Abstract
CXA-101, previously designated FR264205, is a new antipseudomonal cephalosporin. The objective of this study was to determine the penicillin-binding protein (PBP) inhibition profile of CXA-101 compared to that of ceftazidime (PBP3 inhibitor) and imipenem (PBP2 inhibitor). Killing kinetics, the induction of AmpC expression, and associated changes on cell morphology were also investigated. The MICs for CXA-101, ceftazidime, and imipenem were 0.5, 1, and 1 μg/ml, respectively. Killing curves revealed that CXA-101 shows a concentration-independent bactericidal activity, with concentrations of 1× the MIC (0.5 μg/ml) producing a >3-log reduction in bacterial load after 8 h of incubation. Live-dead staining showed that concentrations of CXA-101 as low as 0.5× the MIC stopped bacterial septation and induced an intense filamentation, which is consistent with the documented high affinity of PBP3. CXA-101 was found to be a potent PBP3 inhibitor and showed affinities ≥2-fold higher than those of ceftazidime for all of the essential PBPs (1b, 1c, 2, and 3). Compared to imipenem, in addition to the obvious inverse PBP2/PBP3 affinities, CXA-101 showed a significantly higher affinity for PBP1b but a lower affinity for PBP1c. Furthermore, CXA-101, like ceftazidime and in contrast to imipenem, was found to be a very weak inducer of AmpC expression, consistent with the low PBP4 affinity documented.
Pseudomonas aeruginosa is intrinsically resistant to several antibiotics due to the low permeability of its outer membrane, the constitutive expression of several efflux pumps, and the production of antibiotic-inactivating enzymes (8, 15-17). Intrinsic resistance to β-lactam antibiotics occurs via the induction of chromosomally encoded AmpC β-lactamase (8-10, 16).
Antipseudomonal penicillins (piperacillin) and cephalosporins (cefepime or ceftazidime) are active against P. aeruginosa because they are very weak inducers of this chromosomal β-lactamase, although these compounds are certainly hydrolyzed by AmpC (15). Furthermore, mutants hyperproducing AmpC are frequently selected during treatment with antipseudomonal β-lactams, leading to the failure of antimicrobial therapy with these antibiotics (3, 7, 12).
The activity of β-lactams against P. aeruginosa (and most other Gram-negative pathogens) will mainly depend in the overall balance of three key attributes: (i) concentration reached in the periplasmic space, dependent on the balance between diffusion through the outer membrane and extrusion by efflux pumps; (ii) resistance to the chromosomally encoded AmpC, dependent on whether the β-lactam is stable to hydrolysis and/or whether it does or does not prevent the induction of the β-lactamase; and (iii) most importantly, the affinity against the main targets of β-lactam antibiotics, the essential penicillin-binding proteins (PBPs), which are PBP1b, PBP1c, PBP2, and PBP3 in the case of P. aeruginosa (25). The PBP inhibition profile is therefore crucial for establishing the qualitative and quantitative dimensions of β-lactam bactericidal activity.
Although high-molecular-weight PBPs (1b, 1c, 2, and 3) carry essential functions, the low-molecular-weight penicillin-binding proteins function as dd-carboxypeptidases and/or dd-endopeptidases and are not essential for cell viability (4, 21, 25, 27). Antipseudomonal cephalosporins such as ceftazidime or cefotaxime bind preferentially to PBP3 (1, 4, 23), leading to filament formation, while carbapenems bind to PBP2 (5, 30), ultimately leading to the conversion of rod-shaped cells into spherical cells.
Recent works, however, show that it might not be wise to inhibit all PBPs, since at least one of them, PBP4, is shown to be a trap target for β-lactams that is connected to the AmpC induction pathway (19).
