Abstract
Integrating conjugative elements (ICEs) are mobile genetic elements that can transfer from the chromosome of a host to the chromosome of a new host through the process of excision, conjugation, and integration. Although SXT/R391-related ICEs, originally demonstrated in Vibrio cholerae O139 isolates, have become prevalent among V. cholerae isolates in Asia, the prevalence of the ICEs among Gram-negative bacteria other than Vibrio spp. remains unknown. In addition, SXT/R391-related ICEs carrying genes conferring resistance to extended-spectrum cephalosporins have never been described. Here we carried out a genetic analysis of a cefoxitin-resistant Proteus mirabilis clinical isolate, TUM4660, which revealed the presence of a novel SXT/R391-related ICE, ICEPmiJpn1. ICEPmiJpn1 had a core genetic structure showing high similarity to that of R391 and carried xis and int genes completely identical to those of R391, while an IS10-mediated composite transposon carrying blaCMY-2 was integrated into the ICE. A nucleotide sequence identical to the 3′ part of ISEcp1 was located upstream of the blaCMY-2 gene, and other genes observed around blaCMY-2 in earlier studies were also present. Furthermore, the nucleotide sequences of hot spot 2 and hot spot 4 in ICEPmiJpn1 showed high similarity to that of hot spot 2 in SXTMO10 and with a part of the nucleotide sequence found in P. mirabilis ATCC 29906, respectively. ICEPmiJpn1 was successfully transferred to Escherichia coli, Klebsiella pneumoniae, Salmonella enterica serovar Typhimurium, and Citrobacter koseri in conjugation experiments. These observations suggest that ICEs may contribute to the dissemination of antimicrobial resistance genes among clinically relevant Enterobacteriaceae, which warrants careful observation of the prevalence of ICEs, including SXT/R391-related ICEs.
Acquired AmpC enzymes have recently been distributed globally among nontyphoid Salmonella spp. (10, 15), Escherichia coli (20, 39), and other species of Enterobacteriaceae (16, 27). Most acquired ampC genes are highly expressed, presumably because of the possession of promoters of greater strength than those for chromosomally encoded enzymes and because of the multiple copy numbers of the relevant plasmid, rendering the host organisms resistant to a variety of β-lactams, including oxyimino-cephalosporins and cephamycins (23). CMY-2 and its derivatives, presumably derived from the chromosomally encoded AmpC protein of Citrobacter freundii, appear to be the most common acquired AmpC enzymes worldwide (23). ISEcp1 has frequently been found upstream of blaCMY-2-like genes in full-length or partially inverted form or with insertions of other transposons, such as IS5, IS10, or IS1294, and is assumed to be involved in mobilization and expression of blaCMY-2-like genes (25, 42).
Proteus mirabilis is a common causative organism of urinary tract infections in patients with indwelling urinary catheters or with anatomic or functional abnormalities of the urinary tract (22). Although P. mirabilis originally lacks a chromosomal β-lactamase gene and is highly susceptible to β-lactams (30), the production of β-lactamases, including AmpC enzymes and extended-spectrum β-lactamases (ESBLs), by P. mirabilis clinical isolates, through acquisition of resistance genes by horizontal gene transfer, has been reported in recent years (16, 36). Chromosomal acquisition of blaCMY has also been demonstrated in some studies; however, the mechanism by which the resistance genes were acquired remains unknown (6, 14, 28).
Integrating conjugative elements (ICEs) are self-transmissible genetic elements with the abilities to excise themselves from the chromosome of their host cell to form a circular intermediate, to transfer to another cell by conjugation, and to integrate themselves into the chromosome of the new host cell (7). SXTMO10 is an ICE carrying genes mediating resistance to chloramphenicol, sulfamethoxazole, trimethoprim, and streptomycin and was originally discovered in Vibrio cholerae O139, a newly recognized epidemic strain which emerged in India in the early 1990s (43). While SXT-related ICEs were rarely found in V. cholerae O1 isolates before the emergence of V. cholerae O139, most V. cholerae O1 isolates in Asia, together with O139 isolates, now seem to carry SXT-related ICEs (2, 19, 21). Furthermore, SXT-related ICEs have been found in V. cholerae isolates outside Asia (8, 13), in Vibrio species other than V. cholerae (1, 40), and in other gammaproteobacteria (35, 38).
