Abstract
The bacterial burden on human health is quickly outweighing available therapeutics. Our long-term goal is the development of antimicrobials with the potential for broad-spectrum activity. We previously reported phthalazine-based inhibitors of dihydrofolate reductase (DHFR) with potent activity against Bacillus anthracis, a major component of Project BioShield. The most active molecule, named RAB1, performs well in vitro and, in a cocrystal structure, was found deep within the active site of B. anthracis DHFR. We have now examined the activity of RAB1 against a panel of bacteria relevant to human health and found broad-spectrum applicability, particularly with regard to Gram-positive organisms. RAB1 was most effective against Staphylococcus aureus, including methicillin- and vancomycin-resistant (MRSA/VRSA) strains. We have determined the cocrystal structure of the wild-type and trimethoprim-resistant (Phe 98 Tyr) DHFR enzyme from S. aureus with RAB1, and we found that rotational freedom of the acryloyl linker region allows the phthalazine moiety to occupy two conformations. This freedom in placement also allows either enantiomer of RAB1 to bind to S. aureus, in contrast to the specificity of B. anthracis for the S-enantiomer. Additionally, one of the conformations of RAB1 defines a unique surface cavity that increases the strength of interaction with S. aureus. These observations provide insights into the binding capacity of S. aureus DHFR and highlight atypical features critical for future exploitation in drug development.
Despite the introduction of antibiotics such as penicillin in the 1940s and approval of new classes, such as cyclic lipopeptides like daptomycin, infectious disease continues worldwide. Antibiotic resistance is increasing, and reliance on existing scaffolds is not sufficient to combat multidrug resistance (23, 33). Staphylococcus aureus has become a primary concern among antibiotic-resistant infectious disease agents, with methicillin-resistant S. aureus (MRSA) infections claiming 19,000 lives in the United States per year and costing the United States $3 to 4 billion dollars per year (18). As the incidence of MRSA has shifted from hospital settings to the community at large, treatment preferences have moved to oral formulations and have impacted drug resistance profiles. Community-acquired MRSA is showing increasing resistance to fluoroquinolones and clindamycin, and in response clinicians are shifting to doxycycline, linezolid, or trimethoprim-sulfamethoxazole as the best options for outpatient treatment (21, 30).
While treatment preferences are adjusting to isolated outbreaks, S. aureus is increasingly resistant to trimethoprim (TMP) (13). TMP targets the enzyme dihydrofolate reductase (DHFR), a critical component of folate metabolism, and is required for continued nucleic acid synthesis (6). DHFR is a good target because of the specificity that can be achieved; however, bacteria have developed resistance mechanisms that include acquisition of plasmid-derived (though catalytically inferior) versions of DHFR, leading to high-level resistance. Alternatively, point mutations of the chromosomal DHFR have been demonstrated to confer intermediate resistance, such as the S. aureus DHFR (saDHFR) residue Phe 98 to Tyr (F98Y) mutation (13). While other mutations are frequently found with F98Y, it alone can increase the MIC of TMP by at least 1 order of magnitude. In efforts to compensate for a lower inhibitory action, TMP is frequently combined with a sulfa inhibitor of the enzyme preceding DHFR, termed dihydropterin synthase (DHPS). Unfortunately, sulfa drugs have notoriously poor pharmacokinetics and can have unpleasant, and sometimes fatal, side effects (15).
RAB1 is a lead compound under investigation for treatment of inhalation anthrax. RAB1's structure is modeled on trimethoprim but is extended by addition of an acryloyl linker and phthalazine moiety, including a propyl group at a stereogenic carbon. This results in approximately 40% more surface area than TMP, which provides critical contact regions for further interaction with the binding site. Specific structural features of RAB1 allow unique contacts with the protein that can overcome natural and induced resistance. For B. anthracis, the half-maximal inhibitory concentration (IC50) of purified DHFR enzyme with TMP is 77 μM, indicating a natural resistance, while with RAB1 it is ≈60 nM (3). Selectivity for the binding site in B. anthracis DHFR is controlled by the large hydrophobic phthalazine moiety, which is embedded within and causes extension of the binding site (4). Concomitant with the current work, the broad-spectrum activities of related RAB1-like molecules have been demonstrated by Basilea Pharmaceutica International AG. These studies also highlighted the difficulty in generating spontaneous resistant mutations of chromosomal DHFR to inhibitors in the dihydrophthalazine series (7, 9).
We have characterized the broad-spectrum applicability of RAB1 and its particular effectiveness against S. aureus, including MRSA and vancomycin-resistant S. aureus (VRSA) strains. Both enantiomers of RAB1 show potent antimicrobial activities, as indicated by favorable MICs and IC50s. The impact of the TMP resistance-conferring mutation F98Y is negligible on RAB1 activity and preferentially decreases binding of the R-enantiomer. The X-ray crystal structures of RAB1 complexed to S. aureus DHFR reveal two conformations for the large phthalazine group. While one conformation resides in the known binding site, the other sits in a shallow surface cavity that has been previously undocumented as susceptible to inhibitor binding. This allows an increase in binding strength and provides an explanation for the lack of enantiomeric preference. These observations provide insights into the binding capacity of S. aureus DHFR and highlight atypical features critical for future exploitation in drug development, as well as insights into the mechanism of action of phthalazine-based inhibitors.
MATERIALS AND METHODS
Synthesis of RAB1 and purification of its component enantiomers have been previously described (4, 5).
Determination of the MIC.