CXA-101, previously designated FR264205, is a new cephalosporin currently under clinical development that shows promising characteristics for the treatment of P. aeruginosa infections. Several recent surveys have revealed a potent in vitro activity of CXA-101 against P. aeruginosa, including cystic fibrosis and multidrug-resistant strains (2, 13, 18, 20, 28, 31). In addition, in vitro studies have shown that CXA-101 appears to be stable against the most common resistance mechanisms driven by mutation in this species, particularly noteworthy the overexpression of the chromosomal cephalosporinase AmpC driven by ampD and/or dacB (PBP4) mutations (18, 20, 28, 29). Recent studies have shown that, consistent with this observation, rates of P. aeruginosa mutation to CXA-101 resistance are extremely low (<5 × 10−11), sharply contrasting with the high mutation rates documented for other antipseudomonal cephalosporins or carbapenems (24). Only certain transferable β-lactamases, such as extended-spectrum β-lactamases or class B carbapenemases, which are still infrequent in P. aeruginosa, appear to compromise CXA-101 activity (6, 13, 18). Thus far, no study has been performed to evaluate the binding affinity of CXA-101 to its target PBPs in P. aeruginosa or other pathogens. The objective of the present study was to determine the PBP inhibition profile of CXA-101 compared to those of ceftazidime (a PBP3 inhibitor) and imipenem (a PBP2 inhibitor). Killing kinetics, associated changes on cell morphology, and AmpC-inducing properties were also investigated.
MATERIALS AND METHODS
Susceptibility testing and killing kinetics.
The MICs of CXA-101, ceftazidime, and imipenem for strain PAO1 were determined by standard CSLI broth microdilution (M100-S18). CXA-101 (lot 3F02) was provided by Calixa Therapeutic, Inc. Other antibiotics were purchased from commercial sources. For killing kinetics studies, overnight Mueller-Hinton broth (MHB) cultures of PAO1 were diluted (1/100) in fresh medium and incubated (37°C, 180 rpm) to an optical density at 600 nm (OD600) of 0.2 (mid-log-phase growth). Killing curves were then initiated by inoculating 1 ml in flasks with 20 ml (initial inoculum OD600 0.01, ca. 107 CFU/ml) of MHB containing concentrations of 0, 0.5×, 1×, 2×, or 4× the MIC of CXA-101, ceftazidime, and imipenem. Killing kinetics were then monitored during 8 h and were analyzed by OD600 quantification, CFU enumeration (plating serial dilutions in MH agar [MHA]) and live-dead staining with the Live/Dead BacLight bacterial viability kit (Molecular Probes/Invitrogen, Carlsbad, CA), following the manufacturer's instructions and using a Nikon Eclipse E400 fluorescence microscope at 1,000× magnification. All experiments were performed in triplicate.
Quantification of AmpC induction.
The induction of AmpC production by CXA-101, ceftazidime, and imipenem was determined measuring the level of expression of ampC by real-time reverse transcription-PCR (RT-PCR) in strain PAO1, with or without incubation (3 h) in the presence of subinhibitory concentrations (1/2 and 1/4 MIC concentrations). Cefoxitin at 50 μg/ml, a potent AmpC inducer commonly used in induction experiments (11, 19), was also included for comparative purposes. Total RNA from logarithmic-phase-grown cultures (LB broth) was obtained with the RNeasy minikit (Qiagen, Hilden, Germany) and treated with 2 U of TURBO DNase (Ambion) for 30 min at 37°C to remove contaminating DNA. The reaction was stopped by the addition of 5 μl of DNase inactivation reagent, and the samples were adjusted to a final concentration of 50 ng/μl. A 500-ng sample of purified RNA was then used for one-step reverse transcription and real-time PCR amplification using the QuantiTect SYBR green RT-PCR kit (Qiagen, Hilden, Germany) in a SmartCycler II (Cepheid, Sunnyvale, CA). The previously described primer pairs ACrnaF/ACrnaR (11) and rpsL-1/rpsL-2 (22) were used for amplification of ampC and rpsL, respectively (used as references to normalize the relative amount of mRNA). In all cases, the mean values of relative mRNA expression obtained in at least three independent duplicate experiments were considered.
PBP assays.