R391 is an ICE discovered in a Providencia rettgeri clinical isolate in South Africa (12). Although R391 was once described as a conjugative plasmid of the “IncJ” incompatibility group, recent genetic analysis revealed that R391 is an 89-kb ICE which has high genetic and functional similarity to SXTMO10 (4, 5). SXT/R391-related ICEs have a highly conserved core set of genes, including int, xis, and tra genes (7). Int and Xis mediate integration and excision of the element. Tra proteins of SXTMO10 have significant homology with those encoded by several conjugative plasmids, and excised circular intermediates of SXT/R391-related ICEs appear to transfer to other cells by a mechanism which is similar to that used by conjugative plasmids. It has been documented that SXT/R391-related ICEs are integrated specifically into the 5′ end of the prfC gene, encoding peptide chain release factor 3 (3). In addition to conserved core genes, each SXT/R391-related ICE has specific genes conferring specific properties of the ICE, such as antimicrobial resistance. Interestingly, comparison of the nucleotide sequences of four fully sequenced SXT/R391-related ICEs revealed that most of such additional genes specific to each ICE are located at several specific sites, namely, four hot spots and several sites between attL and traI (7, 35, 38).
In this study, we describe a P. mirabilis clinical isolate carrying blaCMY-2 on a novel SXT/R391-related ICE, designated ICEPmiJpn1, and analyze the genetic structure of the ICE.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. P. mirabilis TUM4660 was isolated in 2006 from a wound swab culture from a patient with soft tissue infection hospitalized in a tertiary care hospital in Japan. E. coli ML4909 and E. coli ML4901, which were used as recipients in conjugation experiments, were provided by Matsuhisa Inoue of Kitasato University. Klebsiella pneumoniae ATCC 13883, Salmonella enterica serovar Typhimurium ATCC 14028, and Citrobacter koseri ATCC 27028 were purchased from the American Type Culture Collection, and their in vitro-obtained rifampin-resistant mutants, TUM5459, TUM5460, and TUM5461, respectively, were used as recipients in conjugation experiments.
TABLE 1.
Bacterial strains used in this study
| Strain | Characteristics | Reference or source |
|---|---|---|
| P. mirabilis TUM4660 | Clinical isolate showing reduced susceptibility to extended-spectrum cephalosporins | This study |
| E. coli ML4909 | F−galK2 galT22 hsdR metB1 relA supE44; rifampin resistant | M. Inoue |
| E. coli ML4901 | F−galK2 galT22 hsdR lacY1 metB1 relA supE44; nalidixic acid resistant | M. Inoue |
| K. pneumoniae TUM5459 | Rifampin-resistant mutant of K. pneumoniae ATCC 13883 | This study |
| S. Typhimurium TUM5460 | Rifampin-resistant mutant of S. Typhimurium ATCC 14028 | This study |
| C. koseri TUM5461 | Rifampin-resistant mutant of C. koseri ATCC 27028 | This study |
| E. coli TUM4670 | ICEPmiJpn1 transconjugant of ML4909 | This study |
| E. coli TUM4672 | ICEPmiJpn1 transconjugant of ML4901 | This study |
Antibacterial agents.
The following antimicrobials were used for antibiotic susceptibility testing and/or selection of transconjugants. Amoxicillin, cefotaxime, ceftazidime, cefoxitin, gentamicin, trimethoprim, nalidixic acid, rifampin, and ciprofloxacin were purchased from Sigma Chemical Co. (St. Louis, MO). Cefepime was from Bristol-Myers Squibb (Tokyo, Japan). Imipenem was from Banyu Pharmaceutical Co., Ltd. (Tokyo, Japan). Clavulanic acid was from GlaxoSmithKline K.K. (Tokyo, Japan). Tazobactam and piperacillin were from Taisho Toyama Pharmaceutical Co., Ltd. (Tokyo, Japan).
Susceptibility testing.
MICs were determined by broth dilution methods according to the guidelines of the Clinical and Laboratory Standards Institute (11). MICs of several β-lactams were determined alone or in combination, with a fixed concentration of clavulanic acid (2 μg/ml) or tazobactam (4 μg/ml). Disk potentiation tests, using Kirby-Bauer (KB) disks (Eiken Chemical Co., Ltd., Tokyo, Japan) containing 30 μg of ceftazidime alone or with 300 μg of 3-aminophenylboronic acid (APB) (Sigma Chemical Co.), were performed to identify the production of AmpC β-lactamase (44).
DNA preparation and identification of β-lactamase gene.
Isolation of genomic DNA was carried out with a Sepagene kit (Sanko Junyaku, Tokyo, Japan). The ampC gene was amplified by PCR with primers ampC1 and ampC2 (Table 2) (26). The PCR product was purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and subsequently sequenced with an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, CA).
TABLE 2.
