Abstract
Brequinar is an inhibitor of dihydroorotate dehydrogenase, an enzyme that is required for de novo pyrimidine biosynthesis. Here we report that brequinar has activity against a broad spectrum of viruses. The compound not only inhibits flaviviruses (dengue virus, West Nile virus, yellow fever virus, and Powassan virus) but also suppresses a plus-strand RNA alphavirus (Western equine encephalitis virus) and a negative-strand RNA rhabdovirus (vesicular stomatitis virus). Using dengue virus serotype 2 (DENV-2) as a model, we found that brequinar suppressed the viral infection cycle mainly at the step of RNA synthesis. Supplementing the culture medium with pyrimidines (cytidine or uridine) but not purines (adenine or guanine) could be used to reverse the inhibitory effect of the compound. Continuous culturing of DENV-2 in the presence of brequinar generated viruses that were partially resistant to the inhibitor. Sequencing of the resistant viruses revealed two amino acid mutations: one mutation (M260V) located at a helix in the domain II of the viral envelope protein and another mutation (E802Q) located at the priming loop of the nonstructural protein 5 (NS5) polymerase domain. Functional analysis of the mutations suggests that the NS5 mutation exerts resistance through enhancement of polymerase activity. The envelope protein mutation reduced the efficiency of virion assembly/release; however, the mutant virus became less sensitive to brequinar inhibition at the step of virion assembly/release. Taken together, the results indicate that (i) brequinar blocks DENV RNA synthesis through depletion of intracellular pyrimidine pools and (ii) the compound may also exert its antiviral activity through inhibition of virion assembly/release.
Dengue, a mosquito-borne viral disease in humans, is caused by dengue virus (DENV). DENV causes disease globally, with an estimated 50 million to 100 million human infections, 500,000 hospitalizations, and 24,000 deaths per year (8). Besides DENV, many other flaviviruses, such as West Nile virus (WNV), yellow fever virus (YFV), Japanese Encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV), also cause significant human diseases (8). No effective antiflavivirus therapy is currently available. Development of DENV vaccines has been challenging because of the need to simultaneously immunize and induce long-lasting protection against all four serotypes of DENV; an individual with incomplete immunization may be sensitized to dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS), a potentially life-threatening disease (35). Therefore, development of safe and effective therapeutics for infections caused by DENV and other flaviviruses is urgently needed.
DENV belongs to the genus Flavivirus in the family Flaviviridae. The viral genome is a single-stranded, positive-sense RNA of about 11 kb in length. The genomic RNA contains a 5′ untranslated region (UTR), a single open reading frame, and a 3′ UTR. The single open reading frame encodes a polyprotein precursor that is cotranslationally processed by host and viral proteases into three structural proteins (the capsid [C], premembrane [PrM], and envelope [Env] proteins) and seven nonstructural proteins (NSs) (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (15). The structural proteins form viral particles and mediate virus entry, and the nonstructural proteins act as components of replication complexes. NS3 functions as a protease with its cofactor (NS2B), a nucleotide triphosphatase, an RNA triphosphatase, and a helicase (6, 33, 34). NS5 exhibits a methlytransferase and an RNA-dependent RNA polymerase (RdRp) (1, 5, 28, 31). Besides participating in viral replication, the NSs also function in virion assembly (13, 18) and invasion of the innate immune response (9, 17, 21, 22).
Both viral and host proteins could be targeted for antiviral development (24). Since viruses rely on host cells for the supply of nucleosides for replication, enzymes involved in nucleoside metabolic pathways are potential cellular targets for antiviral development. Ribavirin, a guanosine analogue that has been used for treatment of hepatitis C virus (HCV) infection, inhibits cellular IMP dehydrogenase (IMPDH), an enzyme that is essential for the de novo biosynthesis of guanine nucleotides (7). The compound exhibits activity against a broad spectrum of viruses, including flaviviruses (11). Depletion of intracellular GTP pools was shown to be the primary antiviral mechanism of ribavirin (14), although the compound could also function as a mutagen (4) or an inhibitor of RNA cap methylation (3). Along a similar line, brequinar (BQR) was reported to inhibit the replications of YFV, Kunjin virus, and DENV in human and monkey cells (10). This compound was discovered in the mid-1980s at DuPont as an antimetabolite in cancer and immunosuppression therapies (2). It exerts its primary antiproliferative activity by inhibiting dihydroorotate dehydrogenase (DHODH), the fourth enzyme in the de novo pyrimidine biosynthesis pathway (16, 19). The inhibition of DHODH prevents the synthesis of DNA and RNA and therefore interferes with rapidly proliferating lymphocytes, which rely heavily upon de novo nucleotide synthesis. Because viral replication relies on nucleotides inside cells, the observed antiviral activity of BQR could be due to its effect on lowering the intracellular level of pyrimidine. However, experimental evidence to support this mechanism of inhibition is lacking.
In this paper, we characterize the antiviral properties of BQR against DENV. We showed that BQR has activity against a broad spectrum of viruses in vitro. A mode-of-action study showed that the compound reduced the level of viral RNA synthesis, and the inhibitory effect could be reversed by addition of exogenous uridine or cytidine. Mutant DENV serotype 2 (DENV-2) resistant to BQR could be selected in cell culture. Mutagenesis analyses using an infectious cDNA clone and a replicon of DENV-2 demonstrated that a mutation in the envelope protein or NS5 gene confers BQR resistance. Mechanistically, we found that the envelope protein mutation renders the mutant virus less susceptible to BQR inhibition at the step of virion assembly/release, whereas the NS5 mutation confers resistance through enhancement of viral RNA synthesis.
MATERIALS AND METHODS
Viruses, compound, antibodies, and cell culture media.
We used the following viruses: WNV (strain 3356), YFV (17D vaccine strain), DENV-2 (strains New Guinea C and TSV01), Powassan virus (PWV; strain 64-7062), Western equine encephalitis virus (WEEV; strain Cova 746), and vesicular stomatitis virus (VSV; New Jersey serotype). The sources of these viruses were reported previously (37). BQR was synthesized in-house and was dissolved in 90% dimethyl sulfoxide (DMSO) for antiviral experiments. DENV-specific monoclonal antibody 4G2 against Env protein (27) was prepared from a hybridoma cell line purchased from the American Type Culture Collection (ATCC; Manassas, VA), and mouse monoclonal antitubulin was purchased from Sigma. Vero cells were cultured in Dulbecco modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS) and infected with virus or virus-like particles (VLPs) in the presence of DMEM plus 2% FBS. A549 cells (human alveolar basal epithelial cells) were maintained in F-12 medium plus 10% FBS, and the infections were performed in F-12 medium plus 2% FBS. C6/36 cells (C6/36 cells are derived from the mosquito, the transmission vector of DENV) were cultured in RPMI 1640 medium with 10% FBS, and virus infection was performed in RPMI 1640 medium with 5% FBS.
CFI assay.