Membranes containing the PBPs of P. aeruginosa PAO1 were obtained according to described protocols (4, 5, 26, 32). Briefly, 500-ml late-log-phase (OD600 = 1) Luria-Bertani (LB; Sigma-Aldrich, St. Louis, MO) cultures of PAO1 were collected by centrifugation (4,400 × g, 10 min) and then washed and suspended in 50 ml of 20 mM KH2PO4-140 mM NaCl (pH 7.5) (buffer A). Cells were then sonicated by using a Digital Sonifier Unit model S-450D (Branson Ultrasonics Corp., Danbury, CT) at 20 W for three 30-s bursts (while immersed in an ice bath) and centrifuged at 12,000 × g for 10 min. Membranes containing the PBPs were isolated from the supernatant through two steps of ultracentrifugation at 150,000 × g for 1 h at 4°C using an Optima L-XP Series Preparative Ultracentrifuge (Beckman Coulter, Inc., Palo Alto, CA) and suspension in buffer A. Total protein content was measured through the Bradford method using the Quick Start Bradford protein assay kit with bovine serum albumin as a standard (Bio-Rad Laboratories, Hercules, CA), according to the manufacturer's instructions. Portions (20 μl, final volume) of PBPs (P. aeruginosa) containing solution were then incubated (30 min, 37°C) in the presence of increasing concentrations of CXA-101, ceftazidime, or imipenem (range of concentrations tested, 0.0156 to 2 μg/ml) and were afterward labeled with a 25 μM concentration of the fluorescent penicillin Bocillin FL (32). The reaction mixtures were then denatured with 20-μl portions of sodium dodecyl sulfate (SDS) denaturing solution (14) at 100°C for 3 min. PBPs were then separated through 10% SDS-polyacrylamide gel electrophoresis (Bio-Rad). The protein gels were rinsed in water immediately after electrophoresis. Labeled PBPs were visualized using a Bio-Rad Molecular Imager FX Pro (Bio-Rad; excitation at 488 nm and emission at 530 nm) and 50% inhibitory concentrations (IC50) of CXA-101, ceftazidime, and imipenem for the different PBPs were determined from triplicate experiments using the QuantityOne software (Bio-Rad).
RESULTS AND DISCUSSION
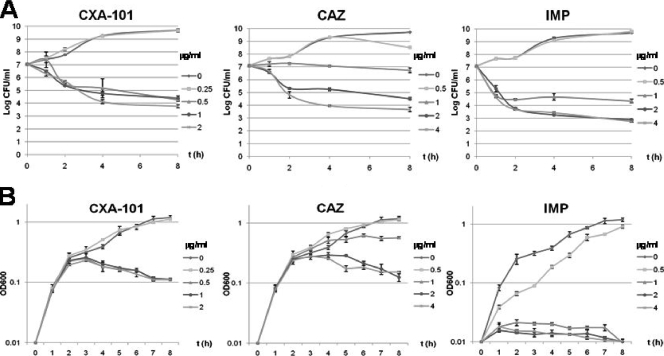
MICs for CXA-101, ceftazidime, and imipenem were 0.5, 1, and 1 μg/ml, respectively. The results for killing kinetics experiments are shown in Fig. 1; Figure 1A shows the results of the killing curves measured in terms of reduction of viable CFU/ml over time, whereas Fig. 1B shows the results in terms of OD600 change. Figure 2 shows the results for Live/Dead staining at concentrations 0.5× and 4× the MIC of each antibiotic.
FIG. 1.
Results for killing kinetics experiments. (A) Results of the killing curves measured in terms of reduction of viable CFU/ml over time. (B) Results of the killing curves in terms of the OD600 change. The concentrations tested were 0×, 0.5×, 1×, 2×, and 4× the MICs (0.5 μg/ml for CXA-101 and 1 μg/ml for ceftazidime [CAZ] and imipenem [IMP]). Mean values for three experiments ± the standard deviation are shown.
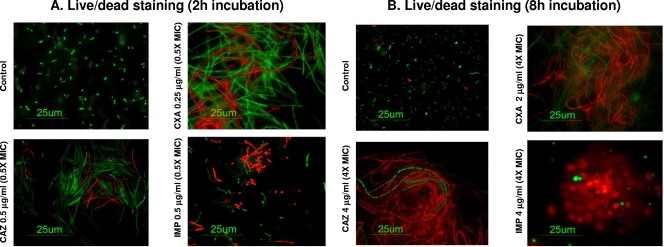
FIG. 2.