Primers used in this study
| Primer | Nucleotide sequence (5′ to 3′) | Region amplified | Reference |
|---|---|---|---|
| ampC1 | ATGATGAAAAAATCGTTATGC | ampC | 26 |
| ampC2 | TTGCAGCTTTTCAAGAATGCGC | ampC | 26 |
| 23S-For | AATGATGGCCAGGCTGTCTCC | 23S rRNA | This study |
| 23S-Rev | CCGCCGTCGATATGAACTCTTG | 23S rRNA | This study |
| LE4 | GTACACACTTTCCGAGGTTACG | attL | 8 |
| LE1 | GTTTCTTCGTTGCACGAACTGG | attL | 8 |
| PMLE1 | ACAACGACAACAGAGCATTGG | attL | This study |
| RE4 | CCGCAATACCCTGCAATACCGA | attR | 8 |
| RE1 | CGGTCTGAATGGCCTGTCCGAA | attR | 8 |
| PMRE1 | TGCACGTTGGATAGCTTGTCCG | attR | This study |
| intFor1 | AAACTAGGGCTGGGCTTATAACATGGCG | int | 32 |
| intRev1 | AAAGATGGCAGCTTGCCGCAACCTC | int | 32 |
| orf14-Rev2 | CTGGTAGCAATGGGAGCATC | int-orf14 | This study |
| orf14-For1 | TTCGTCTCTGCCCGCATCATTG | orf14-blaCMY-2 | This study |
| CMY-Rev | ACGTCATCGGGGATCTGCAGCG | orf14-blaCMY-2 | This study |
| CMY-For | TTATTTCACCTGGGTAAAGCC | blaCMY-2-orf14 | This study |
| orf14-Rev1 | TTTCTGGCCTTGATAGAAGCTC | blaCMY-2-orf14 | This study |
| orf14-For2 | GCTAGGCGTGTTCGAGAGAATG | orf14-traI | This study |
| traI-Rev | GGCAATTAGCTCGTCTTGTGTG | orf14-traI | This study |
| HS1F | GGCTATTCCACCGGTGGTG | Hot spot 1 | 8 |
| HS1R | TGCCGATCACTAGCCCCAAC | orf36-traL | 8 |
| HotS2F | TTCCAGCAATCAGCGCCG | Hot spot 2 | 8 |
| HotS2R | CAGTTGTCCTATGTGGACTCGG | traA-orf55 | 8 |
| HotS4F | ATTGAACGGCTAAACGGAATGG | Hot spot 4 | This study |
| HotS4R | GCACGAAAATCAGCCCAAGC | traN-orf65 | This study |
| HotS3F2 | CCTAAGCATCCTTGGAAGGCT | Hot spot 3 | 8 |
| TraF2R | TGGGATGGTCACCCATAGGA | orf79-traF | 8 |
Conjugation experiments.
Conjugation experiments were performed by filter mating methods. Transconjugants were selected on bromothymol blue-lactose agar supplemented with cefotaxime (4 μg/ml) and either rifampin (50 μg/ml) or nalidixic acid (100 μg/ml).
PCR for genetic structures of SXT/R391-related ICEs.
The involvement of SXT/R391-related ICEs in conjugative transfer of blaCMY-2 from P. mirabilis TUM4660 to E. coli ML4909 was analyzed by molecular methods as described by McGrath et al. (32, 33). PCR amplification of attL and attR junctions in E. coli was attempted with primer pair LE1 and LE4 and primer pair RE1 and RE4, respectively (Table 2). Primer pair PMLE1 and LE4 and primer pair PMRE1 and RE4 were used to amplify attL and attR junctions in P. mirabilis, respectively (Table 2). The presence of an excised circular form of SXT/R391-related ICE was examined by PCR with primers LE4 and RE4. The int gene of SXT/R391-related ICEs was amplified with the primers IntFor1 and IntRev1 (Table 2).
Plasmid analysis and I-CeuI experiments.
Plasmid DNAs were extracted by alkaline lysis according to the method described by Kado and Liu (24). After Southern transfer to a Hybond-N+ membrane (GE Healthcare, Little Chalfont, United Kingdom), the extracted plasmid of TUM4660 was hybridized with a blaCMY-2-specific probe generated by PCR with primers ampC1 and ampC2 (Table 2). Labeling, hybridization, and immunological detection were performed with a DIG High Prime DNA labeling and detection starter kit I (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. A possible chromosomal location of the ampC gene was analyzed with the I-CeuI technique (29, 34). After digestion of whole-cell DNA with I-CeuI (New England Biolabs, Hertfordshire, United Kingdom), the resultant fragments were separated with a CHEF-Mapper apparatus (Bio-Rad, Hercules, CA) at 14°C, 6 V/cm, and a 120° switch angle, with a nonlinear switch time ramp of 5.3 to 49.9 s for 19.7 h. The sizes of the fragments were determined by comparison with a yeast chromosome pulsed-field gel electrophoresis marker (New England Biolabs). After Southern transfer to a Hybond-N+ membrane, the fragments were hybridized with three different PCR-generated probes: the probe for ampC, described above; a probe for the 23S rRNA gene, generated with primers 23S-For and 23S-Rev (Table 2); and a probe for int, generated with primers IntFor1 and IntRev1.
Genetic characterization of ICEPmiJpn1.
