Abstract
The widespread emergence of antibiotic-resistant bacteria and a lack of new pharmaceutical development have catalyzed a need for new and innovative approaches for antibiotic drug discovery. One bottleneck in antibiotic discovery is the lack of a rapid and comprehensive method to identify compound mode of action (MOA). Since a hallmark of antibiotic action is as an inhibitor of essential cellular targets and processes, we identify a set of 308 essential genes in the clinically important pathogen Staphylococcus aureus. A total of 446 strains differentially expressing these genes were constructed in a comprehensive platform of sensitized and resistant strains. A subset of strains allows either target underexpression or target overexpression by heterologous promoter replacements with a suite of tetracycline-regulatable promoters. A further subset of 236 antisense RNA-expressing clones allows knockdown expression of cognate targets. Knockdown expression confers selective antibiotic hypersensitivity, while target overexpression confers resistance. The antisense strains were configured into a TargetArray in which pools of sensitized strains were challenged in fitness tests. A rapid detection method measures strain responses toward antibiotics. The TargetArray antibiotic fitness test results show mechanistically informative biological fingerprints that allow MOA elucidation.
The emergence and widespread development of antibiotic-resistant bacterial pathogens continue to drive the medical need for discovering new antibiotics (48). Despite intensive research efforts within the pharmaceutical and biotechnology industries, the number of promising antibiotic candidates has actually decreased in recent years. Traditionally, the most productive approach for antimicrobial lead discovery has been whole-cell screens for growth inhibitors, followed by optimization of the lead to improve compound antimicrobial and pharmaceutical properties. Most antibiotic classes were derived from natural product extracts during the 1940s and through the 1960s. However, this strategy has produced diminishing returns as the readily identifiable natural products are reisolated (52). In recent years, the advancement of microbial genomics has renewed prospects for identifying novel antibiotics acting on underexplored targets (12, 25, 28). However, despite intensive efforts, the antibacterial drug development pipeline has little to show, due to various challenges in antibiotic drug discovery (39).
One bottleneck in antibiotic drug discovery is the lack of a rapid and systematic approach to identify compound mode of action (MOA). This point is accentuated because whole-cell screening for antimicrobials is highly effective and potent antimicrobial hits are rapidly identified from compound libraries. However, a major limitation is that many hit compounds kill by nonspecific and therefore uninteresting mechanisms. Moreover, in the case of natural products, the vast majority of screening hits represent known antibiotics. Therefore, the development of research tools to profile antimicrobial MOA has great value. In this regard, progress has been made with the developments of cell-based and genomic strategies (4, 16, 57). However, these approaches are limited, as they do not provide comprehensive target coverage or are not a facile and accurate means to elucidate antibiotic MOA.
A few years ago, we developed a genome-wide strategy to identify essential genes in Staphylococcus aureus using regulated antisense (AS) RNA expression (12, 19). The principle behind this method is that conditional expression of an AS RNA will result in binding to the cognate mRNA and block its expression. If the target mRNA encodes an essential function, then bacterial growth is blocked (11, 12, 28). One advantage that this approach offers is that conditional AS strains can be directly configured into a cell-based assay for target-specific inhibitors (11, 12, 19). Specifically, AS RNA expression is titratable by addition of the inducer xylose, which exerts a range of growth defects, from no phenotype to partial or complete blockage of cell growth. The inducer concentration that elicits a mild growth defect is determined, generating a strain hypersensitized to that target. This strategy is highly selective and has successfully been employed with various targets to identify known and novel inhibitors, including the FabF inhibitors cerulenin, phomallenic acids, platensimycin, and platencin (37, 53, 54). Although this is a proven cell-based approach for antibiotic discovery, the strategy has a severe limitation, in that only one cellular target can be screened at time.
An elegant and comprehensive approach to simultaneously screen the activities of antifungal agents against cellular targets has been developed. This approach, called haploinsufficiency profiling or genome-wide yeast fitness testing, was first developed by Davis and colleagues (15). The principle behind this approach relies on the observation that reducing the copy number of the gene target of an antifungal agent specifically hypersensitizes that strain in fitness tests. For yeast, a complete collection of heterozygous deletion mutants was systematically constructed and genetically bar coded, allowing rapid strain quantification in fitness tests by microarray hybridization (45). We have successfully adapted and developed an analogous haploinsufficiency profiling strategy in the clinically important fungal pathogen Candida albicans (42, 56). Given that bacteria are obligate haploids and essential genes cannot be knocked out, the strategy described for yeast is not applicable. However, in principle, conditional alleles of essential genes that are the targets for antibiotics can be similarly constructed and pooled to profile the antibiotic MOA in fitness tests. Such an approach is described here for the clinically importantly pathogen S. aureus (48). The assemblage of sensitized strain sets, termed TargetArray, is formatted for fitness tests to quantifiably profile bacterial responses toward antibiotics. This TargetArray has recently been validated for elucidating the MOA of novel peptidoglycan and protein synthesis inhibitors (5, 23).
MATERIALS AND METHODS
Bacterial strains, plasmids, and antibiotics.
Genomic DNA was obtained from S. aureus RN450 (36). S. aureus AS, integrative promoter replacement (I-PR), and plasmid-borne overexpression (P-OE) clones were constructed in RN4220 (32) because this strain lacks a DNA restriction system. The 236 AS strains were derived from a prior screen (12). All plasmids were constructed in Escherichia coli DH5α and are listed in Table S1 in the supplemental material. Antibiotics were purchased from Sigma-Aldrich (St. Louis, MO) and include actinomycin D, anhydrotetracycline (ATc), azithromycin, carbenicillin, cefotaxime, cerulenin, chloramphenicol, ciprofloxacin, clindamycin, cloxacillin, coumermycin, doxycycline, levofloxacin, lincomycin, mupirocin, novobiocin, oxacillin, phosphomycin, rifampin, roxithromycin, triclosan (i.e., irgasan), trimethoprim, and vancomycin. Indolmycin was isolated and purified from the producing strain by Merck scientists. TR000006 (18) and iclaprim (TR000010) were provided by Trius Therapeutics (San Diego, CA).
Construction of plasmid-borne overexpression strains.
To overexpress essential S. aureus genes, full-length copies of each gene were amplified from RN450 genomic DNA by PCR using Pfu DNA polymerase (Stratagene, San Diego, CA) and cloned into the multiple-cloning site of pTET10. This expression plasmid is an E. coli-S. aureus shuttle vector derived from pEPSA5 (12) by swapping the xylR PT5X cassette for a tetR PTOX cassette. Each clone was verified by DNA sequencing analysis.
Construction of S. aureus chromosomal promoter replacement strains.
The native promoters of selected essential genes were replaced in the chromosome with heterologous ATc-inducible promoters. I-PR strains were made to either overexpress (I-OE) or underexpress (I-UE) target genes or operons. To construct I-PRs, the pEVP3 suicide plasmid, which contains the cat gene and which is unable to replicate in S. aureus (40), was engineered by inserting a tetR cassette expressed from its own promoter and is one of a suite of synthetic promoters/operators divergently positioned from tetR. The regulatory regions consist of a synthetic promoter coupled to one tetO operator (PTOS and PTOX) or two tetO operators (PTOOW and PTOOS) to form the pEVP3RM plasmid-derived series. The PTOS and PTOOS promoters were derived from the bacteriophage T5 PN25 promoter (14), while the other promoters were chimeric derivatives. To quantify promoter activity and ATc regulation, these promoters were transcriptionally fused to the E. coli lacZ reporter gene. To allow homologous recombination into the RN4220 chromosome, the amiC gene was also cloned into these plasmids. Vector DNA was electroporated (31) into RN4220, and transformants were selected as Cmr colonies. Transformant genotypes were verified by PCR. Importantly, pEVP3 had a very low rate of illegitimate recombination in RN4220 (data not shown).