Results of Live/Dead staining. (A) Images obtained at 2 h of incubation with 0.5× MIC concentrations of CXA-101 (CXA), ceftazidime (CAZ), or imipenem (IMP). (B) Images obtained at 8 h of incubation with 4× MIC concentrations of each antibiotic. Live cells are green stained, and dead cells are red stained.
As shown in Fig. 1A, CXA-101 displayed a concentration-independent bactericidal activity; concentrations of 1× MIC (0.5 μg/ml produced a sharp reduction of bacterial load [>3 log]) during the 8 h of incubation, but bactericidal activity was not increased at higher antibiotic concentrations. Ceftazidime bactericidal activity was much lower at 1× MIC and required at least 2× MIC concentrations (2 μg/ml) to reach a bactericidal activity equivalent to that of CXA-101. On the other hand, imipenem bactericidal activity was similar to that of CXA-101 at 1× MIC (1 μg/ml) concentrations, although in contrast to that documented for CXA-101, imipenem bactericidal activity was still increased further at 2× MIC concentrations.
As shown in Fig. 1B, CXA-101 and ceftazidime did not modify OD increase, compared to control without antibiotic, during the first 2 to 3 h of incubation at any of the tested concentrations. This apparent discrepancy between CFU enumeration and OD measurement has been previously documented for ceftazidime and is a consequence of filamentation (filaments increase the OD but not CFU) driven by PBP3 inhibition (1). Nevertheless, after 3 h of incubation, a 1× MIC of CXA-101 concentration sharply reduced the OD, and again this activity was concentration independent. Ceftazidime required a 2× MIC concentration to reduce the OD. In contrast to CXA-101 and ceftazidime, imipenem produced a sharp reduction of OD since the initiation of the killing curves. Live/Dead staining results were consistent with these findings, since CXA-101, as previously described for ceftazidime, inhibited cell septation, producing long filaments even at subinhibitory concentrations (0.5× MIC, 0.25 μg/ml) (Fig. 2A), and most of them were killed after prolonged incubation at concentrations above the MIC (Fig. 2B).
The results of AmpC induction are shown in Table 1. As expected, imipenem produced an extremely high AmpC induction, increasing the ampC expression, even at 1/4 MIC concentrations, by 200-fold. In contrast, CXA-101, like ceftazidime, had very little effect on ampC expression, either at 1/4 or 1/2 MIC concentrations.
TABLE 1.
Levels of ampC expression after incubation of P. aeruginosa PAO1
| Condition (concn [μg/ml])a | Mean ampC expression ± SDb |
|---|---|
| Basal (no antibiotic) | 1 |
| FOX (50) | 50 ± 14 |
| CAZ | |
| 1/4 MIC (0.25) | 1.2 ± 0.2 |
| 1/2 MIC (0.5) | 1.2 ± 0.3 |
| CXA | |
| 1/4 MIC (0.125) | 1.6 ± 1.3 |
| 1/2 MIC (0.25) | 1.3 ± 0.7 |
| IMP | |
| 1/4 MIC (0.25) | 193.2 ± 20.2 |
| 1/2 MIC (0.5) | 269.5 ± 73.1 |
P. aeruginosa PAO1 was incubated in the presence of subinhibitory concentrations of CXA-101 (CXA), ceftazidime (CAZ), imipenem (IMP), and cefoxitin (FOX) as indicated.
The relative amount of ampC mRNA compared to the PAO1 basal level is given.