A cell-based flavivirus immunodetection (CFI) assay was performed as described previously (32). Briefly, 2 × 104 of A549 cells per well in a 96-well plate were infected with DENV-2 (multiplicity of infection [MOI], 0.3) in the presence of 2-fold serial dilutions of BQR. After incubation at 37°C for 48 h, virus antigen Env-protein production was quantified by immunodetection using the 4G2 antibody and goat anti-mouse IgG conjugated with horseradish peroxidase as primary and secondary antibodies, respectively. The concentration of BQR that decreased the level of Env-protein production by 50% (the 50% effective concentration [EC50]) was calculated by nonlinear regression analysis.
Viral titer reduction assay.
Vero cells were seeded in a 12-well plate (4 × 105 cells per well). At 24 h postseeding, the cells were infected with WNV, DENV-2, YFV, WEEV, PWV, or VSV at an MOI of 0.1. For VSV, samples of culture medium were collected at 16 h postinfection (p.i.). For the other four viruses, samples were collected at 42 h postinfection. All viral titers were determined by a double-layer plaque assay on Vero cells, as described previously (25).
3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS), assay.
Cell viability was measured using the CellTiter 96 aqueous one solution cell proliferation assay (Promega, Madison, WI), according to the manufacturer's protocol. Approximately 1 × 104 Vero or A549 cells in 100 μl medium were seeded in a 96-well plate. After 16 h of incubation, the cells were treated with BQR. After another 48 h of incubation, 20 μl of CellTiter 96 solution was added to 100 μl of medium. After 2 h of incubation at 37°C with 5% CO2, the absorbance was measured at 490 nm in a Tecan microplate reader.
Time-of-addition analysis.
Approximately 2 × 105 Vero and A549 cells were seeded in each well of a 24-well plate. At 24 h postseeding, the cells were infected with DENV-2 at an MOI of 2 for 1 h at 4°C, followed by three rounds of phosphate-buffered saline (PBS) washes to remove unabsorbed virus. At different time points postinfection, BQR (5 μM) was added to the infected cells. Culture medium was collected at 24 h p.i. and assayed for viral titers using plaque assays (1). As negative controls, DMSO was added to the infected cells at a final concentration of 0.9% at 0, 10, and 20 h p.i. to estimate its effect on viral production.
Transient replicon assay.
Luciferase-reporting replicon RNA (10 μg) of strain TSV01 was electroporated into 8 × 106 A549 cells (25 μF and 450 V with three pulsings at 3-s intervals). The cells were seeded in a 12-well plate (4 × 105 per well) and immediately treated with 5 μM BQR, 5 μM BQR plus 20 μM uridine, or 0.9% DMSO as a control. At various time points, the cells were washed once with cold PBS and added to 200 μl of 1× lysis buffer (Promega). The plates containing the lysis buffer were sealed with Parafilm and stored at −80°C. Once samples had been collected for all time points, 20 μl of the cell lysates was transferred to a 96-well plate and assayed for luciferase signals in a Clarity luminescence microplate reader (BioTek, Winooski, VT).
Plasmid construction.
The DENV-2 genome-length cDNA clones with specific mutations were constructed by using an infectious cDNA clone of strain TSV01 (pFLTSV01) and two shuttle vectors. Shuttle vector A was constructed by engineering the SacII-XhoI fragment from pFLTSV01 (representing the upstream end of the T7 promoter [for the RNA transcription of genome-length RNA] to nucleotide position 5426 of the viral genome) into the pACYC177 vector (New England BioLabs, Ipswich, MA). A QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to engineer the envelope protein mutation into shuttle vector A. The mutated DNA fragments were pasted back into the pFLTSV01 plasmid at the SacII site (located in the plasmid vector) and BsrGI site (located at nucleotide position 1840 of the viral genome). Shuttle vector B was constructed by engineering an XhoI-ClaI fragment (representing nucleotide position 5426 through the 3′ end of the genome) into a TA cloning vector (Invitrogen, Carlsbad, CA). The NS5 mutation was engineered into shuttle vector B using a QuikChange II XL site-directed mutagenesis kit. The mutant NS5 DNA fragment was pasted back into the pFLTSV01 plasmid at the NruI (nucleotide position 7737) and ClaI sites. For construction of the NS5 mutant replicon, the mutant NS5 fragment from shuttle vector B was digested with XhoI (nucleotide position 5426) and ClaI; the resulting DNA fragment was ligated into a TSV01 replicon cDNA plasmid (predigested with the same restriction enzymes). For VLP packaging experiments, the complete CprMEnv fragment containing the envelope protein mutation was PCR amplified using the mutant genome-length cDNA plasmid described above as a template; the resulting fragment was then cloned into an alphavirus Semliki Forest virus (SFV) expression vector (pSFV1; Invitrogen, Carlsbad, CA) (25) at a unique XmaI site, resulting in plasmid SFV-CprMEnv. All cDNA constructs were verified by DNA sequencing.
In vitro transcription and RNA transfection.
Both genome-length and replicon RNAs of DENV-2 were transcribed in vitro from the corresponding cDNA plasmids that were linearized with ClaI. A T7 mMessage mMachine kit (Ambion, Austin, TX) was used for RNA synthesis, as described previously (30). The SFV-CprMEnv RNA was transcribed in vitro from a linear DNA (predigested with restriction enzyme SapI) using an SP6 mMessage mMachine kit (Ambion). The RNA transcripts were electroporated into BHK-21 cells as described previously (30). After the transfection of genome-length RNA, the cells were incubated at 37°C for the first 24 h, followed by incubation at 30°C. Culture fluids were collected every 24 h until a cytopathic effect was observed from days 5 to 6 posttransfection (p.t.). The culture medium containing viruses was aliquoted and stored at −80°C.
VLP assay.
DENV-2 VLPs were prepared by the in trans supply of viral structural proteins to replicon RNAs (26). Briefly, 8 × 106 BHK-21 cells were electroporated with 10 μg of DENV-2 luciferase replicon RNA (Rluc2ARep) in a 0.4-cm cuvette with the GenePulser apparatus (Bio-Rad, Hercules, CA) using settings of 0.85 kV and 25 μF and with three pulsings at 3-s intervals. The transfected cells were resuspended in DMEM with 10% FBS and incubated at 37°C for 24 h. The cells were again electroporated with 10 μg of SFV-CprMEnv RNA at settings identical to those used for the first transfection. After the second electroporation, the cells were incubated at 30°C. At 48 h and 72 h after replicon RNA input, the culture supernatants were centrifuged to remove cell debris. The supernatants containing VLPs were aliquoted and stored at −80°C. The VLPs were quantified by measuring the replicon RNA copy numbers using real-time reverse transcription-PCR (RT-PCR). Specifically, the replicon RNA was extracted from 140 μl VLPs and dissolved in 50 μl RNase-free water; 5 μl of resuspended RNA extract was used to quantify the replicon RNA copies by real-time RT-PCR. An iScriptTM One-Step RT-PCR kit with SYBR green (Bio-Rad) was used to quantify the viral RNA using primers targeting the NS5 region (primers 5′-GAAGGAGAAGGGCTGCACAAAC-3′ and 5′-GCACACGCACCACCTTGTTTTG-3′), following the manufacturer's protocol. For VLP infection assays, a monolayer of Vero cells (4 × 104 cells per well in a 96-well plate) was infected with VLPs. At the indicated time points, the cells were washed once with cold PBS, lysed in 20 μl of 1× Renilla luciferase lysis buffer for 20 min, and assayed for luciferase activities using a Renilla luciferase assay kit (Promega). The luciferase signals were measured using a Clarity luminescence microplate reader (BioTek).