To make PR strains, a typical clone was constructed by cloning, via PCR, the first 250 to 500 bp of the 5′ coding region of the target essential gene. The upper size limit, ∼500 bp, was employed only for large genes; most clones contained the first ∼300 bp of the coding region. The length of each insert was chosen to allow efficient homologous recombination and to minimize the likelihood that partial-gene duplications would create a truncated gene product that complemented the PR allele. The lower limit, determined empirically, for efficient homologous recombination was ∼250 bp. The PCR fragments were cloned into the multiple-cloning sites (BamHI, EcoRI, AvaI, SmaI, XmaI, and PpuM1 restriction sites) of the pEVP3RM plasmid series and were immediately ligated downstream of the ATc-regulated promoter and a ribosomal binding site (AGGAGG). To create a PR strain, 10 μg of a validated plasmid DNA was electroporated into RN4220. Plasmid chromosomal integration occurred via a single crossover event; transformants were selected as Cmr colonies in the presence of 25 ng/ml ATc to ensure target gene expression. Validated clones were then tested for growth defects by withdrawing ATc for 1 h, followed by plating of serial dilutions on Luria-Bertani (LB) agar plates with 0, 5, 10, or 25 ng/ml ATc. A loss or significant reduction in bacterial growth in the absence of ATc was deemed confirmatory evidence for gene essentiality. The parental vector integrated at the nonessential amiC locus was used as a positive control for growth. The mRNA levels of most PR strains were measured and compared to native gene expression levels by quantitative reverse transcription-PCR (qRT-PCR) with a TaqMan instrument and Sybr green probes. It was generally found that target mRNA levels were significantly lower in or absent from the PR strains grown without ATc than in wild-type cells.
Strain detection strategy and method development.
Primers with calculated melting temperatures of 51°C were designed. These primers were designed to work in multiplex PCR sets, ranging from 10 to 16 primer pairs per group. A common set of forward primers was designed to be specific to the synthetic promoter sequences found either on plasmids (AS or P-OE) or in the chromosome and were labeled with an appropriate fluorophore (VIC or 6-carboxyfluorescein; Perkin-Elmer). Depending on the element, the reverse primers were specific either to each antisense fragment, to the cloned gene (P-OE), or to the chromosomal gene target (I-OE or I-UE). Reverse primers were designed to produce fragments different in length by at least 5 bp from the lengths of other amplicons within the same multiplex PCR set. Amplicons were also designed to be within a 150-bp range to help minimize preferential amplification. PCR fragments were resolved by ABI 3700 capillary electrophoresis with ABI GeneMapper software. When the test peak percentages were smaller than those for the control, the calculation would serve as an indication of strain depletion (fitness loss). Alternatively, when the test strain peak percentages were higher than the percentage for the control, e.g., P-OE, the ratio would serves as an indication of strain enrichment (improved fitness or resistance).
Statistical analyses of fitness results.
The variabilities of the individual markers for the AS and I-UE elements of the TargetArray were normalized, as described previously (5). The strain depletion or enrichment ratio calculations and the subsequent statistical significance (Z score) analysis using an error model were performed as described previously (5). A Z score of >2 was deemed a significant difference.
TargetArray assembly.
A collection of 236 antisense clones was chosen for TargetArray inclusion. Since AS RNA can potentially destabilize the expression of other genes on polycistronic mRNA, the predicted operon structure for each gene target is listed in Table S2 in the supplemental material to facilitate experimental interpretations.
Multiple AS clones were isolated for most essential genes (12). Consequently, clone selection was based on several criteria. (i) AS cloned fragments that were contained within the open reading frames (ORFs) of interest were prioritized over fragments that spanned multiple ORFs. (ii) Cloned AS fragments that spanned to the 5′ ends of their cognate mRNAs were prioritized over fragments that spanned the 3′ ends, because the former fragments were found to be more effective. (iii) Many AS clones were also tested for specific product depletion. The validation methods included measurement of target mRNA destruction by qRT-PCR and/or Northern blot analysis, measurement of target protein depletion by Western analysis, and/or determination of sensitivities to antibiotics that target the cognate protein or metabolic pathway in specific cell-based assays. In sum, optimized AS cloned fragments were selected for TargetArray inclusion.
For each selected AS strain, xylose dose-response measurements were made to group strains according to their xylose sensitivities. In general, we found that the effective concentration that inhibits growth by 20% (EC20) for xylose elicited target-specific sensitization that allowed robust cell growth. To allow similar growth kinetics, strains were binned or grouped according to their xylose EC20 values. Thus, although the xylose concentrations varied between bins, the growth kinetics of all strains and bins remained uniformly constant. In total, the TargetArray consisted of 24 bins (∼10 AS clones per bin) with 17 different xylose levels, ranging from 0.75 to 22.5 mM.
To assemble a bin, representative strains were grown to early log phase (1 × 107 CFU/ml), chilled on ice, and combined with other strains that had similar xylose EC20 values. Since the numbers of strains per bin varied between 5 and 14, the inoculating density for each strain was 1 × 104 CFU/ml, thus ensuring that all bins started with the same numbers of cells. The resulting 24 bins were then replica transferred into 96 deep-well plates and stored at −80°C.
For the I-PR strains, an analogous protocol was used to bin strains according to their ATc EC20 values. Accordingly, the I-UE strains were divided into three bins with ATc EC20s of 4, 8, and 12.5 ng/ml, respectively. For P-OE clones, no ATc was added, as leaky expression from the PTOX promoter ensured overexpression.
TargetArray growth conditions.
To maintain plasmids or integration constructs, LB medium (43) was supplemented with 34 μg/ml chloramphenicol. All growth experiments were performed in a 384-well plate (Costar 3684; Corning, Lowell, MA) in 50 μl at 37°C in a Kendro Cytomat 6110 incubator. Prefrozen aliquots of cells representing bins of various types of strains (AS, I-UE, or P-OE) were thawed on ice and diluted 100-fold into LB medium to a starting optical density at 600 nm of 0.002. Inducer (xylose or ATc) was added at an appropriate concentration to each bin with or without test drug (2% dimethyl sulfoxide [DMSO] was used as a control). Bins were then incubated for ∼4.5 h, or seven generations (1 × 107 CFU/ml), and then backdiluted to a density of ∼6.7 × 104 CFU/ml. After an additional 4.5 h growth, a third backdilution was done to ensure that the strains had grown ∼20 generations. The inclusion of test antibiotic in the fitness tests was done at three concentrations (25% inhibitory concentration [IC25], IC37.5, or IC50s for underexpression tests and 1×, 2×, and 3× MIC values for overexpression tests). Since antibiotic-treated samples grew at lower rates, the incubation times were correspondingly increased to allow ∼20 generations of growth.
RESULTS AND DISCUSSION
Essential gene identification and validation.
Our initial S. aureus screen identified growth-inhibitory AS clones for 658 unique ORFs (12). Many of these ORFs represented expected essential genes or were genes that contained orthologs that overlapped the minimal Mycoplasma genitalium essential gene set (25). However, this gene set also contained hypothetical genes or genes whose essentiality could not be easily corroborated from information in the literature. In addition, some essential genes may have been missed. To independently validate gene essentiality and to ensure a comprehensive essential gene set, gene essentiality was tested by an independent method. Since a rapid test was not available for S. aureus, we selected another model organism and method (33). Here, a second clinically important Gram-positive pathogen, Streptococcus pneumoniae, was chosen to conduct rapid gene knockouts. In this approach, PCR gene knockout amplicons were directly and efficiently transformed and homologously recombined into the chromosome. Using this approach, ∼700 prioritized conserved genes were tested for essentiality, and the gene was considered essential when no viable transformants were obtained (R. J. Haselbeck and J. D. Trawick unpublished data). Thus, through combinatorial efforts with S. aureus AS and S. pneumoniae gene knockouts and a literature review (M. genitalium [25] and the Profiling of E. coli Chromosome database [http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp]), a comprehensive list of conserved essential genes was identified (see below). On the basis of these and other results, 236 AS strains were selected for inclusion in the TargetArray (see Table S2 in the supplemental material).