The MIC determination experiments were carried out through the NIH/NIAID In vitro Assessment for Antimicrobial Activity Resource for Researchers program. Bacterial strains were obtained through the Biodefense and Emerging Infectious Research Repository, NIAID, NIH, or from the American Type Culture Collection. Experiments with Bacillus anthracis Ames, Brucella abortus 2308, Francisella tularensis Schu4, and Yersinia pestis CO92 were carried out in a CDC-registered and approved biosafety level 3 (BSL-3) laboratory with appropriate biosafety and security measures. Experiments with other organisms/strains were carried out in a BSL-2 laboratory with appropriate biosafety and security measures. The MICs were determined using a broth microdilution assay in accordance with the Clinical and Laboratory Standards Institute (CLSI) recommendations (10). Each MIC was determined in duplicate for two replicates; 96-well plates containing 2-fold serial dilutions of test compounds or commercial antibiotics (used for quality control as directed by CLSI guidelines [11]) were prepared in cation-adjusted Mueller-Hinton broth for all agents (with the addition of 2% IsoVitaleX for F. tularensis), except B. abortus, which was grown in Brucella broth. Ten microliters of a standardized inoculum was used to infect wells containing 100 μl of medium with or without drug. Inoculum concentrations were verified by plating for CFU using the CLSI recommended protocol (10). Experimental controls included growth control wells, sterility control wells, and uninoculated drug wells to verify that test compounds remained soluble under experimental conditions. Antimicrobial susceptibility testing was performed for single isolates under the following conditions: B. anthracis for 16 h, Y. pestis for 24 h, B. abortus for 48 h, F. tularensis for 48 h with 5% CO2, and vancomycin-resistant enterococci (VRE) for 20 h, all at 37°C. Four MRSA strains were grown for 18 h, and three VRSA strains were grown for 20 h, all at 35°C. After the appropriate incubation, plates were allowed to equilibrate to room temperature for 30 min. They were then sealed, and the absorbance at 600 nm was spectrophotometrically measured to determine the MIC, defined as the lowest compound concentration that inhibited growth of the microorganism. Visual confirmation of growth was also performed. When variation was obtained in experimental values the MIC is reported as a range. Growth patterns with some S. aureus strains exhibited trailing, as has been previously noted for folate pathway inhibitors (10). In these instances, MIC values were calculated as the concentration of compound resulting in an 80% reduction in growth of the microorganism. However, this did not result in a shift of a MIC from that obtained using the lowest concentration of compound that inhibited growth of the microorganism.
Antibiotic susceptibility profiles for S. aureus isolates were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus website and are listed in Table 1.
TABLE 1.
Antibiotic susceptibility profiles for S. aureus isolatesa
| Drugb | MIC (μg/ml)/CLSI interpretationc |
||||||
|---|---|---|---|---|---|---|---|
| NRS1 | NRS70 | NRS71 | NRS123 | VRS1 | VRS2 | VRS3 | |
| VAN | 8/I | 0.5/S | 0.5/S | 1/S | >256/R | 64/R | >32/R |
| TEC | 8/S | 0.25/S | 0.5/S | 1/S | 64/R | 8/S | 16/I |
| CIP | 16/R | 0.5/S | >16/R | 0.5/S | >16/R | >16/R | >16/R |
| DAP | 2/NA | 0.5/NA | |||||
| CLI | >4/R | >4/R | 0.12/S | >4/R | >4/R | >4/R | |
| ERY | >8/R | >8/R | 0.5/S | >8/R | >8/R | >8/R | |
| GEN | >16/R | 1/S | 0.5/S | 0.5/S | >16/R | >16/R | >16/R |
| LNZ | 1/S | 1/S | 2/S | 2/S | 1/S | 2/S | |
| OXA | >32/R | >16/R | >16/R | >16/R | >16/R | >16/R | >16/R |
| PEN | >8/R | >2/R | >2/R | >2/R | >2/R | >2/R | >2/R |
| SYN | 0.12/S | 0.5/S | 1/S | 0.25/S | 0.5/S | 0.5/S | 0.5/S |
| TET | 32/R | 32/R | |||||
| SXT | ≤0.12/2.28/S | ≤0.25/4.75/S | ≤0.25/4.75/S | ≤0.25/4.75/S | 2/38/S | 1/19/S | 0.25/4.75/S |
Data taken from www.narsa.net.
Abbreviations of drugs: VAN, vancomycin; TEC, teicoplanin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; LNZ, linezolid; OXA, oxacillin; PEN, penicillin; SYN, quinupristin-dalfopristin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole.
S, susceptible; R, resistant; I, intermediate; NA, not available.
Enzyme activity and calculation of the IC50.
Purified recombinant wild-type (wt) and F98Y S. aureus DHFR proteins were stored at −80°C in 10% glycerol (see the description of protein production and purification methods, below); the presence of the histidine tag did not affect activity. The assay was adapted from the standard format (2) to a high-throughput 96-well plate platform with a 200-μl total reaction volume and was carried out with a Biomek 2000 liquid handling robot interfaced with a DTX880 plate reader. Enzyme, saturating NADPH, and inhibitor in dimethyl sulfoxide were preincubated at 30°C; the reaction was initiated by the addition of dihydrofolic acid (DHF) and monitored for 3 min, during which time the reaction remained linear. The protein concentration (4.5 nM) was adjusted to yield an activity of 1.4 nmol DHF reduced per minute for the wild-type DHFR. Detection utilized the redox-sensitive tetrazolium dye 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS). MTS is reduced by the product tetrahydrofolate to yield an increased absorbance at 450 nm, in contrast to the standard assay, which follows the loss of cofactor (oxidation of NADPH) at 340 nm. The increasing signal from MTS reduction is greater than the loss of signal upon NADPH oxidation, facilitating measurement of smaller volumes using shorter path lengths. Reactions were performed in triplicate. The change in signal was calculated as a percentage of a reaction with no inhibitor over 2.8 min of reaction and was used to calculate an absolute IC50 from the fit of a four-parameter logistic model using the KC Jr. plate reader software. The Michaelis-Menten constants (Km) for the DHF substrate were calculated by fitting the Michaelis-Menten equation in Microsoft Excel using the solver plug-in. The IC50 and Km values were used to compute the equilibrium inhibition constant (Ki) by using the formalism of Cheng-Prusoff (8). The Ki value allows for direct comparison between the wild-type DHFR protein and the less catalytically active F98Y mutant. Values for Km of DHF were 43.7 ± 5.9 μM for wild-type saDHFR and 11.0 ± 1.7 μM for the F98Y mutant. These values reflect differences arising from experimental modification, but trends are consistent with previously published values of 5.1 to 14.5 μM for the wild-type enzyme and 0.86 to 7.3 μM for the F98Y mutant (13, 14, 20, 26).
Protein production, purification, and crystallization.