Viral growth kinetics and plaque assay.
Vero cells (4 × 105 cells per well) and C6/36 cells (8 × 105 cells per well) were seeded in a 12-well plate. At 24 h postseeding, the cells were infected with wild-type (WT) and mutant DENV-2 at an MOI of 0.1. Culture medium was collected every 24 h until 96 h. Viral titers were quantified using a plaque assay on BHK-21 cells.
Generation and sequencing of DENV-2 resistant to BQR.
BQR-resistant DENV-2 was generated by passaging the DENV-2 on Vero cells in the presence of BQR. For each round of passaging, Vero cells (2 × 105 per well) in 24-well plates were infected with 100 μl of DENV-2 (derived from the previous passaging, the first round of infection at an MOI of 0.1) in the presence of increasing concentrations of BQR or 0.9% DMSO (a negative control). Passage 1 (P1) to passage 6 were selected with 0.5 μM BQR, passages 7 to 12 were selected with 1.0 μM BQR, and passages 13 to 21 were selected with 2.0 μM BQR. For each passage, viral supernatants were harvested at 72 h p.i., viral titers were quantified by plaque assays, and viruses were analyzed in resistance assays to monitor the improvement of resistance. The selections were terminated at passage 21, when no further improvement of the resistance was observed (see details in Results). Viral RNAs from passage 21 were extracted from the culture supernatants using a QiAamp viral RNA minikit (Qiagen, Valencia, CA). The RNAs were subjected to amplification using SuperScript III one-step RT-PCR kits (Invitrogen). The PCR products were gel purified and sequenced using the Sanger method.
RESULTS
Antiviral activity of BQR.
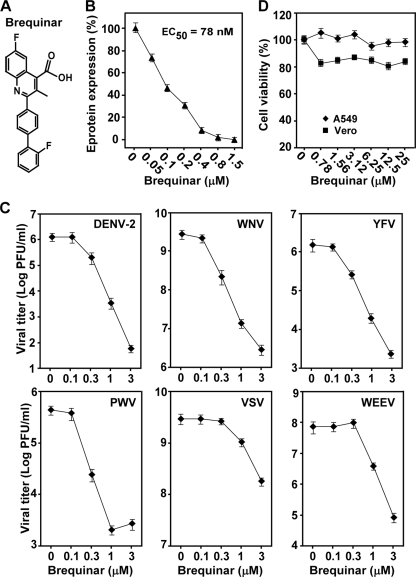
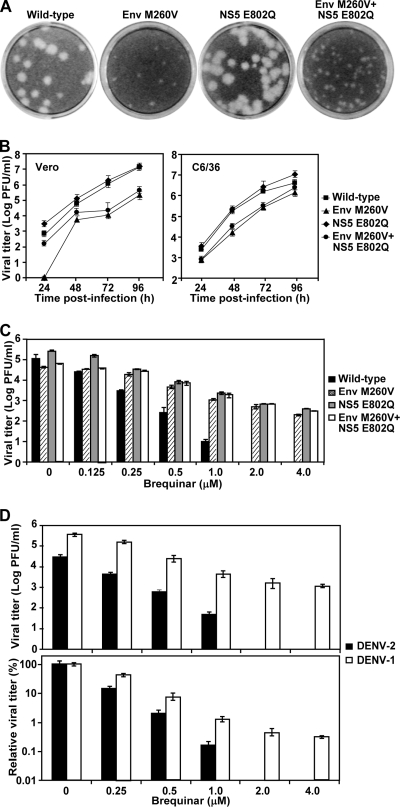
To examine the antiviral activity of BQR (Fig. 1A), we initially performed a CFI assay with A549 cells (Fig. 1B) and a viral titer reduction assay with Vero cells using DENV-2 (Fig. 1C). The CFI assay quantified the expression of viral envelope protein in infected cells (see details in Materials and Methods). Treatment of cells with BQR reduced the level of envelope protein production and the viral titer in a dose-dependent manner (Fig. 1B and C, respectively). The EC50 of BQR was calculated to be 78 nM in the CFI assay. Treatment with 3 μM compound reduced the viral titers >4 log units. Next, we examined the spectrum of activity of BQR against other flaviviruses (YFV, WNV, and PWV), a plus-strand RNA alphavirus (Western equine encephalitis virus), and a negative-strand RNA rhabdovirus (vesicular stomatitis virus). As shown in Fig. 1C, BQR potently inhibited all viruses tested. Cytotoxicity assays using both Vero and A549 cells showed that the observed antiviral activities were not due to compound-mediated toxicity (Fig. 1D). These results demonstrate that BQR has activity against a broad spectrum of viruses.
FIG. 1.
Cytotoxicity and spectrum of BQR antiviral activity. (A) Structure of BQR. (B) CFI assay. A549 cells were infected with DENV-2 (MOI, 0.3) in the presence of 2-fold serial dilutions of BQR. After incubation at 37°C for 48 h, the expression of viral envelope protein was quantified by an immunodetection method (see Materials and Methods). Average results from three experiments are shown. (C) Spectrum of BQR antiviral activity. Vero cells were infected with the indicated viruses at an MOI of 0.1; the infected cells were immediately treated with BQR. For DENV-2, WNV, YFV, PWV, and WEEV, culture media were collected at 42 h p.i. and viral titers were measured using plaque assays. For VSV, culture medium was collected at 16 h p.i. and the viral titer was measured. Average results and standard deviations (n = 3) are presented. (D) Cytotoxicity of BQR in Vero and A549 cells. Cytotoxicity was examined by incubation of Vero and A549 cells with the indicated concentrations of BQR. Cell viability was measured by an MTS assay and is presented as a percentage of the colorimetric absorbance derived from the compound-treated cells compared with that derived from the mock-treated (0.9% DMSO) cells. Average results from three experiments are shown.
BQR inhibits viral RNA synthesis.
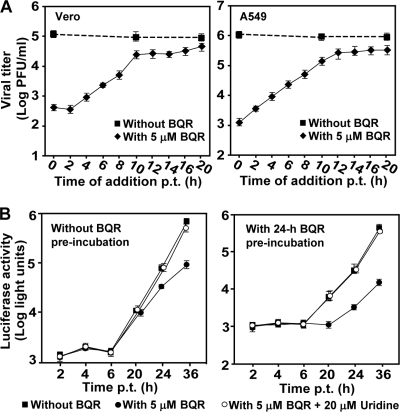
Since viral replication relies on intracellular nucleosides, inhibition of DHODH activity by BQR is expected to block viral RNA synthesis. A time-of-addition experiment was performed to test this hypothesis. As shown in Fig. 2A, Vero and A549 cells were synchronously infected with DENV-2 (MOI, 2). BQR (5 μM) was added to the infected cells at various time points after infection. Virus production was determined at 24 h postinfection. As a control, 0.9% DMSO was added to the infected cells at 0, 10, and 20 h p.i. for estimation of its effect on virus yield. The results showed that the inhibition of BQR on virus production gradually diminished when the compound was added at time points later than 10 h p.i., indicating that the compound blocks a late stage(s) of the viral life cycle (RNA synthesis or virion assembly/release). Similar results were obtained from both Vero and A549 cells, indicating that the observation is not cell type specific.