Although the AS technology proved to be a highly effective means to knock down the expression of S. aureus genes, there are limitations. First, some essential genes were not captured by screening for AS expression. For example, the dnaB gene was missed. This may be due to a combination of an extremely low level of DnaB expression (10 to 20 monomers per cell) (51) and leaky AS vector expression, precluding dnaB AS clone isolation (12). Second, the majority of bacterial genes are grouped in operons. Consequently, AS induction may result in polar effects on downstream genes, complicating phenotype analysis. Therefore, to circumvent these problems, promoter replacement (PR) strains were sought to supplement AS strains.
Construction of a promoter suite to allow physiologically relevant gene expression.
Regulatable promoters that allow essential gene expression over biologically relevant expression ranges were sought. To help ensure that the levels of gene expression in engineered strains would recapitulate those in native strains, a panel of promoters that covers low, middle, and high expression levels was developed. To regulate expression, the tetracycline system derived from transposon Tn10 was selected. The tet system allows a wide range of dynamic expression of the ATc inducer, which does not inhibit bacterial growth, at nanomolar concentrations (30, 59).
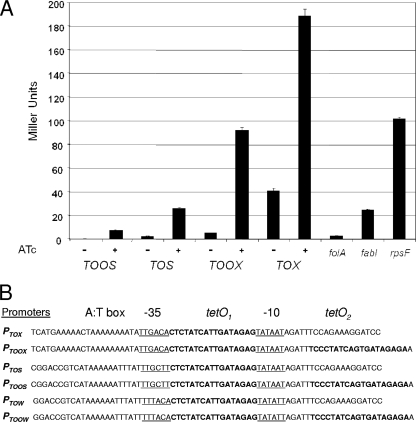
To develop a baseline for endogenous gene expression, a series of transcriptional fusions between S. aureus essential gene promoters and the E. coli lacZ reporter were constructed. Example levels of expression from three promoters covering an ∼50-fold dynamic range are shown in Fig. 1A. Thus, folA represents a promoter expressed at a low level, while fabI and rpsF represent promoters expressed at moderate and high levels, respectively. Efforts to construct and test synthetic promoters resulted in the selection of three core promoters. To differentially regulate expression, each promoter was engineered with one or two tet operators (tetO1 and tetO2) (49), resulting in six promoter-operator configurations (Fig. 1B). These promoters ranged from having very tight low-level expression (PTOOW) to having high-level expression (PTOX). In fact, the PTOOW (weak) promoter did not drive detectable levels of β-galactosidase activity, a poor reporter in S. aureus. However, as described below, PTOOW provided tight repression of gene expression; hence, PTOOW was effective for PR construction, as bacterial growth was often dependent on ATc (see Table S3 in the supplemental material). In addition, the levels of expression of PTOX and PTOS are significantly enhanced when they are placed on multicopy plasmids (data not shown). In sum, this promoter suite allows a wide dynamic expression range in S. aureus.
FIG. 1.
S. aureus promoters. (A) Representative spectra of activity for the native folA, fabI, and rpsF promoters fused to the lacZ reporter are shown. The activities of four synthetic promoters under repressed (−) and ATc-induced (+) conditions are shown. The PTOW and PTOOW promoters did not elicit detectable activity (data not shown). All constructs are integrated at the amiC locus. Promoter activity was measured in Miller units (35). (B) Sequence and regulatory elements for a suite of synthetic promoters. The −10 and −35 boxes are underlined, and tetO sequences are in boldface.
Construction of chromosomal I-UE promoter replacement strains.
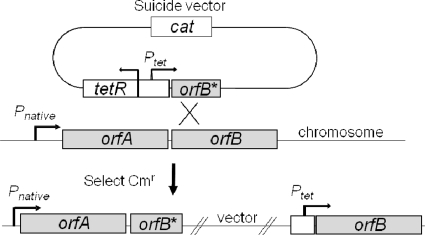
Gene prioritization for PR construction, discussed above, included genes that were not present in the AS collection and thus represent important new additions, while other genes were represented by AS. In general, the development of a built-in redundancy between AS and PR strains was deemed prudent to ensure that key genes were represented by different elements. In addition, I-PR of downstream genes within operons allows operon gene expression to be decoupled, whereby the expression of the upstream gene(s) was not perturbed, while the downstream gene(s) was conditionally expressed (Fig. 2), as the suicide vector contains a transcriptional terminator. This process, called “operon depolarization,” was not possible with AS.
FIG. 2.
Schematic representation of promoter replacement construction. One of the tet-regulated synthetic promoters, generically indicated Ptet, was fused to the 5′ end of a target essential gene. Upon transformation, the suicide plasmid homologously recombines into the chromosome. A two-gene operon where the expression of the downstream gene was decoupled from the native promoter and is under heterologous ATc control is illustrated.
In our effort, approximately 150 genes were originally targeted for PR. Since it typically was not known what promoter should be used for a given gene, we routinely constructed three different PRs per gene (>400 total constructs). Surprisingly, over half of these broadly conserved candidate essential genes targeted for I-UE PR did not elicit a growth dependence on ATc, suggesting that those genes are not essential or, alternatively, that residual leaky expression from the PR was sufficient to support growth. In either case, such strains were generally set aside, as there was no phenotypic validation of gene essentiality. However, in a few cases these genes were studied in greater detail. For instance, I-UE constructs targeting asnS, a gene that encodes asparaginyl-tRNA synthetase and that at the time of construction was predicted to be essential, did not elicit a growth defect in the absence of ATc. Furthermore, asnS was not hit by our global AS screen (12). Interestingly, as was found in a number of bacteria, S. aureus has two alternative pathways for charging tRNA molecules with asparagine and glutamine; therefore, the asparaginyl-tRNA synthetase is dispensable because an amidotransferase can convert Asp-tRNAAsn to Asn-tRNAAsn in S. aureus (9).
Nearly half of the selected broadly conserved genes with predicted essential functions (50) elicited a growth defect in the absence of ATc. Specifically, for 101 individual I-UE PR strains representing 64 different genes, a clear growth defect was observed when no or low levels of ATc were present in the medium, supporting the assertion that these genes are essential. These genes, cloning primers, and promoter types are listed in Table S3 in the supplemental material. The ATc growth dependence of a representative glmS (encodes aminotransferase) I-UE PR strain is shown in Fig. S1 in the supplemental material. As shown, the PTOS-glmS engineered strain does not grow in the presence of ATc at low levels (2.35 ng/ml); however, as the ATc concentrations increase, there was a corresponding increase in the growth rate, until the growth kinetics plateaued when ATc was present at ∼37.5 ng/ml. Lastly, for a subset of I-UE PR strains, qRT-PCR was used to measure mRNA levels. As expected, the target mRNA was depleted as ATc levels were lowered, and this was tightly correlated with strain growth (data not shown).
Construction of chromosomal I-OE promoter replacement strains.
In a complementary approach to strain sensitization, strains that overexpressed essential targets were also constructed as a means to confer antibiotic resistance and, hence, to serve as a reciprocal MOA tool (34, 38). Two approaches were pursued; integrative overexpression via PR and P-OE. Important distinctions exist between these strategies. First, I-OE can drive complete or partial operon overexpression (depending on the insertion site), while the plasmid-borne system described below allows overexpression of only a single gene. For proteins assembled into multiprotein complexes, operon overexpression can be advantageous, as the corresponding genes are often organized in operons. Second, multicopy P-OE allows inherently higher levels of expression than single-copy I-PR. As described above (Fig. 1), the PTOX promoter expresses at exceptionally high levels and was used for the construction of 54 I-OE PR strains (see Table S4 in the supplemental material). PTOX PR strains typically allowed bacterial growth, albeit poorly, in the absence of ATc because of leaky expression (Fig. 1). Upon induction with a modest amount of ATc, e.g., 3 ng/ml, bacterial growth was fully restored to a prototypic PTOX gyrB PR strain (see Fig. S2 in the supplemental material). Overexpression of the target gene or operon mRNA could therefore be achieved at 90 ng/ml ATc. The I-OE constructs were experimentally tested by qRT-PCR to compare the mRNA levels driven by PTOX and those driven by the native promoters. In nearly all cases tested, target mRNA levels were found to be higher when expression was driven by PTOX (90 ng/ml ATc) than when it was driven by the native promoters (data not shown).