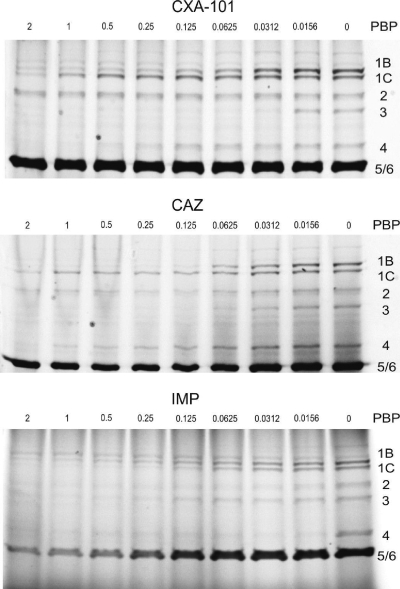
Results for PBPs IC50 of CXA-101, ceftazidime, and imipenem are shown in Table 2, whereas Fig. 3 shows a representative example of the SDS-polyacrylamide gels labeled with Bocillin FL and incubated in the presence of CXA-101, ceftazidime, or imipenem (range, 0.0156 to 2 μg/ml). CXA-101 was found to be a very potent PBP3 inhibitor and showed affinities ≥2-fold higher than those of ceftazidime for all of the essential PBPs (1b, 1c, 2, and 3). Compared to imipenem, in addition to the obvious inverse PBP2/PBP3 affinities, CXA-101 showed a significantly higher affinity for PBP1b but a lower affinity for PBP1c. There are few studies addressing binding affinities of β-lactams to P. aeruginosa PBPs. It is worth mentioning, among these studies, the results for the recently introduced carbapenem doripenem, which was found to show higher affinities than imipenem for PBP2 and PBP3 but lower affinities for PBP1b and PBP1c (5). The affinity of CXA-101 against the nonessential PBP4, recently shown to be a trap target for β-lactams involved in AmpC induction (19), was 15-fold lower than that of imipenem (the most potent AmpC inducer). Although the affinity of CXA-101 against PBP4 was 4-fold higher than that of ceftazidime, the data above from the induction experiments suggest that the affinity may still be too low to significantly affect AmpC expression.
TABLE 2.
IC50 of CXA-101, ceftazidime, and imipenem for P. aeruginosa PAO1 PBPs
| PBP | Mean IC50 (μg/ml) ± SDa |
||
|---|---|---|---|
| CAZ | CXA | IMP | |
| 1b | 0.12 ± 0.03 | 0.07 ± 0.01 | 0.13 ± 0.01 |
| 1c | >2 | 0.64 ± 0.17 | 0.08 ± 0.005 |
| 2 | >2 | 1.36 ± 0.56 | 0.08 ± 0.01 |
| 3 | 0.04 ± 0.01 | 0.02 ± 0.007 | 0.12 ± 0.2 |
| 4 | 1.23 ± 0.49 | 0.29 ± 0.05 | 0.02 ± 0.01 |
| 5/6 | >2 | >2 | 0.2 ± 0.09 |
IC50, 50% inhibitory concentration; CXA, CXA-101; CAZ, ceftazidime; IMP, imipenem.
FIG. 3.
Representative example of the SDS-polyacrylamide gels labeled with Bocillin FL and incubated in the presence of CXA-101, ceftazidime (CAZ), or imipenem (IMP). Range of concentrations tested: 0.0156 to 2 μg/ml.
Concluding remarks.
In the present study, a comprehensive evaluation of the bactericidal mode of action of CXA-101 was performed. CXA-101 was found to show a 2- to 4-fold-higher potency than ceftazidime based on all of the parameters examined, including MICs, killing kinetics, and binding affinities to essential PBPs. Among the essential PBPs, CXA-101 was the most potent inhibitor of PBP3 and PBP1b, whereas imipenem was the most potent inhibitor of PBP2 and PBP1c. CXA-101 showed a 2-fold-lower MIC than imipenem, and both antibiotics showed a similar killing kinetics at 1× MIC concentrations. Furthermore, in contrast to imipenem, CXA-101 was found to be a very weak inducer of AmpC expression, a valuable property, added to previous data showing its high stability against hydrolysis by AmpC, in contrast to ceftazidime (20, 28, 29). Interestingly, CXA-101 showed a totally concentration-independent bactericidal activity, while ceftazidime and imipenem required a 2-fold MIC concentration for maximum bactericidal activity. These results further validated the mechanism of CXA-101 bactericidal activity against P. aeruginosa by targeting essential PBPs and inhibiting bacterial cell wall synthesis featured by cell filamentation. Altogether, these data show that CXA-101 is a potent antipseudomonal agent in vitro and, in conjunction with other studies showing its low MICs against diverse collections of clinical isolates, including multidrug-resistant strains (2, 13, 18, 20, 31), and its stability against β-lactam, carbapenem, and fluoroquinolone resistance mechanisms (2, 13, 18, 20, 28, 29), suggest that CXA-101 could be, if successfully developed, a valuable alternative for the treatment of P. aeruginosa infections, one that needs to be further evaluated in pertinent clinical trials.