The genetic structure of the R391/SXT-related ICE in P. mirabilis TUM4660, ICEPmiJpn1, was analyzed by PCR amplification of variable regions of the ICE with primers designed according to the nucleotide sequences conserved among R391/SXT-related ICEs (Table 2). Extracted genomic DNAs from P. mirabilis TUM4660 and E. coli TUM4670 were used as DNA templates to confirm that they produced the same amplicon by PCR. PCR products were purified and sequenced. Nucleotide sequences were analyzed with BLAST at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Nucleotide sequence accession numbers.
The nucleotide sequences described in this article have been deposited in GenBank under accession numbers AB525688 (attL to traI), AB525225 (hot spot 1), AB525226 (hot spot 2), AB525227 (hot spot 4), AB525228 (hot spot 3), and AB525230 (attR).
RESULTS
Properties of P. mirabilis TUM4660 isolate.
P. mirabilis TUM4660 exhibited resistance to cefoxitin, ceftazidime, and cefotaxime. Clavulanic acid showed no synergistic effect on the susceptibilities to ceftazidime and cefotaxime (Table 3). The addition of 300 μg of APB to KB disks containing ceftazidime resulted in enlargement of the inhibitory zone diameter by >5 mm compared with that on KB disks alone, so the production of a class C β-lactamase by TUM4660 was suggested. Amplification of genomic DNA of TUM4660 with primers ampC1 and ampC2 was successful, and sequencing of the PCR product confirmed the presence of blaCMY-2 in TUM4660.
TABLE 3.
Results of antibiotic susceptibility testing
| Antimicrobial agenta | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| P. mirabilis TUM4660 | E. coli TUM4670 | E. coli TUM4672 | E. coli ML4909 | E. coli ML4901 | |
| Amoxicillin | >128 | >128 | >128 | 2 | 4 |
| Amoxicillin-CLA | >128 | >128 | >128 | 2 | 4 |
| Piperacillin | >128 | >128 | >128 | 2 | 2 |
| Piperacillin-TZB | 16 | >128 | >128 | 2 | 2 |
| Cefotaxime | 8 | 16 | 32 | ≤0.06 | ≤0.06 |
| Cefotaxime-CLA | 16 | 32 | 64 | ≤0.06 | ≤0.06 |
| Ceftazidime | 16 | 128 | 128 | 0.25 | 0.25 |
| Ceftazidime-CLA | 16 | 32 | 64 | 0.12 | ≤0.06 |
| Cefepime | 2 | 1 | 2 | ≤0.06 | ≤0.06 |
| Cefoxitin | 128 | 128 | >128 | 4 | 2 |
| Imipenem | 8 | 0.25 | 0.25 | 0.12 | 0.12 |
| Gentamicin | 1 | 1 | 0.5 | 2 | 2 |
| Trimethoprim | 4 | ≤0.06 | ≤0.06 | 0.12 | ≤0.06 |
| Ciprofloxacin | 0.12 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
Transfer of β-lactam resistance.
An E. coli ML4909 transconjugant of P. mirabilis TUM4660, TUM4670, was obtained at a frequency of 10−9 per donor and showed a broad-spectrum β-lactam resistance phenotype similar to that of TUM4660 (Table 3). A conjugation experiment with E. coli TUM4670 and E. coli ML4901 was successful at a higher frequency (10−5 per donor), and E. coli TUM4672 was obtained as a transconjugant. Conjugative transfer of blaCMY-2 was also successful from E. coli TUM4672 to K. pneumoniae TUM5459, S. Typhimurium TUM5460, and C. koseri TUM5461.
Determination of the location of the blaCMY-2 gene in P. mirabilis TUM4660.
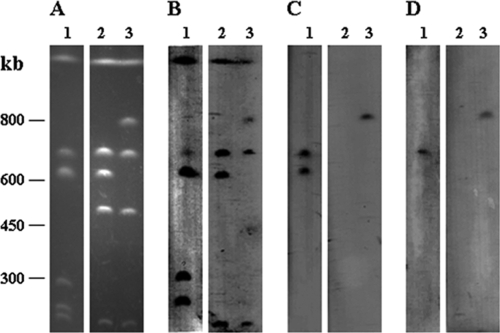
Although TUM4660 carried a 7-kb plasmid, no plasmids were extracted from E. coli TUM4670 by the same procedure. Furthermore, the plasmid of TUM4660 did not hybridize with a blaCMY-2-specific probe (data not shown). Hybridization experiments after I-CeuI digestion of genomic DNA demonstrated that two copies of blaCMY-2 were located on the chromosome of TUM4660 and that blaCMY-2 was also located on the chromosome of TUM4670 (Fig. 1). Therefore, it was suggested that transfer of blaCMY-2 from the chromosome of TUM4660 to the chromosome of E. coli ML4909 occurred in the conjugation experiment.
FIG. 1.