Construction of plasmid-borne overexpressers.
A set of P-OE strains was constructed to extend the repertoire of overexpressers. Target gene selection intentionally excluded gene products involved in large multiprotein complexes, e.g., ribosomal proteins. Here, a total of 55 genes were cloned into the pTET10 E. coli-S. aureus shuttle vector containing the powerful PTOX promoter. The essential gene set and PCR primers used for amplification are listed in Table S5 in the supplemental material. Each plasmid contains a full-length ORF, determined on the basis of the PathoSeq annotation (55). S. aureus recombinant clones were tested for a dominant negative inhibition of growth by spot dilution on LB agar plates with ATc concentrations increasing from 5 to 90 ng/ml. Interestingly, ∼90% of these clones elicited an S. aureus growth inhibition phenotype at high ATc levels. Growth inhibition was independent of ATc or a vector effect because an empty plasmid control did not inhibit growth. These results corroborate those of lacZ expression studies (Fig. 1) that PTOX is indeed a strong S. aureus promoter.
Comprehensive collection of strains differentially expressing essential gene elements.
A total of 446 strains validated to differentially express genes were made in strain RN4220, and 308 essential genes or essential genes within operons were targeted (see Table S6 in the supplemental material). All together these four promoter elements target 131 predicted operons. To help ensure an accurate and robust platform, many of these targets are represented by more than one element. AS strains represent the largest element class (236 strains, or 53%). These AS clones both directly and indirectly (operon) target 277 essential genes, of which 225 are direct AS hits. I-PR strains represent the second largest group (101 strains, or 23%). These element classes allow target-specific knockdown expression. A reciprocal set of strains allows target overexpression. Overexpressers consist of I-OE (54 strains, or 12%) and P-OE (55 strains, or 12%).
Selective AS strain sensitization.
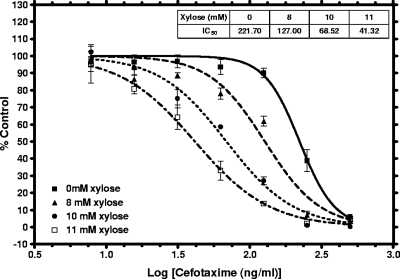
Growth-inhibitory AS clones were identified from a screen conducted at high xylose (2%) concentrations (12). For a given AS strain, the growth kinetics were inhibited to various extents by simply titrating xylose concentrations (11). Next, cell sensitization tests were conducted with antibiotics that target a cognate AS clone. As a representative example, an fmhB AS strain was titrated against xylose. The FmhB transferase catalyzes the incorporation of the first residue of the pentaglycine bridge of peptidoglycan (1). Strain sensitization to a cell wall inhibitor, cefotaxime, was tested in the presence of various concentrations of xylose. The fmhB AS strain was incrementally sensitized to cefotaxime at increasing xylose concentrations (Fig. 3), demonstrating AS-mediated sensitization.
FIG. 3.
Seven-point dose-response to cefotaxime tested against an S. aureus fmhB AS strain grown without xylose or in the presence of xylose at various concentrations. The sensitivity toward cefotaxime is titratable as a function of the xylose concentration. The maximal IC50 shift was >5-fold at 11 mM xylose.
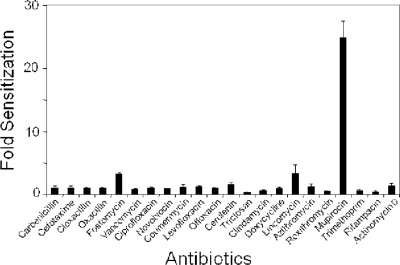
Prior studies showed target-specific drug sensitization of AS clones to the rpsR and fabF genes (11, 12), which, importantly, were also employed to discover new antibiotics (27, 46, 54). To expand these studies, other AS clones were tested. Here, AS clones for the gyrB and ileS genes were chosen and tested against a panel of 22 antibiotics. Indeed, a gyrB AS clone rendered a selective hypersensitivity phenotype (>8-fold) toward a known inhibitor of the β subunit of DNA gyrase, novobiocin (see Fig. S3 in the supplemental material) (13). Hypersensitivity of the gyrB AS strain to several quinolones was not detected, possibly reflecting the possibility that quinolone-target complexes actively poison cells. Thus, target underexpression may actually lead to quinolone resistance (26). Similarly, selective sensitization of the ileS AS clone to mupirocin (>20-fold), an inhibitor of isoleucine-tRNA synthetase (IleRS), was found (Fig. 4). Thus, AS cell sensitization elicits a highly specific and selective response toward cognate antibiotics.
FIG. 4.
Expression of AS RNA toward ileS preferentially sensitizes S. aureus to mupirocin (∼25-fold). Twenty-two different antibiotics were tested. Fold sensitization represents the change in antibiotic IC50s upon AS induction.
Proof-of-concept fitness tests with differentially expressing strains.
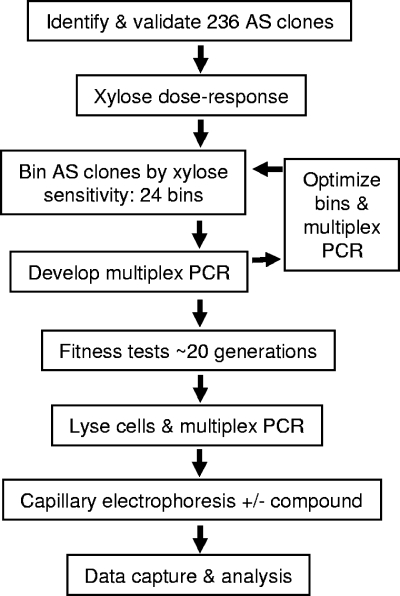
As shown above, induction with the AS sequence elicits strain sensitization to specific antibiotics (Fig. 4). Next, pools of differentially expressing strains were challenged in fitness tests to determine their specific responses. In these tests, sensitized strains would be depleted, while resistant strains would be enriched. An outline of how the TargetArray was built and run is depicted in Fig. 5. A schematic representation of how the PCR amplicons were amplified is shown in Fig. 6.
FIG. 5.
Flowchart for AS TargetArray development and implementation.
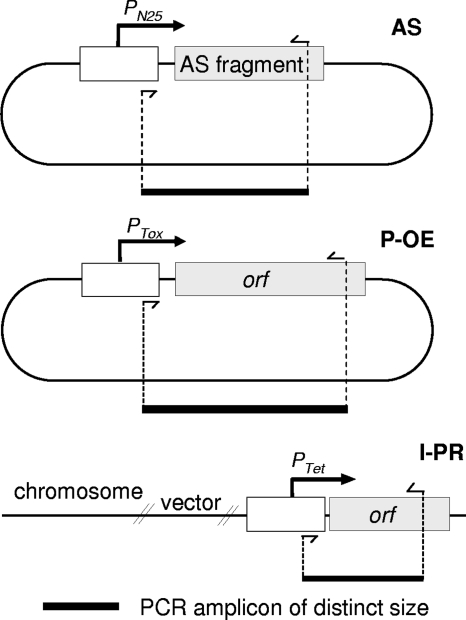
FIG. 6.
Design strategy for detecting AS, P-OE, and I-PR elements in recombinant strains. All markers within a marker set are produced by a common forward primer and uniquely designed reverse primers for multiplex PCR amplification. Markers are resolved by both their unique size and fluorescent label via capillary electrophoresis.
