Abstract
The Klebsiella pneumoniae multidrug resistance plasmid pKP048 was completely sequenced. This plasmid carries several important resistance determinants, such as blaKPC-2, blaDHA-1, qnrB4, and armA, which confer resistance to carbapenems, cephalosporins, fluoroquinolones, and aminoglycosides, respectively. Analysis of the finished 151,188-bp sequence data revealed 163 putative genes, 108 of which were assigned functions such as replication, stable inheritance, antibiotic resistance, a mobile element, conjugal transfer, and a restriction-modification system, showing the strong phylogenetic mosaicism and plasticity of the plasmid.
Multidrug-resistant Klebsiella pneumoniae presents a severe clinical problem, mainly because it harbors plasmids encoding many virulence or resistance determinants. Plasmids play an important role in the horizontal transfer of genetic information and the evolution of the bacterial genome, and in recent years, they have been particularly involved in the dissemination of antibiotic resistance (10).
Plasmid pKP048, belonging to incompatibility (Inc) group F, was detected in carbapenem-resistant K. pneumoniae in 2006 (15). pKP048 contains many plasmid-mediated resistance determinants, such as blaKPC-2, blaDHA-1, qnrB4, and armA, conferring resistance to carbapenems, cephalosporins, fluoroquinolones, and aminoglycosides, respectively (17, 4, 8, 16). Furthermore, the 2′-phosphotransferase Mph2, conferring resistance to macrolides (12), and a mercury resistance protein cluster are also located on this plasmid. The association of many important antibiotic resistance determinants on the same plasmid is of concern because of the risk of dissemination due to coselection by various drugs (13). In order to determine the genetic structure of the multidrug resistance plasmid pKP048, the complete plasmid was sequenced in this study. The components of the plasmid backbone involved in general maintenance and conjugation are also discussed.
K. pneumoniae KP048 was isolated from a sputum specimen at the First Affiliated Hospital, College of Medicine, Zhejiang University, in China in 2006. Species identification was confirmed using a Vitek Gram-Negative Plus identification card (bioMerieux Vitek Inc., Hazelwood, MO). Plasmid pKP048 was extracted from the transconjugant of clinical strain KP048 by using a Qiagen Midi kit (Qiagen, Germany) according to the manufacturer's instructions. The conjugation experiment was carried out in mixed-broth cultures as previously described (15), and rifampin-resistant Escherichia coli EC600 (LacZ− Nalr Rifr) was used as the recipient strain. The MICs of meropenem, imipenem, ertapenem, colistin, tigecycline, ceftazidime, ciprofloxacin, piperacillin-tazobactam, cefoxitin, and amikacin were determined by the Etest technique (AB bioMerieux, Sweden) according to the manufacturer's instructions, and the susceptibility breakpoints were interpreted as recommended by the Clinical and Laboratory Standards Institute (CLSI) and previous reports (5, 9). Plasmid pKP048 from strain KP048 was completely sequenced by using a shotgun approach (15).
The MICs of the antimicrobial agents mentioned above against isolate KP048 and its transconjugant are shown in Table 1. The transconjugant isolate CKP048 inherited resistance to ertapenem, ceftazidime, piperacillin-tazobactam, cefoxitin, and amikacin, as well as reduced susceptibility to meropenem, imipenem, and ciprofloxacin, whereas colistin and tigecycline retained almost their original efficacies.
TABLE 1.
MICs for clinical strain KP048 and its transconjugant CKP048
| Isolate | Species | Plasmid harbored | MIC (μg/ml)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | IPM | ETP | CST | TGC | CAZ | TZP | CIP | FOX | AMK | |||
| KP048 | K. pneumoniae | pKP048 | >32 | >32 | 32 | 0.38 | 0.38 | 96 | >256 | >32 | 256 | >256 |
| CKP048 | E. coli | pKP048 | 2 | 3 | 8 | 0.38 | 0.125 | 32 | >256 | 0.5 | 128 | >256 |
| EC600 | E. coli | None | 0.094 | 0.25 | 0.016 | 0.25 | 0.125 | 0.064 | 2 | 0.012 | 2 | 1 |
MEM, meropenem; IPM, imipenem; ETP, ertapenem; CST, colistin; TGC, tigecycline; CAZ, ceftazidime; TZP, piperacillin-tazobactam; CIP, ciprofloxacin; FOX, cefoxitin; AMK, amikacin.