Recombinant S. aureus DHFR was expressed from a pET 101D vector (Invitrogen) containing a C-terminal 6His affinity tag. The F98Y point mutant was generated using the QuikChange II site-directed mutagenesis kit (Stratagene). Purification utilized immobilized metal ion affinity chromatography (GE Life Sciences), followed by cleavage of the affinity tag using an engineered thrombin site. The final step used an S100 size exclusion column (GE Life Sciences) and exchanged the buffer to 20 mM Tris (pH 8), 150 mM NaCl, and 5% glycerol. Crystallization was carried out in a hanging drop format at room temperature. Protein was concentrated to 10 to 12 mg/ml and incubated with 1 mM NADPH (Sigma) and a 10-fold molar excess of RAB1 for at least 2 h prior to initiation of crystallization. The crystallization well contained 15% polyethylene glycol 6000, 0.15 M sodium acetate, and 0.1 M morpholineethanesulfonic acid (pH 6.5); 2 μl of this solution was mixed with 3 μl of protein solution and sealed over the well. Crystals appeared within a few days and grew to maximum size in 1 to 2 weeks.
X-ray data collection, processing, and refinement.
A single crystal was used for each data collection after cryoprotection in mother liquor with 15% glycerol and plunging into liquid nitrogen. Data were collected with a Rigaku RU3HR rotating anode generator coupled with an Raxis 4++ image plate detector; data were indexed with Mosflm (24), scaled with Scala (17), phased by molecular replacement with Phaser (25), and analyzed with PHENIX (1). Refinement was carried out with Phenix and included a protocol for TLS parameterization with three groups defined per molecule (28) and was alternated with real-space refinement and visualization using COOT (16). The initiating methionine residue was not visible and, following the convention for saDHFR, the next residue (Thr) was numbered as residue 1. In both structures, residues 71 (Glu), 143 (Glu), and 144 (Lys) had poor side chain density and were refined as alanine residues. Four additional C-terminal residues resulted from affinity tag cleavage, are visible in the structures, and make intimate crystal packing contacts. The structures of the wild type and the F98Y point mutant were completed in space group P6122 with one molecule per asymmetric unit. The wild-type structure was refined to an R value of 17.5% (21.3% Rfree), while the F98Y mutant was refined to an R value of 17.0% (20.7% Rfree); crystallographic statistics are listed in Table 2. Ligand binding, including buried surface area, was analyzed with the LPC server (32). Molecular graphics images were produced using the UCSF Chimera package (29).
TABLE 2.
Crystallographic data for S. aureus DHFRa
| Evaluation phase and parameter | Value for ternary complex with RAB1, NADPH, and: |
|
|---|---|---|
| Wild-type DHFR | F98Y mutant DHFR | |
| Data collection | ||
| Space group | P6122 | P6122 |
| Cell dimensions | ||
| a = b, c (Å) | 79.2, 107.9 | 79.2, 107.9 |
| Resolution (Å) | 42.4-2.01 (2.12-2.01) | 42.4-2.01 (2.12-2.01) |
| Rsym | 0.037 (0.118) | 0.093 (0.403) |
| I /σ | 23.7 (10.9) | 18.9 (7.1) |
| Completeness (%) | 99.6 (98.2) | 100.0 (100.0) |
| Redundancy | 5.1 (5.1) | 7.3 (7.2) |
| Refinement | ||
| Resolution (Å) | 31.9-2.01 | 39.6-2.01 |
| No. of reflections | 13,579 | 13,953 |
| Rwork/Rfree | 0.175/0.213 | 0.170/0.207 |
| No. of atoms | ||
| Protein | 1,296 | 1,296 |
| Ligand/ion | 104 | 104 |
| Water | 171 | 166 |
| B-factors (Å2) | ||
| Protein | 19.0 | 19.3 |
| Ligands | 16.2 (RAB1) | 27.0 (RAB1) |
| 17.0 (NADPH) | 15.2 (NADPH) | |
| Water | 32.2 | 31.4 |
| RMS deviations | ||
| Bond lengths (Å) | 0.005 | 0.005 |
| Bond angles (°) | 1.034 | 1.018 |
Values in parentheses are for the highest-resolution shell. RMS, root mean square.
Protein Data Bank accession numbers.
Coordinates and structure factors have been deposited with the Protein Data Bank and given accession codes 3M08 for the ternary complex with wild-type DHFR and 3M09 for the ternary complex with the F98Y mutant.
RESULTS
Broad-spectrum activity of RAB1.
The MIC of RAB1 was assessed in a broth microdilution assay against a panel of pathogens relevant to human health. RAB1 inhibited growth of both Gram-negative (F. tularensis, Y. pestis, and B. abortus) and Gram-positive (B. anthracis, VRE, and S. aureus) organisms, indicating a broad-spectrum profile of activity (Table 3). Despite initial selection of RAB1 (previously BAL17662) for its inhibitory activity against B. anthracis, the present results demonstrate a potent antistaphylococcal profile (3). Among the organisms tested, F. tularensis, B. abortus, B. anthracis, VRS1, and VRS2 have an MIC for TMP of ≥16 μg/ml, which is classified as resistant by CLSI (11). B. anthracis is naturally resistant to TMP, while VRS1 and VRS2 contain a plasmid-encoded TMP-resistant DHFR. Application of the same interpretative standard to MICs with RAB1 would yield only B. abortus as resistant among this panel. The inhibition of plasmid-derived DHFR by RAB1 in the VRSA strains is particularly promising, as TMP is not capable of inhibiting the plasmid-derived DHFR enzymes.
TABLE 3.
RAB1 has broad-spectrum activity
| Strain | MIC (μg/ml) |
|
|---|---|---|
| (S,R)-RAB1 | TMPa | |
| F. tularensis Schu4 | 1-2 | 16 |
| Y. pestis CO92 | 4-8 | ≤1 |
| B. abortus 2308 | 16 | 32 |
| B. anthracis Ames | 1-4 | >256 |
| VRE ATCC 700802 | 0.03 | ≤1 |
| MRSA NRS1 | ≤0.0625-0.125 | 2 |
| MRSA NRS70 | ≤0.0625-0.25 | 2-4 |
| MRSA NRS71 | ≤0.0625-0.125 | 2 |
| MRSA NRS123 | ≤0.0625-0.25 | 2 |
| VRSA VRS1 | 8 | >256 |
| VRSA VRS2 | 4 | >256 |
| VRSA VRS3 | ≤0.125 | 2 |
The CLSI interpretive standard for TMP is susceptibility at an MIC of ≤8 μg/ml and resistance at an MIC of ≥16 μg/ml.