Localization of blaCMY-2 and int in P. mirabilis TUM4660 and its transconjugant. (A) Whole genomic DNAs of P. mirabilis TUM4660 (lane 1), E. coli ML4909 (lane 2), and E. coli TUM4670 (lane 3) were digested with I-CeuI, and the restricted fragments were subjected to pulsed-field gel electrophoresis. DNA fragments were transferred to a nylon membrane and hybridized with probes specific to the 23S rRNA gene (B), blaCMY-2 (C), and int (D).
Involvement of R391/SXT-related ICE in the conjugative transfer of blaCMY-2.
PCR amplification of the genomic DNA of TUM4670 with primer pair LE1/LE4 or RE1/RE4 yielded a 347-bp amplicon or a 454-bp amplicon, respectively, while the results for PCR amplification of the genomic DNA of ML4909 were negative. These results suggested that an SXT/R391-related ICE was integrated into the 5′ end of the prfC gene in TUM4670 during the conjugation experiment. Integration of an SXT/R391-related ICE into the 5′ end of the prfC gene in TUM4660 was also suggested by the positive results of PCRs with primers PMLE1 and LE4 and primers PMRE1 and RE4. PCR with int-specific primers and sequencing of the amplicon demonstrated the presence of an int gene that was 100% identical to that of R391 in TUM4660. PCR with primers LE4 and RE4 yielded a 542-bp product when the genomic DNA of TUM4660 or TUM4670 was used as template DNA, whereas the result was negative for ML4909. This suggested the presence of the excised circular form of an ICE in TUM4660 and TUM4670.
The results of hybridization studies with an int-specific probe supported the hypothesis of the conjugative transfer of blaCMY2 involving an SXT/R391-related ICE (Fig. 1). The SXT/R391-related ICE responsible for the transfer of blaCMY-2 was designated ICEPmiJpn1 according to the nomenclature proposed by Burrus et al. (7), indicating the first SXT/R391-related ICE of P. mirabilis found in Japan. An additional copy of blaCMY-2 unrelated to the SXT/R391-related ICE was also located on the chromosome of TUM4660.
Genetic characterization of ICEPmiJpn1.
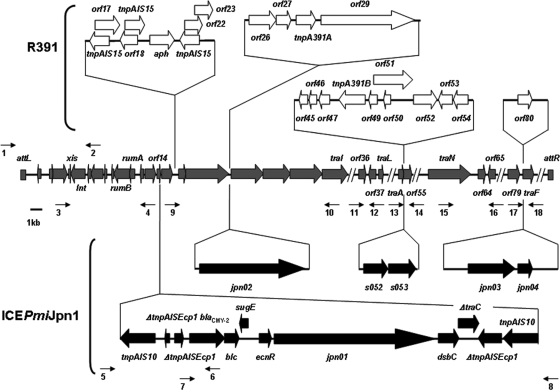
The nucleotide sequences of the attL-traI region and the four hot spots of ICEPmiJpn1 were determined (Fig. 2). Although the nucleotide sequence of ICEPmiJpn1 from attL to traI showed high similarity to that of R391, several open reading frames (ORFs) which did not exist in R391 were found. ICEPmiJpn1 carried an IS10-mediated composite transposon inserted into orf14 of R391. This transposon was composed of two copies of IS10 accompanied by target site 9-bp duplications on both ends and several ORFs, including that encoding the CMY-2 enzyme. A nucleotide sequence identical to the 3′ part of ISEcp1 was located 117 bp upstream from blaCMY-2. On the other hand, a composite transposon in R391 with a kanamycin resistance gene surrounded by three copies of IS15 did not exist in ICEPmiJpn1. Although the nucleotide sequences of hot spot 1 and hot spot 4 showed high similarities with those in R391 (96% and 99%, respectively), the nucleotide sequence of hot spot 2 was almost identical (98%) to that of SXTMO10. Hot spot 3 possessed 2 ORFs, Jpn03 and Jpn04, which have high similarities with ORFs encoding the 5-methylcytosine-specific restriction enzyme B and a conserved hypothetical protein of P. mirabilis ATCC 29906 (98% and 97%, respectively).
FIG. 2.
Schematic representation of genes conserved between ICEPmiJpn1 and R391 and genes specific to ICEPmiJpn1 and R391. The gray arrows represent genes conserved between ICEPmiJpn1 and R391. The black and white arrows represent genes specific to ICEPmiJpn1 and R391 (not to scale), respectively. Thin black arrows represent primers used for PCR amplification of variable regions. Primers: 1, PMLE1; 2, intFor1; 3, intRev1; 4, orf14-Rev2; 5, orf14-For1; 6, CMY-Rev; 7, CMY-For; 8, orf14-Rev1; 9, orf14-For2; 10, traI-Rev; 11, HS1F; 12, HS1R; 13, HotS2F; 14, HotS2R; 15, HotS4F; 16, HotS4R; 17, HotS3F2; 18, TraF2R.
