee, T. Ohta, and K. Nakayama. 1999. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 42:4928-4931. [DOI] [PubMed] [Google Scholar]
- 26.Rotman, B. 1977. On the rate limiting step in downhill transport via the LacY permease of Escherichia coli. J. Supramol. Struct. 7:29-35. [DOI] [PubMed] [Google Scholar]
- 27.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295-1298. [DOI] [PubMed] [Google Scholar]
- 28.Sennhauser, G., P. Amstutz, C. Briand, O. Storchenegger, and M. G. Grutter. 2007. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS. Biol. 5:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Symmons, M. F., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2009. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. U. S. A. 106:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tal, N., and S. Schuldiner. 2009. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl. Acad. Sci. U. S. A. 106:9051-9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takatsuka, Y., C. Chen, and H. Nikaido. 2010. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]