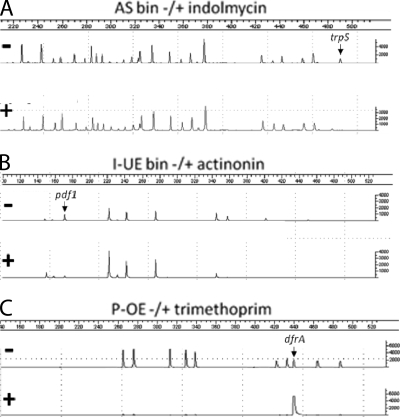
In a first, early fitness test, a pool of 23 AS strains, which constituted a bin of strains with similar xylose sensitivities, was grown for ∼20 generations, followed by multiplex PCR and capillary electrophoresis to measure the relative abundance of each strain grown in the presence of an antibiotic (indolmycin) or mock treated (DMSO) (Fig. 7A). Indolmycin is a known inhibitor of TrpS, the tryptophanyl-tRNA synthetase (24). As predicted, the trpS AS strain was specifically depleted by indolmycin treatment compared to the amount of growth for a no-drug control. This and related experiments provided the first evidence that AS cell sensitization could be used to profile specific antibiotic responses in fitness tests.
FIG. 7.
Proof-of-concept S. aureus fitness tests. Following growth in the presence of various antibiotics, S. aureus strains were detected by multiplex PCR and capillary electrophoresis. The ABI 3700 signal intensity is graphed on the vertical axis, and the migration length (in base pairs) is shown on the horizontal axis. (A) AS fitness test with a mixture of 23 strains grown in 6 mM xylose. Samples were mock treated with DMSO or were treated with indolmycin at 0.5× IC50. Indolmycin treatment depleted the target trpS AS strain. (B) I-UE fitness tests with a mixture of seven strains grown with 12.5 ng/ml ATc. Samples were mock treated or treated with actinonin at 0.5× IC50. Actinonin treatment partially depleted the pdf1 strain. (C) P-OE fitness test with a mixture of 10 strains that overexpress different genes. Samples were mock treated or treated with trimethoprim at the MIC value (6 μg/ml). Trimethoprim treatment resulted in the loss of all strain markers except for the target dfrA-overproducing strain.
In a second round of fitness tests, I-UE PR strains were used in lieu of AS clones. Although conceptually similar, it was experimentally found that I-UE PR fitness tests were more challenging. The first technical problem was developing robust primer sets that allowed reproducible multiplex PCR amplification from a single-copy template. This problem was mitigated from AS pools as template DNA was derived from plasmids containing ori pC194, present at ∼40 copies per cell (17). A second problem was that I-UE PR strains were typically more difficult to grow and stably maintain than AS strains. This problem was attributed to the fact that native promoter strength and temporal cell cycle regulation were difficult to predict and recapitulate with heterologous promoters. This problem was compounded when many PR strains were processed in parallel. Given these caveats, I-UE PR fitness tests were nevertheless pursued. The results from an I-UE PR fitness test are shown in Fig. 7B, where actinonin, an antibiotic that inhibits peptide deformylase (Pdf1) (3), was used. As illustrated, a moderate level of depletion was observed for the pdf1 I-UE PR strain. This experiment was conducted at 0.5× the IC50 for actinonin (2.6 μg/ml). At higher concentrations of actinonin, the pdf1 I-UE PR strain was completely depleted (data not shown). However, as was generally found, high antibiotic concentrations elicit nonspecific PR strain depletions. In sum, although I-UE strains offer limited utility in fitness tests, they clearly have utility when used in secondary single strain assays to confirm compound MOA (4, 59).
Lastly, we tested target overexpression as a means to enrich strains in fitness tests. First, a set of 10 P-OE strains was pooled, grown in the presence of increasing concentrations of the antibiotic trimethoprim, and again assayed by multiplex PCR and capillary electrophoresis analysis. This strain pool includes a clone of dfrA (folA), encoding dihydrofolate reductase, the target of trimethoprim (22). When this strain pool was grown in the presence of the MIC level (6 μg/ml) or above, the dfrA-overexpressing strain was strongly enriched (Fig. 7C). However, when the concentration of trimethoprim was less than the MIC level, the dfrA strain was not enriched (data not shown). Second, in an experimental reciprocal whose results are shown Fig. 7A, where a trpS AS strain was sensitized to indolmycin, a P-OE trpS strain conferred the opposite phenotype and the strain was strongly enriched (data not shown). These results show that target overexpression can confer a selective advantage in S. aureus fitness tests to profile antibiotics.
Building an AS TargetArray and fitness tests.
Proof-of-concept experiments established that AS-sensitized and P-OE-resistant strains elicit robust, predictable, and informative responses in antibiotic fitness tests. To further build an array, efforts focused on the AS clones because those strains behave robustly and a comprehensive collection was in hand. The AS TargetArray was built in three stages, where increasing numbers of validated and reliably behaving AS clones were incorporated into the array (Fig. 5).
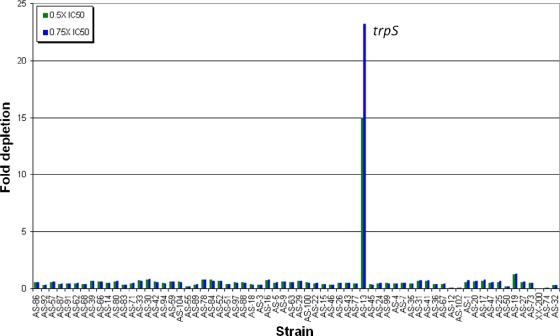
The stage I AS TargetArray consisted of seven xylose bins that contained a total of 66 AS clones that were reliably detected by multiplex PCR. A number of fitness tests were conducted, and a representative result is shown in Fig. 8. Here, indolmycin was added to the seven AS bins at 0.5× and 0.75× the IC50. After ∼20 cell generations, the pools were assayed and the depletion ratios of the treated strains to mock treatment were calculated. The fold depletion values for each AS strain were plotted, and as is evident, the AS clone to trpS was depleted 15- to 23-fold. Importantly, no other strain was depleted more than 2-fold.
FIG. 8.
Fitness tests with stage I AS TargetArray. The array consists of 66 AS clones in seven bins. The array was treated with indolmycin extract at two concentrations, 0.5× or 0.75× the IC50s. The fold strain depletion is color coded and plotted for each strain.
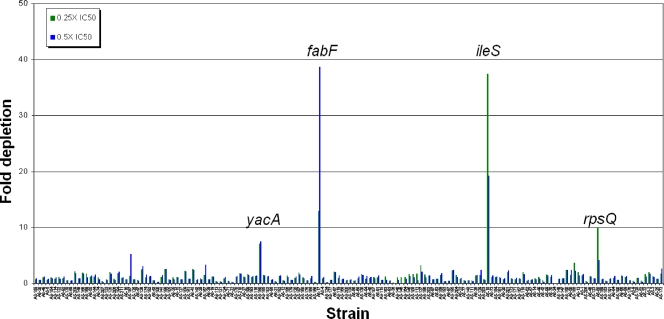
The stage II TargetArray consisted of 160 AS clones in 12 xylose bins. A number of fitness tests were again conducted. To simulate a more complex condition, e.g., the use of a natural product extract with multiple antibiotics, a fitness test was done where two antibiotics were simultaneously added. Here, both cerulenin, which targets FabF (β-ketoacyl-acyl carrier protein synthase II) in fatty acid biosynthesis (41), and mupirocin, were each added at 0.25× and 0.5× their respective IC50s. The fitness results reveal a highly specific response (Fig. 9). Both of the expected AS target strains, the fabF and ileS AS clones, were reproducibly depleted by between 12- and nearly 40-fold. Minor depletion of two other AS strains, the yacA and rpsQ AS clones, was also detected. Interestingly, the yacA (tilS) gene product, involved in tRNAIle modification, was independently found to be essential in S. aureus (2, 47); thus, a yacA AS strain may sensitize cells to mupirocin by depleting the pool of functionally modified tRNAIle, the enzymatic target of isoleucine tRNA synthetase. In addition, the rpsQ AS clone, whose target gene lies within a large ribosomal operon predicted to consist of 29 genes (see Table S6 in the supplemental material), was weakly depleted by the mupirocin-cerulenin mixture. Here, mupirocin, which acts as a protein synthesis inhibitor, might act in an additive manner with an rpsQ AS strain that is depleted in ribosome function. In any event, these results again show that AS fitness tests are highly reproducible and selective. These results also suggest that the TargetArray has utility to deconvolute natural product extracts.