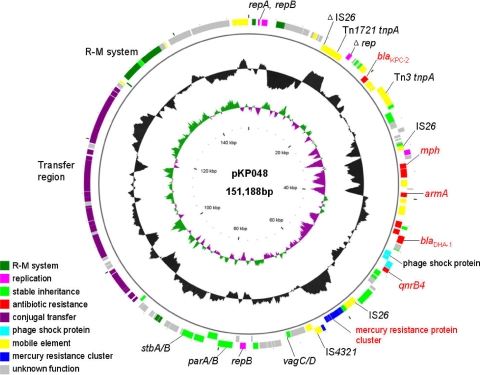
Plasmid pKP048 is a 151,188-bp closed circular DNA sequence with an average G+C content of 51.3% (Fig. 1). The G+C content ranges from 31.7% to 69.3% depending on a sliding window of 1,000 bp and a step size of 10 bp. Annotation of the finished sequence data revealed that pKP048 contained 163 open reading frames (ORFs), 108 of which encoded proteins with homology to proteins with known functions and were assigned to several modules with functions as follows: replication, stable inheritance, antibiotic resistance, mobile element, conjugal transfer, and restriction-modification (R-M) system. More information regarding these ORFs is given in Table S1 in the supplemental material.
FIG. 1.
Circular map of plasmid pKP048. The two outer circles represent ORFs in the plus (outside) and minus (inside) orientations, respectively. Functions are color coded as explained in the key. The two inner circles represent the G+C content plotted against the average G+C content of 51.3% (black circle) and GC skew information (green and purple circles).
The overall genetic structure of plasmid pKP048 was considered a mosaic of functional modules with various phylogenetic origins. The replication, stable inheritance, and conjugal transfer modules constituting the backbone of pKP048 were similar to those of plasmids pKPN3 (GenBank accession no. NC_009649), pKPN4 (NC_009650), and pKPN5 (NC_009651), all of which were found on Klebsiella pneumoniae subsp. pneumoniae MGH 78578 by a K. pneumoniae genome-sequencing project. Nevertheless, the coherent blocks in these plasmids were discrete. Of the 163 ORFs in pKP048, 46 (28%), 68 (42%), and 33 (20%) ORFs were also present in pKPN3, pKPN4, and pKPN5, respectively. The origins of other modules, such as antibiotic resistance determinants or mobile elements, represented more diversification, indicating a high plasticity of the plasmid.
Plasmid pKP048 contained three replication regions, one of which was truncated with loss of function; the other two both belonged to Inc group F. The putative origin of replication of pKP048 is the second fragment of IncF replication protein RepB (positions 76021 to 76887), flanked by several plasmid maintenance and partitioning modules, such as VagC/VagD, ParA/ParB, and StbA/StbB (3, 14). The transfer region of pKP048 comprised 21 tra genes and 5 trb genes (ordered as follows: traM, traJ, traA, traL, traE, traK, traB, traV, traC, trbI, traW, traU, trbC, traN, trbE, traF, traQ, trbB, trbF, traH, traG, traS, traT, traD, traI, traX), finO, and 8 putative genes. The synteny of this region is conserved in pKPN3 and pKPN4, especially in the gene order.
In contrast to the Tn4401 structure in U.S. isolate YC (11) and other blaKPC-containing plasmids (7), the immediate environment surrounding blaKPC-2 in pKP048 was considered to result from the integration of a Tn3-based transposon and a partial Tn4401 structure, with the ORFs ordered as follows: Tn3-transposase, Tn3-resolvase, ISKpn8, the blaKPC-2 gene, and the ISKpn6-like element (see Fig. 3 in reference 15). Each side of the region contained one of the two segments of the Tn1721 transposon, which was split by the Tn4401-Tn3 integration region.
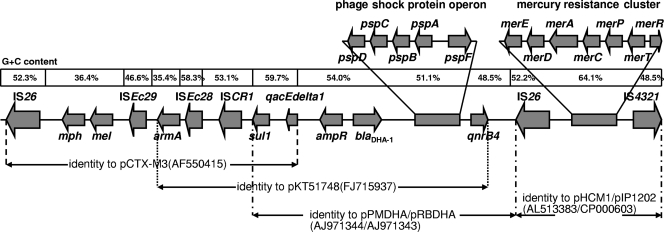
Plasmid pKP048 harbored three copies of an IS26 element. One was a truncated IS26 copy located at positions 11495 to 11797. The other two copies bracketed a 25-kb region that consists of most of the elements involved in mobilization and drug resistance in the plasmid (Fig. 2). Upstream of the first of two IS26 copies was a 12,484-bp segment identified with plasmid pCTX-M3 (GenBank accession number AF550415) (6). This segment ended with an incomplete 3′ conserved sequence (3′-CS) of a sul1-type integron but had lost almost the entire 5′-CS and gene cassettes. The 3′-CS was truncated due to the insertion of an ISCR1 element (positions 41299 to 42840). Downstream of ISCR1 were ISEc28, the aminoglycoside resistance gene armA, ISEc29 (2), and the mel and mph genes, reported previously to be associated with resistance to macrolides (1, 12). The AmpC-type blaDHA-1 gene and its regulator, ampR, were located immediately after the incomplete 3′-CS. Downstream of the blaDHA-1 gene were a so-called phage shock protein operon and quinolone resistance determinant qnrB4.