Both enantiomers of RAB1 are active.
RAB1 is more potent than TMP for inhibiting growth of wild-type S. aureus. This is reflected in a reduced IC50 and a decrease of an order of magnitude in the MIC (Tables 3 and 4). While the F98Y mutation causes a loss in potency of approximately 10-fold for TMP, it has a minimal effect on RAB1 binding (Ki of 2.8 nM versus 0.9 nM for F98Y) (Table 4). Both enantiomers of RAB1 are inhibitory, although the S-enantiomer is more active, as evidenced by a lower IC50 (2.9 nM for wild-type protein versus 11.5 nM for the R-enantiomer) and a much lower MIC (≤0.0625 μg/ml for the wild-type strain versus 1 to 1.6 μg/ml with the R-enantiomer). Interestingly, RAB1 activity seems modestly enhanced by the F98Y mutation for the S-enantiomer but further decreases the affinity for the R-enantiomer. While the activity of the racemic mixture is favorable, purification of the S-enantiomer yields a superior antimicrobial profile and enhanced activity with the F98Y mutation. It should be noted that the S-enantiomer performs an order of magnitude better with the F98Y mutant (Ki of 0.2 nM) than TMP does with the wild-type protein (Ki of 4.4 nM).
TABLE 4.
Both enantiomers of RAB1 possess favorable inhibition profiles
| TMP enantiomer | IC50 (Ki)a |
||
|---|---|---|---|
| wt (Phe 98) | TMPr (Tyr98) | Activity ratio (Y98/F98) | |
| TMP | 14.4 ± 3.6 (4.4 ± 1.1) | 539.7 ± 101.1 (53.6 ± 10.0) | 37.5 (12.3) |
| (S,R)-RAB1 | 9.4 ± 3.9 (2.8 ± 1.2) | 8.8 ± 0.9 (0.9 ± 0.03) | 0.9 (0.3) |
| S-RAB1 | 2.9 ± 0.1 (0.9 ± 0.03) | 1.8 ± 0.3 (0.2 ± 0.03) | 0.6 (0.2) |
| R-RAB1 | 11.5 ± 2.3 (3.5 ± 0.7) | 65.4 ± 3.3 (6.5 ± 0.3) | 5.7 (1.9) |
Values expressed in nM.
Description of the RAB1 binding site in S. aureus DHFR.
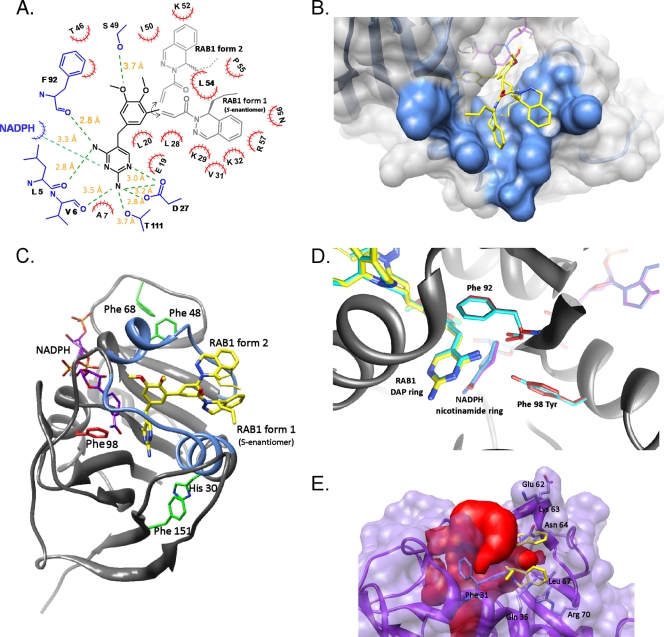
Cocrystallization of saDHFR with RAB1 revealed two conformations for the inhibitor in the binding site irrespective of the identity of the residue at position 98 (Fig. 1). These two conformations of RAB1 are equally populated in the wild-type and F98Y mutant enzyme. In both structures, the regions of RAB1 most similar to other inhibitors, such as TMP, are bound in a single position; the two conformations arise from rotational freedom around the acryloyl linker. This rotation places the phthalazine moiety either embedded within a hydrophobic pocket used to bind the substrate or along a surface cavity. The presence of this secondary binding cavity is a unique observation among saDHFR inhibitor structures and illustrates a new area for future development of inhibitors.
FIG. 1.
RAB1 is found in two conformations in the S. aureus DHFR binding site, which is not affected by the trimethoprim-resistant F98Y mutation. (A) Two-dimensional map of the wild-type S. aureus binding site containing RAB1. Residues are labeled by a single-letter abbreviation and residue number. Red hatched semicircles denote hydrophobic interactions; hydrogen bonds are indicated by green dashed lines, and the corresponding distances are listed. The conformations of RAB1 are listed as form 1 and form 2, as discussed in the text. RAB1 enantiomers are shown; for form 2 either enantiomer can occupy the site, and the orientation of the propyl group was not defined in the crystallographic data. (B) RAB1 form 2 occupies a unique shallow surface cavity, while form 1 is found embedded in the conventional binding site. (C) View of the saDHFR enzyme with secondary structure representation and the two sides of the active site colored blue. Residues important to the overall structure of the binding site are indicated, as discussed in the text. Note that either RAB1 enantiomer can bind in the form 2 conformation. (D) The F98Y mutation has minimal effects on the structure of saDHFR which are limited to movement of the main chain carbonyl of Phe 92 and ≤0.3 Å movement of the nicotinamide ring of NADPH and the DAP ring of RAB1. (E) RAB1 will not fit into the human DHFR binding site due to steric restrictions imposed by a smaller volume, which is contoured in red. Residues with direct clashes to RAB1 are labeled. Coordinates were obtained from PDB ID 2W3A (Leung et al., unpublished).