Acknowledgments
This study was supported by a grant from Calixa Therapeutics and by the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III, through the Spanish Network for the Research in Infectious Diseases (REIPI C03/14 and RD06/0008; PS09/00033).
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.Blázquez, J., J. M. Gómez-Gómez, A. Oliver, C. Juan, V. Kapur, and V. Marin. 2006. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol. Microbiol. 62:84-99. [DOI] [PubMed] [Google Scholar]
- 2.Bulik, C. C., H. Christensen, and D. P. Nicolau. 2010. In vitro potency of CXA-101, a novel cephalosporin, against resistant phenotypes of Pseudomonas aeruginosa, including multi-drug resistant isolates. Antimicrob. Agents Chemother. 54:557-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmeli, Y., N. Troillet, G. M. Eliopoulos, and M. H. Samore. 1999. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Antimicrob. Agents Chemother. 43:1379-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis, N. A., D. Orr, G. W. Ross, and M. G. Boulton. 1979. Competition of beta-lactam antibiotics for the penicillin-binding proteins of Pseudomonas aeruginosa, Enterobacter cloacae, Klebsiella aerogenes, Proteus rettgeri, and Escherichia coli: comparison with antibacterial activity and effects upon bacterial morphology. Antimicrob. Agents Chemother. 16:325-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, T. A., W. Shang, K. Bush, and R. K. Flamm. 2008. Affinity of doripenem and comparators to penicillin-binding proteins of Escherichia coli and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:1510-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giske, C. G., J. Ge, and P. Nordmann. 2009. Activity of cephalosporin CXA-101 (FR264205) and comparators against extended-spectrum-β-lactamase-producing Pseudomonas aeruginosa. J. Antimicrob. Chemother. 642:430-431. [DOI] [PubMed] [Google Scholar]
- 7.Giwercman, B., P. A. Lambert, V. T. Rosdahl, G. H. Shand, and N. Høiby. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in-vivo selection of stable partially derepressed beta-lactamase producing strains. J. Antimicrob. Chemother. 26:247-259. [DOI] [PubMed] [Google Scholar]
- 8.Hancock, R. E. 1999. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 1:S93-S99. [DOI] [PubMed] [Google Scholar]
- 9.Hanson, N. D., and C. C. Sanders. 1999. Regulation of inducible AmpC beta-lactamase expression among Enterobacteriaceae. Curr. Pharm. Des. 5:881-894. [PubMed] [Google Scholar]
- 10.Jacoby, G. A., and L. S. Munoz-Price. 2005. The new beta-lactamases. N. Engl. J. Med. 352:380-391. [DOI] [PubMed] [Google Scholar]
- 11.Juan, C., M. D. Maciá, O. Gutiérrez, C. Vidal, J. L. Pérez, and A. Oliver. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 49:4733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juan, C., O. Gutiérrez, A. Oliver, J. I. Ayestarán, N. Borrell, and J. L. Pérez. 2005. Contribution of clonal dissemination and selection of mutants during therapy to Pseudomonas aeruginosa antimicrobial resistance in an intensive care unit setting. Clin. Microbiol. Infect. 11:8878-8892. [DOI] [PubMed] [Google Scholar]
- 13.Juan, C., L. Zamorano, J. L. Pérez, Y. Ge, and A. Oliver on behalf of the Spanish Group for the Study of Pseudomonas and REIPI. 2010. Activity of a new antipseudomonal cephalosporin, CXA-101 (FR264205), against carbapenem-resistant and multidrug-resistant Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 54:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1979. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lister, P. D., D. J. Wolter, and N. D. Hanson. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22:582-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 18.Livermore, D. M., S. Mushtaq, Y. Ge, and M. Wagner. 2009. Activity of cephalosporin CXA-101 (FR264205) against Pseudomonas aeruginosa and Burkholderia cepacia group strains and isolates. Int. J. Antimicrob. Agents 34:402-406. [DOI] [PubMed] [Google Scholar]