FIG. 2.
Schematic representation of the 25-kb armA, blaDHA-1, and qnrB4 region and the mercury resistance protein cluster region. The G+C content for each of the putative imported DNA sections is given. The average G+C content of the complete plasmid is 51.3%.
Furthermore, a mercury resistance protein cluster containing the mercury-resistance genes merE, merD, merA, merC, merP, merT, and merR (Fig. 2), and several R-M system elements, such as a type I R-M system, all of which facilitated bacterial survival and plasmid evolution, were identified in plasmid pKP048.
The entire sequence of pKP048 provided insight into the genetic structure of this clinical multidrug resistance plasmid and exhibited strong phylogenetic mosaicism. This plasmid chimera might evolve through stepwise integration or recombination events and the evolution might enhance the dissemination of multiple antimicrobial resistance determinants.
Nucleotide sequence accession number.
The annotated sequence of plasmid pKP048 from strain KP048 has been submitted to GenBank under accession number FJ628167.
Supplementary Material
Acknowledgments
This work was funded by grants from the Ministry of Education of China (20070335188), the Ministry of Health of China (200802107), and the Health Bureau of Zhejiang Province (2005QN003).
Footnotes
Published ahead of print on 14 June 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ambrose, K. D., R. Nisbet, and D. S. Stephens. 2005. Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob. Agents Chemother. 49:4203-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berçot, B., L. Poirel, and P. Nordmann. 2008. Plasmid-mediated 16S rRNA methylases among extended-spectrum beta-lactamase-producing Enterobacteriaceae isolates. Antimicrob. Agents Chemother. 52:4526-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernández-Lopez, R., M. P. Garcillan-Barcia, C. Revilla, M. Lazaro, L. Vielva, and F. de la Cruz. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942-966. [DOI] [PubMed] [Google Scholar]
- 4.Gaillot, O., C. Clement, M. Simonet, and A. Philippon. 1997. Novel transferable beta-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J. Antimicrob. Chemother. 39:85-87. [DOI] [PubMed] [Google Scholar]
- 5.Galani, I., F. Kontopidou, M. Souli, P. D. Rekatsina, E. Koratzanis, J. Deliolanis, and H. Giamarellou. 2008. Colistin susceptibility testing by Etest and disk diffusion methods. Int. J. Antimicrob. Agents 31:434-439. [DOI] [PubMed] [Google Scholar]
- 6.Gołebiewski, M., I. Kern-Zdanowicz, M. Zienkiewicz, M. Adamczyk, J. Zylinska, A. Baraniak, M. Gniadkowski, J. Bardowski, and P. Ceglowski. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gootz, T. D., M. K. Lescoe, F. Dib-Hajj, B. A. Dougherty, W. He, P. Della-Latta, and R. C. Huard. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelesidis, T., D. E. Karageorgopoulos, I. Kelesidis, and M. E. Falagas. 2008. Tigecycline for the treatment of multidrug-resistant Enterobacteriaceae: a systematic review of the evidence from microbiological and clinical studies. J. Antimicrob. Chemother. 62:895-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leplae, R., G. Lima-Mendez, and A. Toussaint. 2006. A first global analysis of plasmid encoded proteins in the ACLAME database. FEMS Microbiol. Rev. 30:980-994. [DOI] [PubMed] [Google Scholar]
- 11.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguchi, N., A. Emura, H. Matsuyama, K. O'Hara, M. Sasatsu, and M. Kono. 1995. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob. Agents Chemother. 39:2359-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Périchon, B., P. Bogaerts, T. Lambert, L. Frangeul, P. Courvalin, and M. Galimand. 2008. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob. Agents Chemother. 52:2581-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pullinger, G. D., and A. J. Lax. 1992. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 6:1631-1643. [DOI] [PubMed] [Google Scholar]
- 15.Shen, P., Z. Wei, Y. Jiang, X. Du, S. Ji, Y. Yu, and L. Li. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan, J. J., J. J. Wu, W. C. Ko, S. H. Tsai, C. L. Chuang, H. M. Wu, Y. J. Lu, and J. D. Li. 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob. Chemother. 54:1007-1012. [DOI] [PubMed] [Google Scholar]
- 17.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.