FIG. 2.
Mechanism of BQR-mediated inhibition of DENV-2. (A) Time-of-addition analysis of BQR in DENV-2 infection. Vero and A549 cells were infected with DENV-2 at an MOI of 2 at 4°C for 1 h. The infected cells were washed three times with PBS. BQR (5 μM) was then added to the cells at the indicated time points postinfection. The supernatants were assayed for determination of viral titers at 24 h postinfection. As controls, 0.9% DMSO was added to the infected cells at 0, 10, and 20 h p.i. for estimation of its effect on viral production. (B) Transient replicon assay. (Left panel) A luciferase-reporting replicon (10 μg) was electroporated into A549 cells. The transfected cells were immediately incubated with 5 μM BQR or 0.9% DMSO (as controls) and the luciferase activities were measured at the indicated time points. (Right panel) A549 cells were preincubated with 5 μM BQR for 24 h, after which the replicon (10 μg) was electroporated into these cells. The transfected cells were then incubated with medium containing 5 μM BQR or 0.9% DMSO, and the luciferase signals were measured at the indicated time points. For analyzing the effect of pyrimidine on the antiviral activity of BQR, 5 μM BQR and 20 μM uridine were added to the transfected cells; luciferase activities were measured at the indicated time points. Error bars indicate the standard deviations from three independent experiments.
We then used a luciferase-reporting replicon of DENV-2 to examine the effect of BQR on viral RNA synthesis. In the first set of experiments, we treated A549 cells with BQR (5 μM) immediately after the replicon was transfected into cells. As shown in Fig. 2B (left panel), the compound did not affect the luciferase signals at 2 to 6 h p.t. (representing translation of input replicon RNA), whereas the luciferase activities at 24 h and 36 h p.t. (representing viral RNA synthesis) were reduced by 52% and 85%, respectively. In the second set of experiments, we preincubated A549 cells with 5 μM BQR for 24 h (to deplete the preexisting pyrimidine pool). The preincubation step was done on the basis of a previous result that treatment with BQR at 1 μM for 24 h could deplete 60% of the UTP pools in human T cells (29). After the 24-h preincubation, the cells were electroporated with the DENV-2 replicon, continuously treated with 5 μM BQR or 0.9% DMSO (as a control), and assayed for luciferase at various time points. As shown in Fig. 2B (right panel), no suppression of luciferase signals was detected at 2 to 6 h p.t.; in contrast, the luciferase signals at 20 to 36 h p.t. were decreased by >85%. Taken together, these results demonstrated that BQR acts at the RNA synthesis step.
Pyrimidine reverses the antiviral effect of BQR.
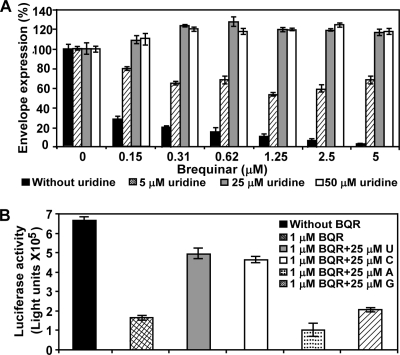
BQR has been shown to inhibit DHODH activity and thereby block the de novo pyrimidine biosynthesis. We asked whether supplementing cytidine or uridine in the culture medium would allow pyrimidine synthesis to proceed via the salvage pathway and thus counteract the inhibitory effect of BQR. In the BQR-treated and DENV-infected A549 cells, addition of uridine to the culture medium rescued viral replication in a dose-responsive manner (Fig. 3A). Supplementing with 5 μM uridine partially restored viral replication; supplementing with 25 or 50 μM uridine completely rescued viral replication. In agreement with the viral infection results, full restoration of replicon replication was also observed when 20 μM uridine was added to the BQR-treated and replicon-transfected cells (Fig. 2B).
FIG. 3.
Pyrimidine reverses the antiviral effect of BQR. (A) Uridine reverses the antiviral effect of BQR in the CFI assay. A549 cells were infected with DENV-2 and treated with different doses of BQR and uridine. At 48 h p.i., viral envelope protein was quantified by the CFI assay. (B) Specificities of pyrimidines to reverse the antiviral effect of BQR. Vero cells (4 × 104 cells/well in a 96-well plate) were infected with DENV-2 VLPs (5 × 105 focus-forming units). The cells were then treated with 1 μM BQR in the presence or absence of 25 μM uridine (U), cytidine (C), adenosine (A), or guanosine (G). Luciferase activities were assayed at 48 h postinfection. Error bars indicate the standard deviations from three independent experiments.
Next, we examined the specificity of the rescuing effect by all four nucleosides. DENV-2 VLPs (prepared by in trans packaging a luciferase-reporting replicon with viral structural proteins) were used to infect Vero cells in the presence of BQR (1 μM) plus U, C, A, or G (25 μM). As expected, treatment of VLP-infected cells with BQR reduced the luciferase signals (Fig. 3B; compare the first two bars). Supplementing with U or C but not A or G significantly rescued lucifrerase activities (Fig. 3B; compare the third and fourth bars with the first bar). These results clearly demonstrate that pyrimidine could reverse the antiviral activity of BQR. The results also suggest that BQR exerts its antiviral activity by reducing the pyrimidine nucleotide pools.
Selection of BQR-resistant DENV-2.
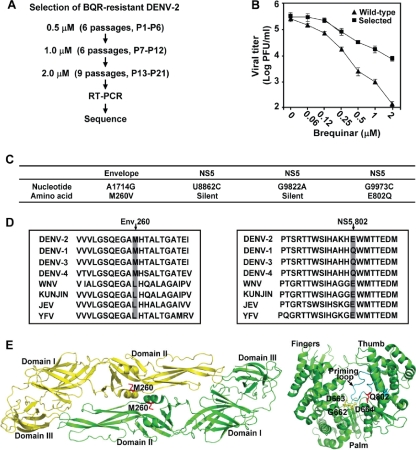
BQR-resistant viruses were selected by culturing WT DENV-2 in the presence of increasing concentrations of BQR for 21 rounds (Fig. 4A). Viruses from P21 were assayed for the resistance phenotype by comparison of the viral titers from the mock-treated infections (containing 0.9% DMSO; the compound-treated infections also contained 0.9% DMSO) with the viral titers from the BQR-treated infections. Compared with the WT, the P21 virus was partially resistant to BQR (Fig. 4B). When it was treated with 2 μM BQR, the P21 virus generated titers that were about 100-fold higher than those of the WT virus. Further passaging of the P21 viruses did not improve the resistance phenotype (data not shown). These results demonstrate that BQR-resistant viruses can be selected in cell culture.
FIG. 4.