FIG. 9.
Fitness tests with stage II AS TargetArray. The array consists of 160 AS elements in 12 bins. The array was challenged with two antibiotics, cerulenin and mupirocin, at 0.25× and 0.5× their respective IC50s. The fold strain depletion is color coded and plotted for each strain.
Application of AS TargetArray for antibacterial target identification.
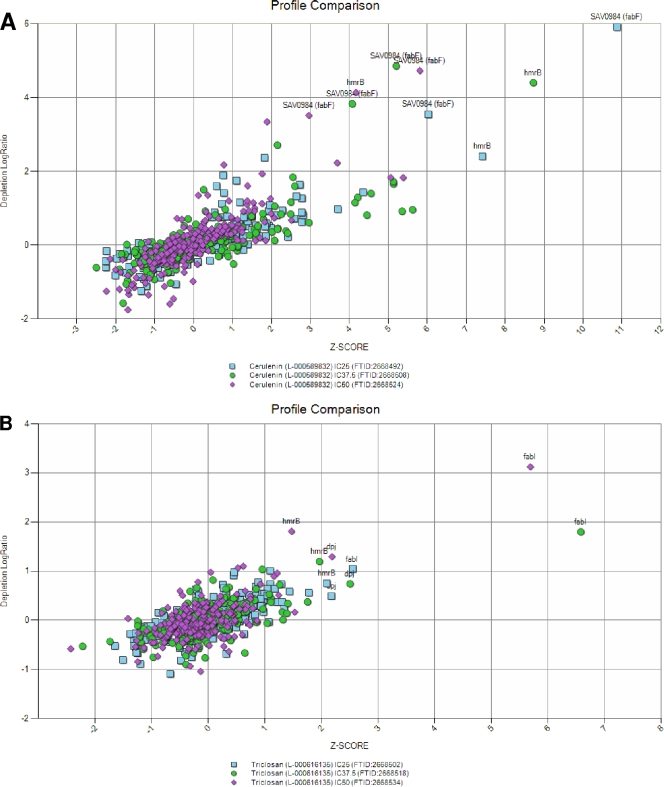
With pilot proof-of-concept results in hand, a fully built AS TargetArray (stage III) that consisted of 236 AS strains placed into 24 xylose bins was made (Fig. 5). This array was then tested as described above with a number of known antibacterial inhibitors as well as novel inhibitors (Fig. 10; see also Fig. S4 and S5 in the supplemental material). In each case, antibiotics were added at three concentrations (IC25, IC37.5, and IC50) to ensure reproducibility and the use of experimentally relevant antibiotic concentrations. In the first test, cerulenin was used, and the fabF (SAV0984) AS clone was again the most prominently depleted strain (∼6 log units; Fig. 10A). This was expected, as FabF is the target of cerulenin, and this result confirms the stage II array findings (Fig. 9). The second AS clone that was prominently depleted was one with AS toward the hmrB gene, an ortholog of the E. coli acpP gene (60), which in S. aureus contributes to methicillin resistance (21). Since hmrB encodes an acyl carrier protein predicted to serve as a FabF substrate in fatty acid biosynthesis in S. aureus, the depletion of the hmrB AS strain by cerulenin is a mechanistically informative response. A second fatty acid synthesis inhibitor, triclosan, which targets FabI (enol-acyl carrier protein reductase) (20), was used in fitness tests. Again, a very specific response was found: the fabI AS clone was the most prominent and reproducibly depleted strain (∼3 log units; Fig. 10B). As was seen with cerulenin, the hmrB AS clone was again depleted by triclosan treatment.
FIG. 10.
Fitness tests with complete AS TargetArray. The results are shown as overlays of Z scores versus depletion ratios. Significantly depleted strains are indicated. The antibiotics used are cerulenin (A) and triclosan (B), both of which were tested at multiple concentrations (IC25, IC37.5, and IC50).
The stage III array was tested against fosfomycin, an antibiotic known to target UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) (8, 29). As shown in Fig. S4A in the supplemental material, the murA AS clone was the most severely (∼5 log units) and reproducibly depleted strain. Interestingly, S. aureus encodes two murA genes, murA1 and murA2 (murZ), which are 59% identical at the nucleic acid level; neither is essential, although a double mutation is lethal (7). The murA AS clone apparently inactivates both murA loci (12), consequently sensitizing cells to fosfomycin. Although other AS strains (infC, hprK, and SAV0248-SAV0249-SAV0250) were also depleted, they were not as significantly or as reproducibly depleted as the murA AS strain.
Potential challenges to the TargetArray technology for elucidating antibiotic MOAs are inhibitors of large macromolecular complexes, such as ribosomes. In principle, the depletion of one subunit within a ribosome, for example, may abolish complex formation and function and the resulting sensitization. With this caveat in mind, a macrolide antibiotic, erythromycin, which targets the ribosomal large subunit (50S) 23S rRNA (6) and the ribosomal proteins L4 (rplD) and L22 (rplV) located around the peptide exit tunnel (58), was profiled. When erythromycin was tested in the TargetArray, only the rpsR (which encodes 30S rRNA subunit S18) AS strain was sharply and reproducibly depleted (see Fig. S4B in the supplemental material). Interestingly, under these experimental conditions, the AS clones with AS sequences directed toward 23S rRNA, rplD, or rplV were not significantly depleted, even though mutant L4 and L22 proteins can confer erythromycin resistance (58). This result suggests a possible direct or indirect interaction between erythromycin and ribosomal protein S18. Alternatively, the rpsR gene product could play a role in intrinsic antibiotic resistance, as rpsR overexpression in E. coli was recently found to confer broad-spectrum antibiotic resistance (38).
Indolmycin was again tested in the fully assembled AS TargetArray. As seen with the stage II array, the trpS AS strain was again depleted (∼5 log units). Not surprisingly, indolmycin treatment also strongly depleted an AS clone targeting an operon rich with tRNA genes (see Table S6 in the supplemental material), including the tRNA gene for tryptophan (Fig. S5A in the supplemental material). Thus, AS depletion of both the direct indolmycin target (TrpS) and its substrate, which would logically further deplete levels of charged tRNATrp, results in fitness loss. A novel tRNA synthetase inhibitor (TR000006) known to specifically target methionyl tRNA synthetase (MetS) was also tested (10, 18). Confirming prior findings, the metS AS strain was the most severely depleted (see Fig. S5B in the supplemental material, which shows the structure).
Iclaprim, a new selective inhibitor of dihydrofolate reductase (DfrA) undergoing clinical trials by Arpida Ltd. (Switzerland) for the treatment of serious hospital infections caused by Gram-positive pathogens and respiratory tract infections, was tested (44). Prior proof-of-concept fitness tests found that dfrA overexpression resulted in strong enrichment of the dfrA P-OE clone in the presence of trimethoprim, a DfrA inhibitor (Fig. 7). When iclaprim (TR000010) was tested, the dfrA AS strain was the most severely and reproducibly depleted over a range of concentrations (see Fig. S5C in the supplemental material). An analog of iclaprim was also tested and found to elicit a nearly identical TargetArray fingerprint, with the dfrA AS strain again being the sole strain depleted and was depleted by 4 to 7 log units (data not shown). These results show that the TargetArray can be used as a tool to confirm that chemical analogs act by a mechanism similar to that for the parent compound. In sum, we demonstrate the utility of the TargetArray for elucidating antibiotic MOA.
Conclusion.