The more conserved areas of RAB1 were bound to the binding site using interactions observed with most inhibitors, as illustrated in Fig. 1. The diaminopyrimidine (DAP) ring is embedded within the site and anchored by hydrogen bonds to residues Leu 5, Val 6, Asp 27, Phe 92, and Thr 111. In addition, hydrophobic contacts are mediated by residue Ala 7 and the cofactor NADPH. The consequence of the F98Y mutation appears to be introduced at the main chain carbonyl of Phe 92, causing a small shift (≤0.3 Å) of the nicotinamide ring of the NADPH cofactor and translating to the DAP ring positioning of both RAB1 conformations (Fig. 1D). The central aromatic ring of RAB1 contains two dimethoxy substituents and is slotted into a hydrophobic groove comprised of residues Glu 19, Leu 20, and Leu 28 on one side and Thr 46 and Phe 92 on the other. The aromatic portion is oriented parallel with this groove, and the polar methoxy groups protrude along the top of the binding site. A long hydrogen bond (3.7 Å) is formed between the side chain oxygen of Ser 49 and one of the methoxy moieties (Fig. 1A).
For discussion purposes the conformation of RAB1 that occupies the substrate binding site will be referred to as form 1, while the conformation that reveals the secondary surface cavity will be termed form 2 (Fig. 1). Form 1 is positioned along the same path as was previously observed for RAB1 binding to DHFR from B. anthracis (4). This fills a major pocket lined with residues Val 31, Leu 54, Pro 55, and the carbon atoms from the side chains of Lys 29, Lys 32, and Arg 57. Residue Asn 56 demarcates the edge of the pocket, but it is over 5 Å away from RAB1. All contacts between the phthalazine moiety and the protein are hydrophobic. The enantiomeric preference for the propyl group in this conformation is the same as previously observed, with the S-enantiomer appearing to be the only occupant (4). While the propyl tail is not in direct contact with protein atoms, its path follows that of residue Lys 29. Form 2 utilizes a surface cavity that appears unique to saDHFR. Residues lining this cavity are Leu 54 and Pro 55 on one side with Ile 50 and Lys 52 on the opposing side. This cavity is relatively exposed and results in almost 100 Å2 less buried surface (form 1 buries ≈680 Å2 while form 2 buries ≈585 Å2 of the ≈775 Å2 available). This increase in solvent exposure is largely limited to the phthalazine moiety and is highest at the stereogenic carbon (Fig. 1B). The electron density for the propyl group at the stereogenic carbon is absent for form 2, indicating that either enantiomer could occupy this secondary site. It is likely that this secondary site is the key to the increased affinity for RAB1 with saDHFR versus other bacterial DHFR enzymes.
For form 1, the phthalazine group is not as buried in saDHFR as was seen with baDHFR, and no major side chain movements are noted in saDHFR as a consequence of RAB1 binding. Residue identities between the binding site of baDHFR and saDHFR are largely conserved, and of the contacting residues in saDHFR five have conservative changes relative to baDHFR (Leu5Met, Gln19Asn, Asp27Gln, Lys29Gln, and Ser49Ala). However, the shape of the site in saDHFR is somewhat more shallow but considerably wider than baDFHR. Outward movement occurs on each side of the binding site and is seen in structures with or without NADPH, indicating this larger shape is intrinsic to saDHFR. On the side comprised of residues 20 to 25 and helix 1 (residues 26 to 37), the protein lacks a critical “aromatic sandwich” interaction between residues His 30 and Phe 151 (Fig. 1C). This unusual ring stacking is observed in all other DHFR enzymes, as first reported for Pneumocystis jirovecii DHFR residues Tyr 35 and Phe 199 (for examples, see Protein Data Bank [PDB] codes 2FZH and 2FZI) and for murine DHFR residues Tyr 33 and Phe 179 (see PDB code 2FZJ) (12). This interaction is also present in baDHFR (residues Tyr 31 and Tyr 155) and human DHFR (Tyr 33 and Phe 179). The face stacking of two aromatic rings effectively tethers the orientation of helix 1 of the active site to the stabilizing β-sheet of the protein's core; loss of this appears to allow more flexibility in the positioning of helix 1 at one side of the active site. The opposite side of the binding site is made up by helix 2 (residues 44 to 51) and a loop region (residues 52 to 56). There appear to be fewer stabilizing interactions for this area, but of note are two Tyr-to-Phe changes in saDHFR at residues 47 and 68 relative to other DHFR enzymes (Fig. 1C). In baDHFR, the polar hydroxyl group of these Tyr residues influences the surrounding regions, including a hydrogen bond to the main chain of Pro 54 (Pro 55 in saDHFR). The combination of these effects results in a wider, perhaps more mobile active site architecture, which in turn allows RAB1 to bind in both the active site (form 1) and the shallow side cavity (form 2). The saDHFR enzyme has a unique capacity for inhibitor binding, and RAB1 fully utilizes this additional flexibility and extra binding surface to yield a very strong interaction.
Superposition of S. aureus DHFR substrate and inhibitors.