Selection and sequencing of BQR-resistant DENV-2. (A) Scheme for selection of BQR-resistant DENV-2. P1 through P6 were selected at 0.5 μM BQR, P7 through P12 were selected at 1.0 μM, and P13 through P21 were selected at 2.0 μM. (B) Resistance analysis. Vero cells were infected with wild-type or P21 viruses (MOI, 0.1) in the presence of BQR or 0.9% DMSO (as a negative control). At 48 h p.i., the viral titers in culture fluids were quantified by plaque assays. Resistance is quantified by comparison of the viral titers from the BQR-treated infections with the viral titers from the DMSO-treated infections. Error bars indicate the standard deviations from three independent experiments. (C) Mutations recovered from the P21 resistant virus. The locations of the nucleotide and amino acid changes are indicated. (D) Amino acid sequence alignment. (Left panel) Sequence alignment of a flavivirus envelope protein region; (right panel) sequence alignment of a flavivirus RdRp region. The sequences of DENV-1, DENV-2, DENV-3, DENV-4, WNV, KUNV, JEV, and YFV are derived from the sequences with GenBank accession numbers U88535, M29095, M93130, AY947539, AF404756, D00246, AF315119, and X03700, respectively. (E) Locations of resistance mutations on crystal structures of DENV-2 envelope protein and DENV-3 RdRp. (Left panel) DENV-2 envelope protein structure (Protein Data Bank code 1OKE [20]). The two envelope protein subunits are in yellow and green; the mutated residue M260 is labeled in red. (Right panel) DENV-3 RdRp structure (Protein Data Bank code 2HFZ [36]). The fingers, palm, and thumb subdomains are indicated; the GDD active site of RdRp is shown in yellow; the priming loop is colored in cyan; the mutated residue Q802 is shown in red. The images were prepared using the PyMol program.
We sequenced the complete genome of the P21 resistant virus. Four nucleotide changes were identified (Fig. 4C): (i) an A → G change at nucleotide position 1714, resulting in an amino acid substitution from Met to Val at position 260 (M260V) in the envelope protein; (ii) two silent mutations in NS5, a U → C substitution at nucleotide position 8862 and a G → A substitution at nucleotide position 9822; and (iii) a G → C change at position 9973, resulting in an amino acid substitution from Glu to Gln at position 802 (E802Q) in the NS5 RdRp domain. We also plaque purified three isolates from the P21 virus pool; sequencing of individual isolates revealed the same mutations described above (data not shown). As a negative control, viruses cultured in 0.9% DMSO for 21 passages did not exhibit these mutations. Sequence alignment showed that M260 is conserved among the four serotypes of the DENV envelope protein, whereas the envelope proteins of WNV, Kunjin virus, JEV, and YFV have a Leu residue at this position (Fig. 4D, left panel). On the crystal structure of the DENV-2 envelope protein, M260 is located at a helix from domain II (Fig. 4E, left panel) (20). For the NS5 E802Q mutation, the WT E802 is shared by DENV-2, WNV, Kunjin virus, JEV, and YFV NS5, whereas mutated residue Q802 is found in NS5 from DENV-1, -3, and -4 (Fig. 4D, right panel). Residue Q802 is located at the priming loop of the DENV-3 RdRp (Fig. 4E, right panel) (36).
Envelope or NS5 mutation alone is sufficient to confer resistance.
An infectious cDNA clone of DENV-2 was used to prepare three recombinant viruses: the envelope (Env) protein M260V mutation (Env M260V), NS5 E802Q, and Env M260V-NS5 E802Q. The recombinant viruses were sequenced to verify the engineered mutations with no other changes (data not shown). Plaque assays showed that the mutant viruses had different plaque morphologies: the NS5 E802Q plaque was similar to the WT virus plaque, whereas the Env M260V and Env M260V-NS5 E802Q plaques were smaller than the WT virus plaque (Fig. 5A). Analysis of the growth kinetics showed that in both Vero and C6/36 cells, the NS5 E802Q virus replicated slightly faster than the WT virus, and the Env M260V and the Env M260V-NS5 E802Q viruses replicated more slowly than the WT virus (Fig. 5B).
FIG. 5.
Analysis of resistance mutations. (A) Plaque morphologies of WT, Env M260V, NS5 E802Q, and Env M260V-NS5 E802Q viruses. Plaques were developed in the absence of BQR. (B) Growth kinetics of WT and mutant viruses. Vero and C6/36 cells were infected with the WT and mutant viruses at an MOI of 0.1. After 1 h of incubation, the cells were washed three times with PBS; the medium was then replenished. Viral titers in culture fluids were quantified at the indicated time points using plaque assays. (C) Resistance analyses of WT mutant viruses. The resistance assays were performed as described in the legend to Fig. 4B. (D) Resistance analyses of WT DENV-1 and DENV-2. Vero cells were infected with WT DENV-1 and DENV-2 at an MOI of 0.1 in the presence of BQR. Viral titers in culture fluids were quantified by plaque assays at 48 h postinfection. (Top panel) Viral titers; (bottom panel) relative viral titers (relative viral titers are the viral titers from the BQR-treated infections/viral titers from the mock-treated infections). Error bars indicate the standard deviations from three independent experiments.
Resistance assays showed that after treatment of infected cells (MOI, 0.5) with BQR, all three mutant viruses yielded viral titers that were higher than the titers derived from the WT virus (Fig. 5C). At a 2.0 μM concentration, the compound reduced the WT virus titer by approximately 5 log units. In contrast, the same concentration of BQR reduced the three mutant virus titers by about 2 to 2.5 log units. Notably, the double mutant viruses (Env M260V-NS5 E802Q) did not exhibit more resistance than the single mutant viruses (Env M260V or NS5 E802Q) (Fig. 5C). These results demonstrate that (i) the Env protein or NS5 mutation alone is sufficient to confer resistance and (ii) the Env protein and NS5 mutations do not have an additive effect on further improvement of resistance.
As described above, the mutated NS5 802Q residue (selected from DENV-2) is the wild-type sequence in DENV-1, -3, and 4 (Fig. 4D). If the Gln residue at position 802 is sufficient for resistance, WT DENV-1, -3, and -4 would be less sensitive to BQR inhibition than DENV-2. We tested this idea by measuring the sensitivity of DENV-1 to BQR. As shown in Fig. 5D, DENV-1 (with Q802 in RdRp) was more resistant to BQR than DENV-2 (with E802 in RdRp). Notably, in the absence of compound, DENV-1 replicated to a titer that was about 10-fold higher than that of DENV-2. These results further confirm that the Gln residue at position 802 of RdRp is responsible for the BQR resistance.
NS5 E802Q mutation confers resistance through enhancement of viral RNA synthesis.