DISCUSSION
In this study, we characterized a novel SXT/R391-related ICE, ICEPmiJpn1, carrying blaCMY-2 on the chromosome of the P. mirabilis clinical isolate TUM4660. To the best of our knowledge, ICEPmiJpn1 is the first SXT/R391-related ICE carrying a gene conferring resistance to extended-spectrum cephalosporins.
ICEPmiJpn1 had int and xis genes completely identical to those of R391, and the nucleotide sequence between attL and traI also showed high similarity to that of R391 (Fig. 2). Nevertheless, several major differences, which characterize the properties of each ICE, were found between the nucleotide sequences of ICEPmiJpn1 and R391. ICEPmiJpn1 lacked the IS15-mediated composite transposon carrying a kanamycin resistance gene, whereas it possessed the IS10-mediated composite transposon of 14.2 kb carrying blaCMY-2. The nucleotide sequence between the two copies of IS10 showed >99% similarity to the nucleotide sequence in the 13-kb type I structure frequently found on blaCMY-2-carrying plasmids in E. coli and Salmonella enterica isolates from humans and animals in the United States (25, 42). This structure contained a 2,823-bp region which appeared to be involved in ISEcp1-mediated mobilization of the blaCMY-2-like gene from the C. freundii chromosome, including ISEcp1, 117 bp upstream from blaCMY-2, and blc, sugE, and ecnR, which are downstream of blaCMY-2 (42). Verdet et al. recently reported that a clinical isolate of P. mirabilis RPCMY isolated in France carried a plasmid containing the 13-kb type I structure flanked by a copy of IS10 at its 5′ end (42). We also identified a P. mirabilis isolate carrying a plasmid with the same genetic structure from retail chicken sampled in Japan (GenBank accession no. AB525078) (unpublished data). These findings suggest that a part of the 13-kb type I structure, once constructed on a relevant plasmid involving the acquisition of a part of the C. freundii chromosome, was employed in the prototypical ICE of ICEPmiJpn1 through IS10-mediated transposition.
In ICEPmiJpn1, the nucleotide sequences of hot spot 1 and hot spot 4 showed high similarities to those in R391, while the nucleotide sequence of hot spot 2 showed high similarity to that of SXTMO10. Hot spot 3 carried nucleotide sequences unrelated to those of R391 or SXTMO10. A similar situation was demonstrated in ICEVchLao1, which carried an R391-like insertion in hot spot 1, an SXTMO10-like insertion in hot spot 2, and the original insertion in hot spot 3 (7, 41). Despite using an identical integration site, namely, the 5′ end of prfC, on the chromosome of the host cell, R391 and SXTMO10 can coexist in a cell and form a tandem array under experimental conditions (18). Formation of a hybrid ICE possessing R391-like nucleotide sequences in one part and SXTMO10-like nucleotide sequences in another part was observed in transconjugants derived from donor cells containing tandem arrays of R391 and SXTMO10 (9). This phenomenon may explain the mechanism that generated the mosaic structure observed in ICEPmiJpn1 and ICEVchLao1 and may contribute to the diversity of SXT/R391-related ICEs.
Although the presence of several IncJ elements was demonstrated for several isolates of Proteus spp. in the 1970s (17, 31), the presence of novel ICEs in Proteus spp. was not reported until recently. Pearson et al. analyzed the complete genome sequence of P. mirabilis HI4320, a representative strain cultured from the urine of a patient with a long-term indwelling urinary catheter, and identified the presence of an ICE similar to R391 (37). Although the unintended detection of an SXT/R391-related ICE in the representative strain of P. mirabilis might suggest that the presence of SXT/R391-related ICEs in P. mirabilis clinical isolates is common, the prevalence of SXT/R391-related ICEs among P. mirabilis isolates has remained undetermined so far. The presence of an ICE could be overlooked if the ICE does not confer specific phenotypic properties, such as antimicrobial resistance, on the host. Even if it provides antimicrobial resistance to the host, one may wrongly assume that the antimicrobial resistance gene is located on a conjugative plasmid, without an active search for the involvement of an ICE, because of the ability to transfer the gene conferring antimicrobial resistance through conjugation.
Host range is one of the critical factors in determining the impact of a mobile genetic element conferring antimicrobial resistance. McGrath et al. identified prfC homologues in many different genera and found by an in silico search that a 12-bp section of the 17-bp integration site of R391 is conserved among 15 members of the Gammaproteobacteria. They demonstrated that up to a 23.5% mismatch between attP and attB was allowed in the integration process of R391, and they succeeded in conjugative transfer of R391 into S. Typhimurium, Enterobacter cloacae, and Serratia marcescens (33). We showed in this study that ICEPmiJpn1 can be transferred to several species of the Enterobacteriaceae. Given this broad potential host range and the selective advantage in the presence of antibiotics, ICEPmiJpn1 or other SXT/R391-related ICEs conferring antimicrobial resistance could play an important role in spreading antimicrobial resistance genes among clinically relevant Enterobacteriaceae in the near future, as already observed among Vibrio species globally (1, 2, 8, 13, 19, 21, 40).