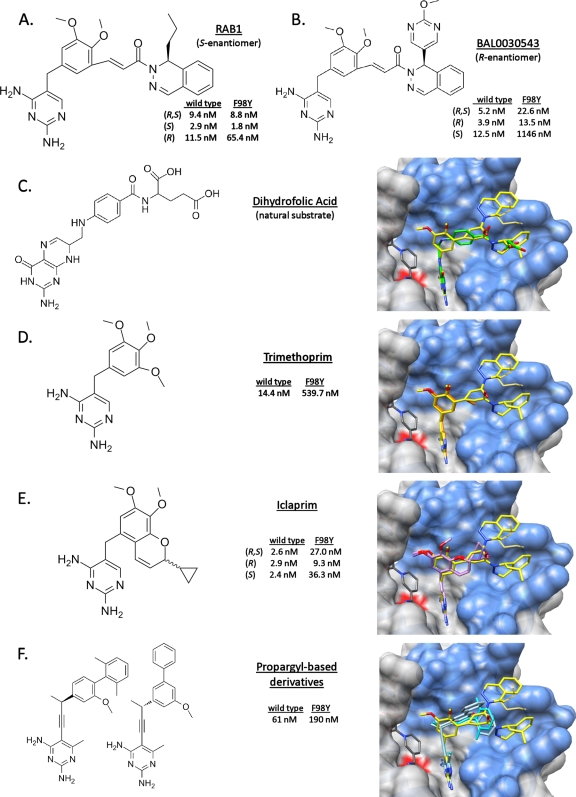
The binding of RAB1 closely mimics that of the natural substrate, DHF (Fig. 2C). The paths of DHF and RAB1 form 1 are essentially identical, and the DAP ring of RAB1 superimposes well with the pterin ring of DHF. RAB1's linker overlays with the peptide-like linker of DHF, and the end of the para-aminobenzyl glutamate tail of DHF closely matches the dihydropyridazine portion of the dihydrophthalazine moiety of RAB1. Finally, the propyl group for the S-enantiomer of RAB1, as seen in form 1, matches the space used by the glutamate-like extension on DHF. Additional surfaces are occupied by RAB1 and include the end of the large hydrophobic pocket used in accommodating form 1 and the shallow surface pocket occupied by form 2. The large overlap of the DHF functionally required binding surface and of RAB1 lessens the likelihood of resistance mutations arising.
FIG. 2.
RAB1 occupies a similar space as the natural substrate and other inhibitors; in addition it utilizes a unique surface cavity. The IC50 of RAB1 is not affected by the F98Y mutation to the same extent as other inhibitors. RAB1 is depicted as yellow in the binding site (blue), residue 98 is marked with a red surface, and the listed small molecules are superimposed. (A) RAB1 with IC50 data from the current study. (B) BAL0030543, a related compound under development against S. aureus, with IC50 data from reference 7. (C) Dihydrofolic acid, with coordinates obtained from PDB ID 3FRD (26) (D) Trimethoprim, with IC50 data from the current study and the coordinates obtained from PDB ID 2W9G with NADPH (22). (E) Iclaprim, with IC50 and coordinates obtained from PDB IDs 3FYV and 3FYW (27). (F) Propargyl-based inhibitors, with IC50 data and coordinates obtained from PDB ID 3F0B (19) and from PDB ID 3F0Q (K. M. Frey et al., unpublished; no IC50 data are publicly available).
Other cocrystal structures have recently been determined with saDHFR and inhibitors, including TMP (22), Iclaprim (26, 27), and a series of propargyl-containing molecules (19, 20). Like RAB1, each of these molecules contains a DAP ring which docks essentially identically in the protein binding site. TMP then contains a trimethoxybenzyl ring surrounded by residues Leu 20, Leu 28, Ile 50, Leu 54, and Phe 92 and the NADPH cofactor, which superimposes with RAB1's dimethoxybenzyl ring (Fig. 2D) and Iclaprim's dimethoxychromene moiety (Fig. 2E). Iclaprim extends from the chromene with a cyclopropyl ring, creating a chiral carbon with equally active isomers. The positions occupied by the two conformations of RAB1's acryloyl linker region superpose markedly well with the enantiomeric cyclopropyl groups of Iclaprim, and these regions all make similar hydrophobic contacts with protein residues Leu 28, Ile 50, and Leu 54.
RAB1 continues along the binding site and fills the length of available space as well as the additional surface cavity. The propargyl-containing molecules present limited similarity to RAB1 beyond the DAP moiety (Fig. 2F). The propargyl-containing molecules do occupy the same main pocket and make similar hydrophobic interactions which terminate at a position equivalent with the linker of RAB1. The most notable features are found in the inhibitors from PDB ID 3F0Q (unpublished) and 3F0B (19), which protrude into a space below the surface cavity occupied by RAB1 in form 2. This is the only other example of utilization of a secondary binding space, although with the propargyl inhibitors the space is continuous with the large hydrophobic pocket.
The positional agreement of these inhibitors and the natural substrate DHF all reflect a high degree of specificity for the enzyme. It is speculated that Iclaprim's equally active isomers would further reduce resistance mutations from occurring (26); the extended nature and larger footprint of RAB1 should heighten this effect. Further, the utilization of a functionally required area in concert with a secondary binding site provides limited opportunity for protein mutations conferring resistance.
Steric hindrance accounts for the bacterial specificity of RAB1.
RAB1 is a very poor inhibitor of human DHFR (hDHFR; IC50, 110 μM) and is at least 4 orders of magnitude more selective for bacterial DHFR (3). The lack of binding to hDHFR was rationalized by using a superimposition of the saDHFR-RAB1 structure with hDHFR (PDB ID 2W3A) (A. K. W. Leung, L. J. Ross, S. Zywno-Van Ginkel, R. C. Reynolds, L. E. Seitz, V. Pathak, W. W. Barrow, E. L. White, W. J. Suling, J. R. Piper, and D. W. Borhani, unpublished data). This revealed an obvious lack of space in the hDHFR site with severe steric clashes of RAB1 and hDHFR residues Phe 31, Phe 34, Gln 35, Leu 67, and Arg 70 (Fig. 1E). As expected, the secondary binding site is not present in hDHFR and is occluded by loop residues 61 to 64 (Pro, Glu, Lys, and Asn). Overall, the hDHFR binding site has approximately the same volume as saDHFR (each ≈1,290 Å3), but in hDHFR this volume is a different shape that does not extend down the length of the binding site as far as saDHFR and bacterial DHFR enzymes in general. This indicates that the dihydrophthalazine moiety confers bacterial specificity to this class of inhibitors due to steric restrictions with the hDHFR enzyme. The structure of RAB1 seems to fill the binding site of bacterial DHFR with limiting flexibility and polar interactions (entropic costs) to present an ideal inhibitor for bacterial enzymes. However, flexibility in the linker region is crucial for accessing the secondary binding site, which in turn confers the potent inhibition observed with S. aureus strains.