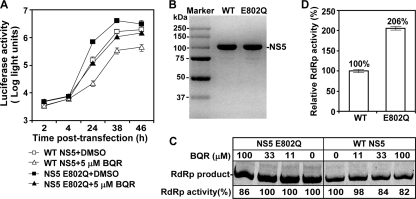
To examine the mechanism of NS5 E802Q in resistance, we engineered the mutation into a luciferase-reporting replicon of DENV-2. The replicon system allowed us to analyze its effect on viral replication without complications from virion assembly/release. After transfection of A549 cells with equal amounts of WT and NS5 mutant replicons, equivalent levels of luciferase signals were observed at 2 to 4 h p.t., indicating that the mutation does not affect viral translation; the replicon RNAs were also observed to have equal transfection efficiencies (Fig. 6A). At 24, 38, and 46 h p.t., the NS5 mutant replicon generated 4.6-, 2.5-, and 1.5-fold higher luciferase signals, respectively, than the WT replicon did. Treatment of the transfected cells with BQR (5 μM) suppressed the luciferase activities from both the WT and NS5 mutant replicons; however, the luciferase signals produced from the BQR-treated mutant replicon was similar to that generated from the mock-treated WT replicon (Fig. 6A), suggesting that the NS5 E802Q mutation confers resistance through enhancement of viral RNA synthesis.
FIG. 6.
Analysis of NS5 E802Q mutation. (A) Luciferase-reporting replicon analysis. A549 cells were preincubated with 5 μM BQR for 24 h, after which an equal amount of WT or NS5 E802Q replicon RNA (10 μg) was electroporated into the cells. The transfected cells were incubated with medium containing BQR (5 μM) or 0.9% DMSO (as a control). Luciferase activities were measured at the indicated time points posttransfection. Average results from three independent experiments are presented. (B) Recombinant proteins of full-length NS5. Full-length WT and mutant E802Q NS5 of DENV-2 (2.5 μg each) were analyzed by SDS-PAGE and stained with Instant Blue (Expedeon Ltd., United Kingdom). The proteins were expressed and purified as reported elsewhere (23). The molecular masses of protein markers are labeled. (C) Inhibition of de novo RNA synthesis by BQR. Subgenomic RNA of DENV-2 (containing the 5′-terminal 169 nucleotides directly connected to the 3′-terminal 462 nucleotides of the genome) was incubated with equal amounts of recombinant NS5 (0.25 μg of WT or E802Q) of DENV-2, as described previously (23). The reaction mixtures were incubated with the indicated concentrations of BQR. The RdRp products were analyzed on a 10% denaturing polyacrylamide gel with 7 M urea and quantified using a PhosphorImager. For both WT and E208Q NS5, the RdRp activity in the absence of BQR was set equal to 100%, and the remaining RdRp activities in the presence of BRQ are indicated. A representative result from three experiments is shown. (D) Enhancement of de novo RNA synthesis by the E802Q mutation. The RdRp products, derived from the WT and mutant NS5 in the absence of BQR, as shown in panel C, were quantified; the RdRp product from the WT NS5 is set equal to 100%. The average results of three experiments are presented, with error bars representing standard deviations.
To further demonstrate the function of the E802Q mutation, we prepared recombinant full-length NS5 proteins of DENV-2 with or without the E802Q mutation (Fig. 6B). An assay for de novo RdRp production was performed using equal amounts of WT and mutant NS5 proteins and a DENV-2 subgenomic RNA as the template. The reactions were analyzed on a denaturing polyacrylamide gel (Fig. 6C). Quantification of the RdRp products showed that the mutant NS5 was about 2-fold more active than the WT NS5 (Fig. 6D). Interestingly, increasing concentrations of BQR weakly inhibited the RdRp activities of both the WT NS5 and the mutant NS5; at 100 M BQR (the highest concentration tested), less than 20% of the RdRp activities were suppressed (Fig. 6C). It should be noted that the compound inhibited the WT and mutant NS5 activities at similar levels. Overall, these results demonstrate that the NS5 mutation exerts its resistance through enhancement of polymerase activity.
Env M260V mutation confers resistance at the step of virus assembly/release.
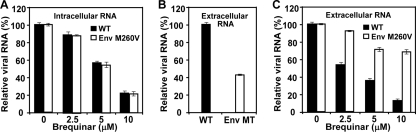
To explore the role of the Env M260V mutation in resistance, we transfected BHK-21 cells with equal amounts of WT and mutant Env protein genome-length RNAs. Real-time RT-PCR was used to quantify the amounts of intracellular and extracellular viral RNAs. At 12 and 16 h p.t., equivalent levels of intracellular viral RNA were detected for the WT and Env protein mutant (data not shown), indicating that the Env protein mutation does not affect RNA synthesis. Treatment of the transfected cells with BQR suppressed intracellular viral RNA levels to a similar extent between the WT and Env protein mutant (Fig. 7A). For the extracellular viral RNA, WT RNA-transfected cells released 2.4-fold more RNA than the Env protein mutant-transfected cells at 24 h p.t. (Fig. 7B), suggesting that the Env protein mutation attenuates virus assembly/release. Treatment of the transfected cells with BQR suppressed the extracellular RNA levels derived from both the WT and Env protein mutant RNAs (Fig. 7C). However, the Env protein mutant RNA was less sensitive to compound inhibition than the WT RNA; for example, 10 μM BQR reduced extracellular WT RNA levels by nearly 90%, whereas the same treatment reduced the Env protein mutant RNA levels by only 30%. These results indicate that the Env M260V mutation contributes to resistance at the step of virus assembly/release.
FIG. 7.
Effects of Env M260V on RNA synthesis, virion assembly/release, and BQR resistance. BHK-21 cells were transfected with WT or Env M260V genome-length RNA. The transfected cells were incubated with different concentrations of BQR. At 24 h p.t., the cells were washed three times with PBS and the intracellular viral RNA was quantified (using the host GAPDH RNA level for normalization). (A) Relative amounts of intracellular viral RNA, using the amount of viral RNA derived from the DMSO-treated sample set equal to 100%. At 24 h p.t., the supernatants were filtered through 0.2-μm-pores-size filters and the extracellular viral RNA was quantified. (B) Relative amounts of extracellular WT RNA (set equal to 100%) and Env protein mutant (MT) RNA without BQR treatment. (C) Relative amounts of extracellular viral RNA for BQR-treated samples; for either WT or Env protein mutant RNA, the amount of RNA derived from the mock-treated sample is set equal to 100%. Average results from three independent experiments are presented.
VLP analysis of NS5 and Env protein mutations.