This paper describes the comprehensive identification of 308 essential genes in the clinically important pathogen S. aureus. Most of these essential genes are broadly conserved in a wide range of bacterial pathogens. Since antibiotics target essential genes and processes, the genes in this gene set represent key targets for antibiotic drug discovery. For these 308 essential genes, a total of 446 conditionally expressing alleles were constructed. For a subset of these elements, the AS clones, a TargetArray that consists of 236 sensitized strains and that allows rapid and global profiling of antibiotic MOAs was built. In most cases, this resulted in accurate identification of the molecular target or at least the cellular pathway that the antibiotics inhibit. Moreover, TargetArray fitness tests proved to be information rich, as not only the molecular targets were identified but also potential interacting proteins were identified. For example, mupirocin treatment identified the ileS and yacA AS clones (Fig. 9). It should also be noted that nonselective compounds, such as membrane disrupters, denaturants, and others, which commonly arise from whole-cell screens can be profiled. Thus, nuisance compounds, which kill by nonselective mechanisms, can be identified and deprioritized. This platform therefore represents a new and powerful tool to aid with antibiotic drug discovery and has indeed been validated with novel agents (23).
We and others have independently found that AS RNA expression is an effective means to inactivate gene expression in S. aureus (12, 28). In order for AS to work, the AS RNA must pair with its cognate mRNA to block expression or in some cases the activity of an RNA gene product. Primary mechanisms of inhibition include degradation of the antisense-mRNA complex and/or steric hindrance of translation. Criteria for successful implementation of AS include a strong inducible promoter and stable antisense RNA that allows high-level accumulation at steady state. These criteria were met in S. aureus, as the resulting AS RNA was readily detected as a distinct band on ethidium bromide-stained agarose gels whose intensity rivals the intensities of the 16S and 23S rRNA bands. We have tried AS knockdown expression in a variety of Gram-positive and Gram-negative bacteria. These efforts were met with mixed results, particularly in Gram-negative bacteria. One striking difference in RNA metabolism between Gram-negative organisms, such as E. coli and S. aureus, is the lack of an RNase E homolog in S. aureus. On the basis of this information, an AS screen was conducted with an E. coli RNase E mutant and resulted in a significant improvement in the frequency of isolation of expected clones with AS RNA toward essential genes (unpublished data). In sum, the combination of our expression system and S. aureus RNA metabolism makes this species particularly amenable to antisense gene inactivation.
Supplementary Material
Acknowledgments
We thank our collaborators from Merck Research Laboratories for their support, for making Elitra data available for publication, and for continued development and utilization of the TargetArray. R.A.F. served as lead scientist for assembly of the 236 AS TargetArray. We thank John Finn of Trius Therapeutics for kindly providing compounds.
Footnotes
Published ahead of print on 14 June 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Arbeloa, A., J. E. Hugonnet, A. C. Sentilhes, N. Josseaume, L. Dubost, C. Monsempes, D. Blanot, J. P. Brouard, and M. Arthur. 2004. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J. Biol. Chem. 279:41546-41556. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri, R. R., A. G. Allen, P. J. Owen, G. Shalom, K. Stone, M. Harrison, T. A. Burgis, M. Lockyer, J. Garcia-Lara, S. J. Foster, S. J. Pleasance, S. E. Peters, D. J. Maskell, and I. G. Charles. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, D. Z., D. V. Patel, C. J. Hackbarth, W. Wang, G. Dreyer, D. C. Young, P. S. Margolis, C. Wu, Z. J. Ni, J. Trias, R. J. White, and Z. Yuan. 2000. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry 39:1256-1262. [DOI] [PubMed] [Google Scholar]
- 4.DeVito, J. A., J. A. Mills, V. G. Liu, A. Agarwal, C. F. Sizemore, Z. Yao, D. M. Stoughton, M. G. Cappiello, M. D. Barbosa, L. A. Foster, and D. L. Pompliano. 2002. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 20:478-483. [DOI] [PubMed] [Google Scholar]
- 5.Donald, R. G., S. Skwish, R. A. Forsyth, J. W. Anderson, T. Zhong, C. Burns, S. Lee, X. Meng, L. LoCastro, L. W. Jarantow, J. Martin, S. H. Lee, I. Taylor, D. Robbins, C. Malone, L. Wang, C. S. Zamudio, P. J. Youngman, and J. W. Phillips. 2009. A Staphylococcus aureus fitness test platform for mechanism-based profiling of antibacterial compounds. Chem. Biol. 16:826-836. [DOI] [PubMed] [Google Scholar]
- 6.Douthwaite, S., and W. S. Champney. 2001. Structures of ketolides and macrolides determine their mode of interaction with the ribosomal target site. J. Antimicrob. Chemother. 48(Suppl. T1):1-8. [DOI] [PubMed] [Google Scholar]
- 7.Du, W., J. R. Brown, D. R. Sylvester, J. Huang, A. F. Chalker, C. Y. So, D. J. Holmes, D. J. Payne, and N. G. Wallis. 2000. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J. Bacteriol. 182:4146-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eschenburg, S., M. Priestman, and E. Schonbrunn. 2005. Evidence that the fosfomycin target Cys115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J. Biol. Chem. 280:3757-3763. [DOI] [PubMed] [Google Scholar]
- 9.Feng, L., K. Sheppard, S. Namgoong, A. Ambrogelly, C. Polycarpo, L. Randau, D. Tumbula-Hansen, and D. Soll. 2004. Aminoacyl-tRNA synthesis by pre-translational amino acid modification. RNA Biol. 1:16-20. [PubMed] [Google Scholar]
- 10.Finn, J., M. Stidham, M. Hilgers, and G. C. K. 2008. Identification of novel inhibitors of methionyl-tRNA synthetase (MetRS) by virtual screening. Bioorg. Med. Chem. Lett. 18:3932-3937. [DOI] [PubMed] [Google Scholar]
- 11.Forsyth, A., and L. Wang. 2008. Techniques for the isolation and use of conditionally expressed antisense RNA to achieve essential gene knockdowns in Staphylococcus aureus. Methods Mol. Biol. 416:307-321. [DOI] [PubMed] [Google Scholar]
- 12.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto-Nakamura, M., H. Ito, Y. Oyamada, T. Nishino, and J. Yamagishi. 2005. Accumulation of mutations in both gyrB and parE genes is associated with high-level resistance to novobiocin in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3810-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentz, R., and H. Bujard. 1985. Promoters recognized by Escherichia coli RNA polymerase selected by function: highly efficient promoters from bacteriophage T5. J. Bacteriol. 164:70-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaever, G., D. D. Shoemaker, T. W. Jones, H. Liang, E. A. Winzeler, A. Astromoff, and R. W. Davis. 1999. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 21:278-283. [DOI] [PubMed] [Google Scholar]
- 16.Girgis, H. S., A. K. Hottes, and S. Tavazoie. 2009. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One 4:e5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruss, A. D., H. F. Ross, and R. P. Novick. 1987. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc. Natl. Acad. Sci. U. S. A. 84:2165-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond, M. L., A. H. Leeman, M. Maletic, G. M. Santorelli, S. T. Waddell, J. Finn, M. Morytko, J. Hill, and D. Keith. April 2003. Catechols as antimicrobial agents. U.S. patent 6,545,015.