In conclusion, this study demonstrated the involvement of a novel SXT/R391-related ICE, ICEPmiJpn1, in the acquisition of blaCMY-2 by a P. mirabilis clinical isolate. Although this is the first report of an ICE conferring resistance to extended-spectrum cephalosporins, extensive surveillance for the presence of ICEs has never been attempted. The mobile nature of ICEs warrants further studies to clarify the impact of ICEs on the spread of antimicrobial resistance genes in the Enterobacteriaceae.
Acknowledgments
We thank Tse Hsien Koh, Singapore General Hospital, for his useful suggestions and comments. We thank Kiyoshi Sugihara for his technical assistance.
This study was supported by a research promotion grant from Toho University Graduate School of Medicine (grant no. 07-01) and by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sport and Technology of Japan (grant no. 19591185). The collection of Proteus mirabilis TUM4660 was supported by a grant from Toyama Chemical Co., Ltd., and Taisho Toyama Pharmaceutical Co., Ltd.
Footnotes
Published ahead of print on 21 June 2010.
REFERENCES
- 1.Ahmed, A., S. Shinoda, and T. Shimamoto. 2005. A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS Microbiol. Lett. 242:241-247. [DOI] [PubMed] [Google Scholar]
- 2.Amita, S. Chowdhury, M. Thungapathra, T. Ramamurthy, G. Nair, and A. Ghosh. 2003. Class I integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae O139 outbreak, Calcutta, India. Emerg. Infect. Dis. 9:500-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaber, J., V. Burrus, B. Hochhut, and M. Waldor. 2002. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell. Mol. Life Sci. 59:2065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaber, J., B. Hochhut, and M. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böltner, D., C. MacMahon, J. Pembroke, P. Strike, and A. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bret, L., C. Chanal-Claris, D. Sirot, E. Chaibi, R. Labia, and J. Sirot. 1998. Chromosomally encoded AmpC-type β-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 42:1110-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrus, V., J. Marrero, and M. Waldor. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173-183. [DOI] [PubMed] [Google Scholar]
- 8.Burrus, V., R. Quezada-Calvillo, J. Marrero, and M. Waldor. 2006. SXT-related integrating conjugative element in New World Vibrio cholerae. Appl. Environ. Microbiol. 72:3054-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrus, V., and M. Waldor. 2004. Formation of SXT tandem arrays and SXT-R391 hybrids. J. Bacteriol. 186:2636-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli, A., F. Tosini, W. Giles, M. Rupp, S. Hinrichs, F. Angulo, T. Barrett, and P. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. Document M7-A7. CLSI, Wayne, PA.
- 12.Coetzee, J., N. Datta, and R. Hedges. 1972. R factors from Proteus rettgeri. J. Gen. Microbiol. 72:543-552. [DOI] [PubMed] [Google Scholar]
- 13.Dalsgaard, A., A. Forslund, D. Sandvang, L. Arntzen, and K. Keddy. 2001. Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48:827-838. [DOI] [PubMed] [Google Scholar]
- 14.D'Andrea, M., E. Nucleo, F. Luzzaro, T. Giani, R. Migliavacca, F. Vailati, V. Kroumova, L. Pagani, and G. Rossolini. 2006. CMY-16, a novel acquired AmpC-type β-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy. Antimicrob. Agents Chemother. 50:618-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egorova, S., M. Timinouni, M. Demartin, S. Granier, J. Whichard, V. Sangal, L. Fabre, A. Delauné, M. Pardos, Y. Millemann, E. Espié, M. Achtman, P. Grimont, and F. Weill. 2008. Ceftriaxone-resistant Salmonella enterica serotype Newport, France. Emerg. Infect. Dis. 14:954-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Empel, J., A. Baraniak, E. Literacka, A. Mrówka, J. Fiett, E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2008. Molecular survey of β-lactamases conferring resistance to newer β-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob. Agents Chemother. 52:2449-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedges, R. 1975. R factors from Proteus mirabilis and P. vulgaris. J. Gen. Microbiol. 87:301-311. [DOI] [PubMed] [Google Scholar]
- 18.Hochhut, B., J. Beaber, R. Woodgate, and M. Waldor. 2001. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 183:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochhut, B., Y. Lotfi, D. Mazel, S. Faruque, R. Woodgate, and M. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins, K., M. Batchelor, E. Liebana, A. Deheer-Graham, and E. Threlfall. 2006. Characterisation of CTX-M and AmpC genes in human isolates of Escherichia coli identified between 1995 and 2003 in England and Wales. Int. J. Antimicrob. Agents 28:180-192. [DOI] [PubMed] [Google Scholar]