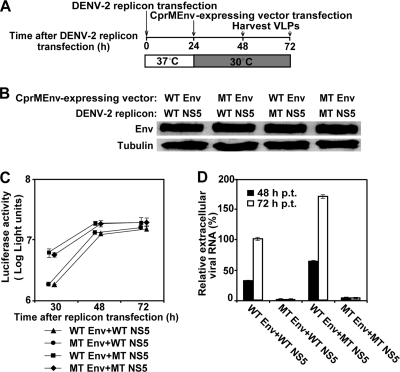
We used a VLP system of DENV-2 to further validate the roles of the mutations in Env protein and NS5 in viral replication and virion assembly/release. The VLPs were prepared by two sequential electroporations of a luciferase replicon of DENV-2 and a structural protein CprMEnv-expressing vector (Fig. 8A). Two types of replicons (WT and NS5 mutant) were individually packaged with two types of structure proteins (WT and mutant Env proteins), resulting in four possible VLPs (WT replicon plus WT Env protein, WT replicon plus mutant Env protein, mutant NS5 replicon plus WT Env protein, and mutant NS5 replicon plus mutant Env protein). Western blotting showed that both the WT and mutant Env proteins were equally expressed in the transfected cells (Fig. 8B). At 30 to 72 h p.t., the mutant NS5 replicon produced higher luciferase signals than the WT replicon; coexpression of the WT or mutant Env structural protein with NS5 did not affect the luciferase activities (Fig. 8C). Quantification of extracellular levels of replicon RNAs showed that more mutant NS5 replicon plus WT Env protein VLPs than WT replicon plus WT Env protein VLPs were produced; however, the packaging efficiencies of the WT replicon plus the Env protein and the NS5 MT replicon plus the mutant Env protein VLPs were reduced by >90%. Notably, the negative effect of the Env protein mutation on virion assembly/release was more dramatic in the VLP packaging system than in the genome-length RNA (compare Fig. 8D with Fig. 7B); the discrepancy is most likely due to the difference in expression of structural proteins in trans and in cis, respectively. Nevertheless, the VLP results confirmed that the NS5 mutation enhances viral RNA synthesis, whereas the Env protein mutation reduces virus assembly/release.
FIG. 8.
VLP analysis of NS5 and envelope protein mutations. (A) Scheme for DENV-2 VLP production. VLPs were prepared by sequential transfection of BHK-21 cells with a luciferase-reporting replicon and a vector expressing viral CprMEnv proteins. See Materials and Methods for details. (B) Western blotting of WT and mutant (MT) envelope proteins. The doubly transfected cells were lysed at 30 h after the first electroporation. The lysates were blotted for envelope proteins using mouse monoclonal 4G2 IgG and horseradish peroxidase-labeled anti-mouse IgG as primary and secondary antibodies, respectively; γ-tubulin was also blotted as a loading control. The combinations of WT and NS5 mutant replicons and WT and Env mutant structural proteins are indicated above the blotting image. (C) Kinetics of replicon replication in VLP packaging cells. Luciferase activities were measured from the doubly transfected VLP packaging cells at the indicated time points after the first replicon transfection. Average results from three experiments are shown. (D) Effects of envelope protein and NS5 mutations on VLP assembly/release. VLP production in culture fluid was measured by determination of the amount of extracellular viral RNA at 48 and 72 h after the first transfection. The relative amounts of RNA are shown; the amount of viral RNA derived from the VLP (collected at 72 h p.t.) containing the WT replicon and WT structural proteins was set equal to 100%. Average results from three independent experiments are presented.
DISCUSSION
The goal of this study is to characterize the anti-DENV activity of BQR and to analyze the resistance profile in cell culture. We found that BQR has activity against a broad spectrum of viruses. Besides flaviviruses, the compound also potently inhibited an alphavirus (Western equine encephalitis virus) and a rhabdovirus (vesicular stomatitis virus). The broad antiviral spectrum indicates that BQR inhibits a host pathway or a cellular factor that is required for the replication of these evolutionarily distant viruses. Mechanistic analysis showed that the inhibitory effect of BQR could be completely reversed by supplementing the culture medium with pyrimidine, whereas supplementing with purine did not rescue viral replication. These results are in agreement with previous results that BQR inhibits DHODH, an enzyme involved in the pyrimidine biosynthesis pathway (16, 19). These observations indicate that a host pathway shared by different viruses could be targeted for panantiviral development.
Antiviral approaches targeting host factors have to overcome two major hurdles. The first hurdle is to achieve a therapeutic window without causing toxicity. Compared with the approach that directly targets viral proteins, targeting host proteins or pathways is more likely to lead to toxicity. The second hurdle is the feasibility to suppress the host pathway sufficiently enough to reduce viral replication. Pharmacokinetic studies showed that BQR could reach a maximum concentration in plasma of 231 μM upon oral dosing of 25 mg/kg of body weight in mice (unpublished results). However, the compound was predicted (using GastroPlus ADMET Predictor software; SimulationsPlus, Inc.) to have high plasma protein binding activity of 99.06%, leading to a free BQR concentration of 2.17 μM. Since the compound has an EC50 of 78 nM in cell culture (Fig. 1B), it was expected to have in vivo efficacy in the mouse. To the contrary, BQR (orally dosed at 3 mg/kg twice per day) did not show any efficacy in the dengue virus AG129 mouse model (data not shown). In line with the in vivo efficacy results, the plasma concentration of uridine from the BQR-treated mice was found to be 6 μM (unpublished data). These results indicate that plasma uridine (derived either from the diet or from residual de novo pyrimidine synthesis) could preclude the efficacy of BQR in vivo.
BQR inhibits DENV replication through suppression of the pyrimidine biosynthesis pathway and depletion of intracellular pyrimidine pools. This mechanism of inhibition is supported by four lines of evidences in the current study. (i) Time-of-addition experiments showed that the compound suppressed at the late stage of DENV infection cycle. (ii) A transient reporting replicon assay demonstrated that BQR directly suppressed the luciferase signals that were representative of viral RNA synthesis. (iii) The compound-mediated inhibition of viral replication could be reversed by supplementing the culture medium with pyrimidine. (iv) DENV-2 partially resistant to BQR contained an adaptive mutation (E802Q) in the viral RdRp domain. Recombinant DENV-2 containing the NS5 E802Q mutation conferred the resistance phenotype. Interestingly, the wild-type amino acid at position 802 of the three other DENV serotypes is Q, suggesting that DENV-1, -3, and -4 would be partially resistant to BQR. Indeed, compared with the WT DENV-2, DENV-1 was less sensitive to BQR inhibition. In the context of DENV-2 genome-length RNA and replicon, the E802Q mutation appeared to confer resistance through enhancing viral RNA synthesis. Since residue 802 is located at the RdRp priming loop (which is assumed to be critical for de novo RNA synthesis) (36), the E802Q change in this region may affect the efficiency of RNA initiation or elongation. Biochemical experiments are ongoing to differentiate these possibilities.
Our results suggest that besides blocking viral RNA synthesis, BQR may also suppress virus assembly/release. Since flavivirus assembly is coupled to viral RNA synthesis (12), it is technically difficult to completely dissociate viral RNA synthesis from virion assembly. Nevertheless, the following results imply the effect of BQR on DENV-2 assembly/release. (i) Time-of-addition study results showed that no matter how late BQR was added, the viral titers from the compound-treated culture were always lower than the titers from the mock-treated culture (Fig. 2A). (ii) A mutation within the viral envelope protein (M260V) was sufficient to confer the resistance observed with resistant DENV-2. Although the envelope protein mutation reduced the efficiency of virion production, the efficiency of mutant virus assembly by BQR was suppressed much less than the efficiency of wild-type virus assembly. The envelope protein mutation is located at a helix of domain II. The underlying mechanism of this residue in modulating virion assembly/release and BQR inhibition remains to be determined.