- 19.Moya, B., A. Döstch, C. Juan, J. Blázquez, L. Zamorano, S. Haussler, and A. Oliver. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moya, B., L. Zamorano, C. Juan, J. L. Pérez, Y. Ge, and A. Oliver. 2010. Activity of a new cephalosporin, CXA-101 (FR264205), against β-lactam-resistant Pseudomonas aeruginosa mutants selected in vitro and after antipseudomonal treatment of intensive care unit patients. Antimicrob. Agents Chemother. 543:1213-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguchi, H., M. Matsuhashi, M. Takaoka, and S. Mitsuhashi. 1978. New antipseudomonal penicillin, PC-904: affinity to penicillin-binding proteins and inhibition of the enzyme cross-linking peptidoglycan. Antimicrob. Agents Chemother. 14:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh, H., J. Stenhoff, S. Jalal, and B. Wretlind. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb. Drug Resist. 9:323-328. [DOI] [PubMed] [Google Scholar]
- 23.Pucci, M. J., J. Boice-Sowek, R. E. Kessler, and T. J. Dougherty. 1991. Comparison of cefepime, cefpirome, and cefaclidine binding affinities for penicillin-binding proteins in Escherichia coli K-12 and Pseudomonas aeruginosa SC8329. Antimicrob. Agents Chemother. 35:2312-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riera, E., M. D. Macià, A. Mena, X. Mulet, J. L. Pérez, Y. Ge, and A. Oliver. 2010. Anti-biofilm and resistance suppression activities of CXA-101 against chronic respiratory infection phenotypes of Pseudomonas aeruginosa strain PAO1. J. Antimicrob. Chemother. 65:1399-1404. [DOI] [PubMed] [Google Scholar]
- 25.Sauvage, E., F. Kerff, M. Terrak, J. A. Ayala, and P. Charlier. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234-258. [DOI] [PubMed] [Google Scholar]
- 26.Sifaoui, F., M. D. Kitzi, and L. Gutmann. 1996. In vitro selection of one-step mutants of Streptococcus pneumoniae resistant to different oral β-lactam antibiotics is associated with alterations of PBP2x. Antimicrob. Agents Chemother. 40:152-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spratt, B. G., and K. D. Cromie. 1988. Penicillin-binding proteins of gram-negative bacteria. Rev. Infect. Dis. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 28.Takeda, S., T. Nakai, Y. Wakai, F. Ikeda, and K. Hatano. 2007. In vitro and In vivo activities of a new cephalosporin, FR264205, against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 51:826-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda, S., Y. Ishii, K. Hatano, K. Tateda, and K. Yamaguchi. 2007. Stability of FR264205 against AmpC β-lactamase of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 30:443-445. [DOI] [PubMed] [Google Scholar]
- 30.Trautmann, M., M. Heinemann, R. Zick, A. Möricke, M. Seidelmann, and D. Berger. 1998. Antibacterial activity of meropenem against Pseudomonas aeruginosa, including antibiotic-induced morphological changes and endotoxin-liberating effects. Eur. J. Clin. Microbiol. Infect. Dis. 17:754-760. [DOI] [PubMed] [Google Scholar]
- 31.Zamorano, L., C. Juan, A. Fernández-Olmos, Y. Ge, R. Cantón, and A. Oliver. 16 February 2010, posting date. Activity of the new cephalosporin CXA-101 (FR264205) against Pseudomonas aeruginosa isolates from chronically infected cystic fibrosis patients. Clin. Microbiol. Infect. doi: 10.1111/j.1469-0691.2010.03130.x. [DOI] [PubMed]
- 32.Zhao, G., T. I. Meier, S. D. Kahi, K. R. Gee, and L. C. Blaszczak. 1999. BOCILLIN FL, a sensitive and commercially available reagent for the detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]