- 21.Iwanaga, M., C. Toma, T. Miyazato, S. Insisiengmay, N. Nakasone, and M. Ehara. 2004. Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob. Agents Chemother. 48:2364-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobsen, S., D. Stickler, H. Mobley, and M. Shirtliff. 2008. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 21:26-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacoby, G. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kado, C., and S. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, M., T. Besser, and D. Call. 2006. Variability in the region downstream of the blaCMY-2 β-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50:1590-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koeck, J. L., G. Arlet, A. Philippon, S. Basmaciogullari, H. V. Thien, Y. Buisson, and J. D. Cavallo. 1997. A plasmid-mediated CMY-2 β-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol. Lett. 152:255-260. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., Q. Li, Y. Du, X. Jiang, J. Tang, J. Wang, G. Li, and Y. Jiang. 2008. Prevalence of plasmid-mediated AmpC β-lactamases in a Chinese university hospital from 2003 to 2005: first report of CMY-2-type AmpC β-lactamase resistance in China. J. Clin. Microbiol. 46:1317-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Literacka, E., J. Empel, A. Baraniak, E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2004. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including novel enzymes CMY-14 and CMY-15, in a Proteus mirabilis clone widespread in Poland. Antimicrob. Agents Chemother. 48:4136-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, S., A. Hessel, and K. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livermore, D. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthew, M., R. Hedges, and J. Smith. 1979. Types of β-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 138:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGrath, B., J. O'Halloran, A. Piterina, and J. Pembroke. 2006. Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J. Microbiol. Methods 66:32-42. [DOI] [PubMed] [Google Scholar]
- 33.McGrath, B., and J. Pembroke. 2004. Detailed analysis of the insertion site of the mobile elements R997, pMERPH, R392, R705 and R391 in E. coli K12. FEMS Microbiol. Lett. 237:19-26. [DOI] [PubMed] [Google Scholar]
- 34.Naas, T., D. Aubert, A. Ozcan, and P. Nordmann. 2007. Chromosome-encoded narrow-spectrum Ambler class A β-lactamase GIL-1 from Citrobacter gillenii. Antimicrob. Agents Chemother. 51:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osorio, C., J. Marrero, R. Wozniak, M. Lemos, V. Burrus, and M. Waldor. 2008. Genomic and functional analysis of ICEPdaSpaI, a fish-pathogen-derived SXT-related integrating conjugative element that can mobilize a virulence plasmid. J. Bacteriol. 190:3353-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagani, L., R. Migliavacca, L. Pallecchi, C. Matti, E. Giacobone, G. Amicosante, E. Romero, and G. Rossolini. 2002. Emerging extended-spectrum β-lactamases in Proteus mirabilis. J. Clin. Microbiol. 40:1549-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson, M., M. Sebaihia, C. Churcher, M. Quail, A. Seshasayee, N. Luscombe, Z. Abdellah, C. Arrosmith, B. Atkin, T. Chillingworth, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, D. Walker, S. Whithead, N. Thomson, P. Rather, J. Parkhill, and H. Mobley. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190:4027-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pembroke, J., and A. Piterina. 2006. A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol. Lett. 264:80-88. [DOI] [PubMed] [Google Scholar]
- 39.Sidjabat, H., D. Paterson, Z. Qureshi, J. Adams-Haduch, A. O'Keefe, A. Pascual, J. Rodríguez-Baño, and Y. Doi. 2009. Clinical features and molecular epidemiology of CMY-type β-lactamase-producing Escherichia coli. Clin. Infect. Dis. 48:739-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taviani, E., D. Ceccarelli, N. Lazaro, S. Bani, P. Cappuccinelli, R. Colwell, and M. Colombo. 2008. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol. Ecol. 64:45-54. [DOI] [PubMed] [Google Scholar]
- 41.Toma, C., N. Nakasone, T. Song, and M. Iwanaga. 2005. Vibrio cholerae SXT element, Laos. Emerg. Infect. Dis. 11:346-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdet, C., V. Gautier, E. Chachaty, E. Ronco, N. Hidri, D. Decré, and G. Arlet. 2009. Genetic context of plasmid-encoded blaCMY-2-like genes in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:4002-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waldor, M., H. Tschäpe, and J. Mekalanos. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yagi, T., J. Wachino, H. Kurokawa, S. Suzuki, K. Yamane, Y. Doi, N. Shibata, H. Kato, K. Shibayama, and Y. Arakawa. 2005. Practical methods using boronic acid compounds for identification of class C β-lactamase-producing Klebsiella pneumoniae and Escherichia coli. J. Clin. Microbiol. 43:2551-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]