In summary, this study showed that BQR, an inhibitor of the pyrimidine biosynthesis pathway, has activity against a broad spectrum of viruses in vitro. Although the compound is very potent (EC50, 78 nM) in cell culture, the efficacy remains to be achieved in vivo. Using DENV-2 as a model, we showed that BQR-resistant viruses could be selected in cell culture. The escape mutant viruses used two mechanisms to confer resistance: a mutation in NS5 RdRp to enhance viral RNA synthesis and a mutation in the viral envelope protein to become less suppressed during virion assembly/release.
Acknowledgments
We thank Yen-liang Chen, Christophe Bodenreider, Wouter Schul, Andy Yip, Suresh Lakshminarayana, Anne Goh, Sui Sum Yeong, Chin Chin Lim, Niyomwattanakit Pornwaratt, Shahul Nilar, Sebastian Sonntag, Christian Noble, and Thomas Keller for support and helpful discussions during the course of the study.
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Batt, D. 1999. Inhibitors of dihydroorotate dehydrogenase. Expert Opin. Ther. Patents 9:41-54. [Google Scholar]
- 3.Benarroch, D., M. P. Egloff, L. Mulard, C. Guerreiro, J. L. Romette, and B. Canard. 2004. A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J. Biol. Chem. 279:35638-35643. [DOI] [PubMed] [Google Scholar]
- 4.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. (Erratum, 7:255, 2001.) [DOI] [PubMed] [Google Scholar]
- 5.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falgout, B., R. H. Miller, and C. J. Lai. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster, G., and P. Mathurin. 2008. Hepatitis C virus therapy to date. Antivir. Ther. 13(Suppl. 1):3-8. [PubMed] [Google Scholar]
- 8.Gubler, D., G. Kuno, and L. Markoff. 2007. Flaviviruses, p. 1153-1253. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1, 5th ed. Lippincott William & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Guo, J., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwee, T., D. Stanford, and M. Sim. January 2005. Dihydroorotate dehydrogenase inhibitors for the treatment of viral-mediated diseases. U.S. patent 6841561.
- 11.Jordan, I., T. Briese, N. Fischer, J. Y. Lau, and W. I. Lipkin. 2000. Ribavirin inhibits West Nile virus replication and cytopathic effect in neural cells. J. Infect. Dis. 182:1214-1217. [DOI] [PubMed] [Google Scholar]
- 12.Khromykh, A. A., A. N. Varnavski, P. L. Sedlak, and E. G. Westaway. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 75:4633-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leyssen, P., J. Balzarini, E. De Clercq, and J. Neyts. 2005. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 79:1943-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., H.-J. Thiel, and C. M. Rice. 2007. Flaviviridae: the virus and their replication, p. 1101-1152. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1, 5th ed. Lippincott William & Wilkins, Philadelphia, PA. [Google Scholar]
- 16.Liu, S., E. A. Neidhardt, T. H. Grossman, T. Ocain, and J. Clardy. 2000. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure 8:25-33. [DOI] [PubMed] [Google Scholar]
- 17.Liu, W., X. Wang, V. Mokhonov, P.-Y. Shi, R. Randall, and A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean, J. E., E. A. Neidhardt, T. H. Grossman, and L. Hedstrom. 2001. Multiple inhibitor analysis of the brequinar and leflunomide binding sites on human dihydroorotate dehydrogenase. Biochemistry 40:2194-2200. [DOI] [PubMed] [Google Scholar]
- 20.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niyomrattanakit, P., Y. L. Chen, H. Dong, Z. Yin, M. Qing, J. F. Glickman, K. Lin, D. Mueller, H. Voshol, J. Y. Lim, S. Nilar, T. H. Keller, and P. Y. Shi. 2010. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J. Virol. 84:5678-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble, C. G., Y. L. Chen, H. Dong, F. Gu, S. P. Lim, W. Schul, Q. Y. Wang, and P. Y. Shi. 2010. Strategies for development of dengue virus inhibitors. Antiviral Res. 85:450-462. [DOI] [PubMed] [Google Scholar]
- 25.Puig-Basagoiti, F., M. Tilgner, B. Forshey, S. Philpott, N. Espina, Wentworth, S. Goebel, P. S. Masters, B. Falgout, P. Ren, Ferguson, and P. Y. Shi. 2006. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob. Agents Chemother. 50:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qing, M., W. Liu, Z. Yuan, F. Gu, and P. Y. Shi. 2010. A high-throughput assay using dengue-1 virus-like particles for drug discovery. Antiviral Res. 86:163-171. [DOI] [PubMed] [Google Scholar]
- 27.Rajamanonmani, R., C. Nkenfou, P. Clancy, Y. H. Yau, S. G. Shochat, S. Sukupolvi-Petty, W. Schul, M. S. Diamond, S. G. Vasudevan, and J. Lescar. 2009. One mouse monoclonal antibody that neutralizes all four dengue virus serotypes. J. Gen. Virol. 90:799-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray, D., A. Shah, M. Tilgner, Y. Guo, Y. Zhao, H. Dong, T. Deas, Y. Zhou, H. Li, and P.-Y. Shi. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruckemann, K., L. D. Fairbanks, E. A. Carrey, C. M. Hawrylowicz, D. F. Richards, B. Kirschbaum, and H. A. Simmonds. 1998. Leflunomide inhibits pyrimidine de novo synthesis in mitogen-stimulated T-lymphocytes from healthy humans. J. Biol. Chem. 273:21682-21691. [DOI] [PubMed] [Google Scholar]
- 30.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Q. Y., S. J. Patel, E. Vangrevelinghe, H. Y. Xu, R. Rao, D. Jaber, W. Schul, F. Gu, O. Heudi, N. L. Ma, M. K. Poh, W. Y. Phong, T. H. Keller, E. Jacoby, and S. G. Vasudevan. 2009. A small-molecule dengue virus entry inhibitor. Antimicrob. Agents Chemother. 53:1823-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wengler, G., and G. Wengler. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707-715. [DOI] [PubMed] [Google Scholar]
- 34.Wengler, G., and G. Wengler. 1993. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265-273. [DOI] [PubMed] [Google Scholar]
- 35.Whitehead, S. S., J. E. Blaney, A. P. Durbin, and B. R. Murphy. 2007. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 5:518-528. [DOI] [PubMed] [Google Scholar]
- 36.Yap, T., T. Xu, Y. Chen, H. Malet, M. Egloff, B. Canard, S. Vasudevan, and J. Lescar. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81:4753-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin, Z., Y.-L. Chen, W. Schul, Q.-Y. Wang, F. Gu, J. Duraiswamy, K. R. Reddy, P. Niyomrattanakit, S. B. Lakshminarayana, A. Goh, H. Y. Xu, W. Liu, B. Liu, J. Y. H. Lim, C. Y. Ng, M. Qing, C. C. Lim, A. Yip, G. Wang, W. L. Chan, H. P. Tan, K. Lin, B. Zhang, G. Zou, K. A. Bernard, C. Garrett, B. Beltz, M. Dong, W. M., H. He, X. Han, A. Pichota, V. Dartois, T. H. Keller, and P.-Y. Shi. 2009. An adenosine nucleoside inhibitor of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 106:20435-20439. [DOI] [PMC free article] [PubMed] [Google Scholar]