DISCUSSION
RAB1 is able to inhibit the growth of both Gram-positive and Gram-negative organisms. With all microorganisms tested, except Y. pestis, it was more effective than TMP, enabling it to be developed for broad-spectrum treatment of bacterial infections. The activity of RAB1 arises from its ability to inhibit the DHFR enzyme. The binding site of saDHFR possesses unique properties, including the shallow surface cavity, likely arising from a lack of stabilizing interactions seen in other DHFR enzymes (such as the “aromatic sandwich”). An intrinsic property of the saDHFR binding site is the ability to accommodate larger ligands, to the extent that multiple inhibitors have been observed in more than one conformation. Visualization of RAB1 binding in the cocrystal structures highlights two possible conformations for the specificity-conferring phthalazine group. While one of these was expected, the second conformation delineates a previously unappreciated shallow surface cavity believed unique to saDHFR. While the expected conformation only accommodates the S-enantiomer, as previously observed with baDHFR, the second conformation appears occupied by either enantiomer. This limited binding mode for the R-enantiomer is consistent with higher IC50s (Table 4 and Fig. 2A). The two conformations of RAB1 allow more variability in the binding mode and account for the increased activity with S. aureus.
The impact of the F98Y mutation was evidenced by a marked decrease in activity of TMP but had essentially no effect on the activity of RAB1. This is in stark contrast to other inhibitors, including Iclaprim, which has an increase in the IC50 from 2.2 nM to 27 nM when the F98Y mutation is present (Fig. 2E) (27). Surprisingly, the F98Y mutation seems to enhance binding of the S-enantiomer of RAB1 (IC50 from 2.9 nM to 1.8 nM and Ki from 0.9 nM to 0.2 nM) while concomitantly decreasing the activity of the R-enantiomer (IC50 from 11.5 nM to 65.4 nM and Ki from 3.5 nM to 6.5 nM) (Fig. 2A). The structural consequence of this is not clear, but the larger size of RAB1 could attenuate the impact of the F98Y mutation by allowing less lateral freedom of movement within the site. Presumably the stress on Phe 92 induced by the mutation (Fig. 1D) must impair binding of form 2 in the surface cavity while enhancing the interaction with form 1.
There was no evidence for NADPH mobility, as was previously observed with propargyl-containing inhibitors (19). These inhibitors were also more sensitive to the F98Y substitutions; both observations are probably related to modifications to the DAP ring in the propargyl inhibitor series (Fig. 2F). While two members of the biphenyl class of propargyl inhibitors are demonstrated to occupy a space below the surface cavity identified here, this propargyl inhibitor series lacks selectivity for bacterial enzymes and possesses IC50 (or Ki) values an order of magnitude larger than RAB1. Their conclusions correctly surmise that extension further into the DHFR site would provide increased affinity and specificity, as is demonstrated by RAB1.
The binding of RAB1 is similar to that of Iclaprim. While the IC50 of RAB1 is greater than that of Iclaprim (14.4 nM versus 2.2 to 7 nM), the MIC values are comparable at ≤0.125 to 0.06 μg/ml (26, 31). The impact of the F98Y mutation on (R)-RAB1 is consistent with recent observations with related phthalazine-based DHFR inhibitors, such as BAL0030543 (Fig. 2B) (7, 9). While the three-dimensional structures with the BAL compounds are not known, they may use a similar binding mode such that the phthalazine group follows the path of RAB1 form 1, while the pyrimidine moiety extending from the phthalazine would occupy the same position as RAB1 form 2. This would present some steric strain that should exacerbate the effect of the F98Y mutation.
While toxicology remains to be tested, predictive calculations indicate RAB1 has a higher than desirable log(P) of 5.45. Future refinements to the structure will seek to optimize the pharmocodynamic profile to further development of RAB1-like candidates as viable antimicrobial therapies in our arsenal against infectious diseases.
Acknowledgments
X-ray data were collected at the Macromolecular Crystallography Laboratory, University of Oklahoma, Department of Chemistry and Biochemistry, Norman, OK. MIC data were obtained through the NIH/NIAID In vitro Assessment for Antimicrobial Activity Resource for Researchers program. We are grateful to Leonard Thomas for maintaining the X-ray facility at the University of Oklahoma and to Paul Cook at the University of Oklahoma for discussion of kinetic parameters.
This work was supported by the Sitlington Endowed Chair in Infectious Disease (W. W. Barrow) and the Chemistry Department at Oklahoma State University (K. D. Berlin and R. A. Bunce). The UCSF Chimera package is made freely available by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, and is supported by NIH P41 RR-01081.
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Adams, P. D., P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, and P. H. Zwart. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrow, E. W., P. C. Bourne, and W. W. Barrow. 2004. Functional cloning of Bacillus anthracis DHFR and confirmation of natural resistance to trimethoprim. Antimicrob. Agents Chemother. 48:4643-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrow, E. W., J. Dreier, S. Reinelt, P. C. Bourne, and W. W. Barrow. 2007. In vitro efficacy of new antifolates against trimethoprim-resistant Bacillus anthracis. Antimicrob. Agents Chemother. 51:4447-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne, C. R., R. A. Bunce, P. C. Bourne, K. D. Berlin, E. W. Barrow, and W. W. Barrow. 2009. Crystal structure of Bacillus anthracis dihydrofolate reductase with the dihydrophthalazine-based trimethoprim derivative RAB1 provides a structural explanation of potency and selectivity. Antimicrob. Agents Chemother. 53:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunce, R. A., C. R. Bourne, P. C. Bourne, E. W. Barrow, W. W. Barrow, and K. D. Berlin. 2009. HPLC detection and enantiomeric separation of (+)- and (−)-(E)-3-[5-(2,4-diaminopyrimidin-5-ylmethyl)-2,3-dimethoxyphenyl]-1-(1-propyl-1H-phthalazin-2-yl)propenone, inhibitors of Bacillus anthracis dihydrofolate reductase, by supercritical fluid chromatography. Proc. Okla. Acad. Sci. 89:87-90. [Google Scholar]
- 6.Burchall, J. J. 1973. Mechanism of action of trimethoprim-sulfamethoxazole. II. J. Infect. Dis. 128(Suppl.):437-441. [DOI] [PubMed] [Google Scholar]
- 7.Caspers, P., L. Bury, B. Gaucher, J. Heim, S. Shapiro, S. Siegrist, A. Schmitt-Hoffmann, L. Thenoz, and H. Urwyler. 2009. In vitro and in vivo properties of dihydrophthalazine antifolates, a novel family of antibacterial drugs. Antimicrob. Agents Chemother. 53:3620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Y., and W. H. Prusoff. 1973. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22:3099-3108. [DOI] [PubMed] [Google Scholar]
- 9.Clark, C., L. M. Ednie, G. Lin, K. Smith, K. Kosowska-Shick, P. McGhee, B. Dewasse, L. Beachel, P. Caspers, B. Gaucher, G. Mert, S. Shapiro, and P. C. Appelbaum. 2009. Antistaphylococcal activity of dihydrophthalazine antifolates, a family of novel antibacterial drugs. Antimicrob. Agents Chemother. 53:1353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed., vol. 29. Clinical Laboratory Standards Institute, Wayne, PA.