- 19.Haselbeck, R., D. Wall, B. Jiang, T. Ketela, J. Zyskind, H. Bussey, J. G. Foulkes, and T. Roemer. 2002. Comprehensive essential gene identification as a platform for novel anti-infective drug discovery. Curr. Pharm. Des. 8:1155-1172. [DOI] [PubMed] [Google Scholar]
- 20.Heath, R. J., Y. T. Yu, M. A. Shapiro, E. Olson, and C. O. Rock. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273:30316-30320. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 22.Hitchings, G. H. 1973. Mechanism of action of trimethoprim-sulfamethoxazole. I. J. Infect. Dis. 128(Suppl.):433-436. [DOI] [PubMed] [Google Scholar]
- 23.Huber, J., R. G. Donald, S. H. Lee, L. W. Jarantow, M. J. Salvatore, X. Meng, R. Painter, R. H. Onishi, J. Occi, K. Dorso, K. Young, Y. W. Park, S. Skwish, M. J. Szymonifka, T. S. Waddell, L. Miesel, J. W. Phillips, and T. Roemer. 2009. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem. Biol. 16:837-848. [DOI] [PubMed] [Google Scholar]
- 24.Hurdle, J. G., A. J. O'Neill, and I. Chopra. 2004. Anti-staphylococcal activity of indolmycin, a potential topical agent for control of staphylococcal infections. J. Antimicrob. Chemother. 54:549-552. [DOI] [PubMed] [Google Scholar]
- 25.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 26.Ince, D., and D. C. Hooper. 2003. Quinolone resistance due to reduced target enzyme expression. J. Bacteriol. 185:6883-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayasuriya, H., K. B. Herath, C. Zhang, D. L. Zink, A. Basilio, O. Genilloud, M. T. Diez, F. Vicente, I. Gonzalez, O. Salazar, F. Pelaez, R. Cummings, S. Ha, J. Wang, and S. B. Singh. 2007. Isolation and structure of platencin: a FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew. Chem. Int. ed. Engl. 46:4684-4688. [DOI] [PubMed] [Google Scholar]
- 28.Ji, Y., B. Zhang, S. F. Van, Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 29.Kahan, F. M., J. S. Kahan, P. J. Cassidy, and H. Kropp. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235:364-386. [DOI] [PubMed] [Google Scholar]
- 30.Kamionka, A., R. Bertram, and W. Hillen. 2005. Tetracycline-dependent conditional gene knockout in Bacillus subtilis. Appl. Environ. Microbiol. 71:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 32.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 33.Lee, M. S., B. A. Dougherty, A. C. Madeo, and D. A. Morrison. 1999. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl. Environ. Microbiol. 65:1883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, X., M. Zolli-Juran, J. D. Cechetto, D. M. Daigle, G. D. Wright, and E. D. Brown. 2004. Multicopy suppressors for novel antibacterial compounds reveal targets and drug efflux susceptibility. Chem. Biol. 11:1423-1430. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 37.Ondeyka, J. G., D. L. Zink, K. Young, R. Painter, S. Kodali, A. Galgoci, J. Collado, J. R. Tormo, A. Basilio, F. Vicente, J. Wang, and S. B. Singh. 2006. Discovery of bacterial fatty acid synthase inhibitors from a Phoma species as antimicrobial agents using a new antisense-based strategy. J. Nat. Prod. 69:377-380. [DOI] [PubMed] [Google Scholar]
- 38.Pathania, R., S. Zlitni, C. Barker, R. Das, D. A. Gerritsma, J. Lebert, E. Awuah, G. Melacini, F. A. Capretta, and E. D. Brown. 2009. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat. Chem. Biol. 5:849-856. [DOI] [PubMed] [Google Scholar]
- 39.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 40.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price, A. C., K. H. Choi, R. J. Heath, Z. Li, S. W. White, and C. O. Rock. 2001. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J. Biol. Chem. 276:6551-6559. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Suarez, R., D. Xu, K. Veillette, J. Davison, S. Sillaots, S. Kauffman, W. Hu, J. Bowman, N. Martel, S. Trosok, H. Wang, L. Zhang, L. Y. Huang, Y. Li, F. Rahkhoodaee, T. Ransom, D. Gauvin, C. Douglas, P. Youngman, J. Becker, B. Jiang, and T. Roemer. 2007. Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem. Biol. 14:1163-1175. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Schneider, P., S. Hawser, and K. Islam. 2003. Iclaprim, a novel diaminopyrimidine with potent activity on trimethoprim sensitive and resistant bacteria. Bioorg. Med. Chem. Lett. 13:4217-4221. [DOI] [PubMed] [Google Scholar]
- 45.Shoemaker, D. D., D. A. Lashkari, D. Morris, M. Mittmann, and R. W. Davis. 1996. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 14:450-456. [DOI] [PubMed] [Google Scholar]
- 46.Singh, S. B., H. Jayasuriya, J. G. Ondeyka, K. B. Herath, C. Zhang, D. L. Zink, N. N. Tsou, R. G. Ball, A. Basilio, O. Genilloud, M. T. Diez, F. Vicente, F. Pelaez, K. Young, and J. Wang. 2006. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J. Am. Chem. Soc. 128:11916-11920. [DOI] [PubMed] [Google Scholar]
- 47.Soma, A., Y. Ikeuchi, S. Kanemasa, K. Kobayashi, N. Ogasawara, T. Ote, J. Kato, K. Watanabe, Y. Sekine, and T. Suzuki. 2003. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell 12:689-698. [DOI] [PubMed] [Google Scholar]
- 48.Taubes, G. 2008. The bacteria fight back. Science 321:356-361. [DOI] [PubMed] [Google Scholar]
- 49.Tovar, K., A. Ernst, and W. Hillen. 1988. Identification and nucleotide sequence of the class E tet regulatory elements and operator and inducer binding of the encoded purified Tet repressor. Mol. Gen. Genet. 215:76-80. [DOI] [PubMed] [Google Scholar]
- 50.Trawick, J. D., and C. H. Schilling. 2006. Use of constraint-based modeling for the prediction and validation of antimicrobial targets. Biochem. Pharmacol. 71:1026-1035. [DOI] [PubMed] [Google Scholar]
- 51.Ueda, K., R. McMacken, and A. Kornberg. 1978. dnaB protein of Escherichia coli. Purification and role in the replication of phiX174 DNA. J. Biol. Chem. 253:261-269. [PubMed] [Google Scholar]
- 52.von Nussbaum, F., M. Brands, B. Hinzen, S. Weigand, and D. Habich. 2006. Antibacterial natural products in medicinal chemistry—exodus or revival? Angew Chem. Int. ed. Engl. 45:5072-5129. [DOI] [PubMed] [Google Scholar]
- 53.Wang, J., S. Kodali, S. H. Lee, A. Galgoci, R. Painter, K. Dorso, F. Racine, M. Motyl, L. Hernandez, E. Tinney, S. L. Colletti, K. Herath, R. Cummings, O. Salazar, I. Gonzalez, A. Basilio, F. Vicente, O. Genilloud, F. Pelaez, H. Jayasuriya, K. Young, D. F. Cully, and S. B. Singh. 2007. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. U. S. A. 104:7612-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, J., S. M. Soisson, K. Young, W. Shoop, S. Kodali, A. Galgoci, R. Painter, G. Parthasarathy, Y. S. Tang, R. Cummings, S. Ha, K. Dorso, M. Motyl, H. Jayasuriya, J. Ondeyka, K. Herath, C. Zhang, L. Hernandez, J. Allocco, A. Basilio, J. R. Tormo, O. Genilloud, F. Vicente, F. Pelaez, L. Colwell, S. H. Lee, B. Michael, T. Felcetto, C. Gill, L. L. Silver, J. D. Hermes, K. Bartizal, J. Barrett, D. Schmatz, J. W. Becker, D. Cully, and S. B. Singh. 2006. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358-361. [DOI] [PubMed] [Google Scholar]
- 55.Wang, L., J. D. Trawick, R. Yamamoto, and C. Zamudio. 2004. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res. 32:3689-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, D., B. Jiang, T. Ketela, S. Lemieux, K. Veillette, N. Martel, J. Davison, S. Sillaots, S. Trosok, C. Bachewich, H. Bussey, P. Youngman, and T. Roemer. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 3:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin, D., B. Fox, M. L. Lonetto, M. R. Etherton, D. J. Payne, D. J. Holmes, M. Rosenberg, and Y. Ji. 2004. Identification of antimicrobial targets using a comprehensive genomic approach. Pharmacogenomics 5:101-113. [DOI] [PubMed] [Google Scholar]
- 58.Zaman, S., M. Fitzpatrick, L. Lindahl, and J. Zengel. 2007. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol. Microbiol. 66:1039-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, L., F. Fan, L. M. Palmer, M. A. Lonetto, C. Petit, L. L. Voelker, A. St. John, B. Bankosky, M. Rosenberg, and D. McDevitt. 2000. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255:297-305. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Y., and J. E. Cronan, Jr. 1996. Polar allele duplication for transcriptional analysis of consecutive essential genes: application to a cluster of Escherichia coli fatty acid biosynthetic genes. J. Bacteriol. 178:3614-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.