- 11.CLSI. 2010. Performance standards for antimicrobial susceptibility testing, 20th information update, vol. 29. Clinical Laboratory Standards Institute, Wayne, PA.
- 12.Cody, V., J. Pace, K. Chisum, and A. Rosowsky. 2006. New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions. Proteins 65:959-969. [DOI] [PubMed] [Google Scholar]
- 13.Dale, G. E., D. Broger, A. D'Arcy, P. G. Hartman, R. DeHoogt, S. Jolidon, I. Kompis, A. M. Labhardt, H. Langen, H. Locher, M. G. Page, D. Stüber, R. L. Then, B. Wipf, and C. Oefner. 1997. A single amino acid substitution in Staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance. J. Mol. Biol. 266:23-30. [DOI] [PubMed] [Google Scholar]
- 14.Dale, G. E., H. Langen, M. G. P. Page, R. L. Then, and D. Stüber. 1995. Cloning and characterization of a novel, plasmid-encoded trimethoprim-resistant dihydrofolate reductase from Staphylococcus haemolyticus MUR313. Antimicrob. Agents Chemother. 39:1920-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dibbern, D. A., Jr., and A. Montanaro. 2008. Allergies to sulfonamide antibiotics and sulfur-containing drugs. Ann. Allergy Asthma Immunol. 100:91-100. [DOI] [PubMed] [Google Scholar]
- 16.Emsley, P., and K. Cowtan. 2004. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 17.Evans, P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72-82. [DOI] [PubMed] [Google Scholar]
- 18.Fischbach, M. A., and C. T. Walsh. 2009. Antibiotics for emerging pathogens. Science 325:1089-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey, K. M., J. Liu, M. N. Lombardo, D. B. Bolstad, D. L. Wright, and A. C. Anderson. 2009. Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance. J. Mol. Biol. 387:1298-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey, K. M., M. N. Lombardo, D. L. Wright, and A. C. Anderson. 2010. Towards the understanding of resistance mechanisms in clinically isolated trimethoprim-resistant, methicillin-resistant Staphylococcus aureus dihydrofolate reductase. J. Struct. Biol. 170:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grim, S. A., R. P. Rapp, C. A. Martin, and M. E. Evans. 2005. Trimethoprim-sulfamethoxazole as a viable treatment option for infections caused by methicillin-resistant Staphylococcus aureus. Pharmacotherapy 25:253-264. [DOI] [PubMed] [Google Scholar]
- 22.Heaslet, H., M. Harris, K. Fahnoe, R. Sarver, H. Putz, J. Chang, C. Subramanyam, G. Barreiro, and J. R. Miller. 2009. Structural comparison of chromosomal and exogenous dihydrofolate reductase from Staphylococcus aureus in complex with the potent inhibitor trimethoprim. Proteins 76:706-717. [DOI] [PubMed] [Google Scholar]
- 23.Keiser, M. J., V. Setola, J. J. Irwin, C. Laggner, A. I. Abbas, S. J. Hufeisen, N. H. Jensen, M. B. Kuijer, R. C. Matos, T. B. Tran, R. Whaley, R. A. Glennon, J. Hert, K. L. Thomas, D. D. Edwards, B. K. Shoichet, and B. L. Roth. 2009. Predicting new molecular targets for known drugs. Nature 462:175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie, A. G. W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EAMCB Newslett. Protein Crystallogr., no. 26.
- 25.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oefner, C., M. Bandera, A. Haldimann, H. Laue, H. Schulz, S. Mukhija, S. Parisi, L. Weiss, S. Lociuro, and G. E. Dale. 2009. Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity. J. Antimicrob. Chemother. 63:687-698. [DOI] [PubMed] [Google Scholar]
- 27.Oefner, C., S. Parisi, H. Schulz, S. Lociuro, and G. E. Dale. 2009. Inhibitory properties and X-ray crystallographic study of the binding of AR-101, AR-102 and iclaprim in ternary complexes with NADPH and dihydrofolate reductase from Staphylococcus aureus. Acta Crystallogr. D Biol. Crystallogr. 65:751-757. [DOI] [PubMed] [Google Scholar]
- 28.Painter, J., and E. A. Merritt. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol. Crystallogr. 62:439-450. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 30.Powell, J. P., and R. P. Wenzel. 2008. Antibiotic options for treating community-acquired MRSA. Expert Rev. Anti Infect. Ther. 6:299-307. [DOI] [PubMed] [Google Scholar]
- 31.Schneider, P., S. Hawser, and K. Islam. 2003. Iclaprim, a novel diaminopyrimidine with potent activity on trimethoprim sensitive and resistant bacteria. Bioorg. Med. Chem. Lett. 13:4217-4221. [DOI] [PubMed] [Google Scholar]
- 32.Sobolev, V., A. Sorokine, J. Prilusky, E. E. Abola, and M. Edelman. 1999. Automated analysis of interatomic contacts in proteins. Bioinformatics (Oxford) 15:327-332. [DOI] [PubMed] [Google Scholar]
- 33.Zinner, S. H. 2007. Antibiotic use: present and future. New Microbiol. 30:321-325. [PubMed] [Google Scholar]