Abstract
The infected cell protein no. 0 (ICP0) of herpes simplex virus 1 (HSV-1) is a promiscuous transactivator shown to enhance the expression of gene introduced into cells by infection or transfection. At the molecular level, ICP0 is a 775-aa ring finger protein localized initially in the nucleus and late in infection in the cytoplasm and mediates the degradation of several proteins and stabilization of others. None of the known functions at the molecular level account for the apparent activity of ICP0 as a transactivator. Here we report that ICP0 functionally interacts with cellular transcription factor BMAL1, a member of the basic helix–loop–helix PER-ARNT-SIM (PAS) super family of transcriptional regulators. Specifically, sequences mapped to the exon II of ICP0 interacted with BMAL1 in the yeast two-hybrid system and in reciprocal pull-down experiments in vitro. Moreover, the enhancement of transcription of a luciferase reporter construct whose promoter contained multiple BMAL1-binding sites by ICP0 and BMAL1 was significantly greater than that observed by ICP0 or BMAL1 alone. Although the level of BMAL1 present in nuclei of infected cells remained unchanged between 3 and 8 h after infection, the level of cytoplasmic BMAL1 was reduced at 8 h after infection. The reduction of cytoplasmic BMAL1 was significantly greater in cells infected with the ICP0-null mutant than in the wild-type virus-infected cells, suggesting that ICP0 mediates partial stabilization of the protein. These results indicate that ICP0 interacts physically and functionally with at least one cellular transcription-regulatory factor.
Most of the products of the α genes of herpes simplex virus 1 (HSV-1) synthesized immediately after infection are highly posttranslationally modified by both viral and cellular kinases, nucleotidylylated by casein kinase II (1–5), and perform multiple functions at various times in the course of the viral replicative cycle (3). Of the five proteins in this group, none present a greater challenge and interest than the infected cell protein no. 0 (ICP0). The major phenotype associated with this protein is enhancement of expression of genes introduced into cells by infection or transfection, although it also has been shown to activate resident cellular genes (3, 6). The gene encoding ICP0, α0, maps in the inverted repeats flanking the long component of HSV-1 DNA (3, 6). The protein contains a ring finger and does not bind DNA (3, 6). In cells infected with mutants lacking both copies of the α0 gene, viral replication is sluggish and viral gene expression beyond that of α genes ensues primarily at high multiplicities of infection (3, 6).
The molecular basis of the phenotype of ICP0 has been the subject of investigation in many laboratories. To date four sets of observations describe the apparent functions of ICP0 in infected cells. In brief, (i) in all cell lines tested, newly synthesized ICP0 is transported into the nucleus. In primary human fibroblasts and in some continuous cell lines, ICP0 is translocated into the cytoplasm from the nucleus (ref. 7 and P. Lopez, C. Van Sant, and B.R., unpublished results). It would appear that ICP0 functions in both nucleus and cytpoplasm.
(ii) In the nucleus, ICP0 localizes with a nuclear structure known as ND10 (8). This structure is associated with proteasomes degradation of proteins and indeed ICP0 mediates the degradation of several cellular proteins. Thus it has been shown that ICP0 binds a ubiquitin-specific protease (USP-7), which deconjugates Sumo-1-conjugated promyelocytic leukemia or Sp100 and targets them for destruction (9). ICP0 also mediates the destruction of CENP-A, CENP-C, DNA-dependent protein kinase (10, 11). With time this list is likely to grow. In recent studies, it has been reported that ICP0 aggregates with ubiquitinated proteins (12).
(iii) ICP0 binds, stabilizes cyclin D3, and translocates it to the ND10 structures (13, 14). The ICP0 mutant containing the D199A substitution colocalizes with ND10 and mediates the degradation of the proteins listed above but no longer binds stabilizes or colocalizes with cyclin D3 (14, 15). The mutant also is not translocated into the cytoplasm (14). At a biological level, the D199A mutant virus grows less well in contact inhibited cells and is less virulent in mice (15). The stabilization of cyclin D3 is not associated with expression of S phase proteins because cdc2 is not activated and E2F proteins appear to be nonfunctional (16, 17). In recent studies, it has been shown that the translocation from the nucleus to the cytoplasm is mediated by cyclin D3 and requires a function expressed by γ2 proteins whose synthesis requires the onset of viral DNA synthesis (ref. 14 and P. Lopez, C. Van Sant, and B.R., unpublished data). At least two lines of evidence suggest that the translocated ICP0 is dynamically associated with proteasomal subunits. First, ICP0 translocated into the cytoplasm of cells infected with wild-type virus is relocated back to the nucleus on exposure of cells to proteasome inhibitors (e.g., MG132) (P. Lopez, C. Van Sant, and B.R., unpublished data). Second, in cells infected with a mutant lacking ICP4, the major regulatory protein of HSV-1, ICP0 is translocated into the cytoplasm and aggregates in vesicle-like structures very early in infection. In these cells, addition of MG132 causes proteasomal subunits to form a shell surrounding the aggregated ICP0.
(iv) The only function associated with the ICP0 translocated into the cytoplasm is its interaction with the translation elongation factor 1δ (EF-1δ) (7). Although the role of ICP0 in this interaction is not known, the data indicate that herpesviruses target EF-1δ for hyperphosphorylation. Thus following the observation that EF-1δ is phosphorylated by the HSV-1 protein kinase UL13, our laboratories reported that members of all three subfamilies of herpesvirus hyperphosphorylate EF-1δ (18, 19). Hyperphosphorylation of EF-1δ occurs in the course of development and is associated with high level synthesis of proteins (20, 21).
With the possible exception of the interaction of ICP0 with EF-1δ, the functions of ICP0 described above are associated with selective stabilization or destruction of cellular proteins. In large part they may be reconciled with blocking cell division and potential cellular responses inimical to viral replication. The link between the known functions of ICP0 and its major characteristic: promiscuous activation of viral and cellular genes introduced by infection or transfection is not apparent. In this report, we provide a potential link: the evidence that ICP0 interacts functionally with the transcriptional factor BMAL1.
Materials and Methods
Cells and Viruses.
HEp-2, Vero, SK-N-SH, rabbit skin, and 143TK− cell lines have been described (13, 22). HSV-1(F), a limited-passage isolate, is the prototype strain used in our laboratories (23). The constructions of HSV-1 recombinant viruses HSV-1(F)Δ305, R7205, R7910, and R7911 have been reported (15, 18, 24, 25). All viruses except R7910 were propagated and titered in Vero cells. R7910 was grown and titered in a cell line (N3) expressing ICP0 (13).
Plasmids.
pBH1010, containing a 2.3-kbp partial cDNA encoding human BMAL1 codons 95–627, was isolated in a yeast two-hybrid screen. pBH1015 and pBH1016 contained BMAL1 codons 95–444 and 95–311, respectively, fused in-frame with GAL4 activation domain in pGADGH. pRB4994 and pRB4995, expressing glutathione S-transferase (GST) fused to amino-terminal and carboxyl-terminal domains of ICP0, respectively, have been described (7). pBH1023 and pBH1024 contained BMAL1 codons 95–450 and 467–627, respectively, fused to GST. Mammalian expression vectors for BMAL1 (PL832) and MOP4 (PL833), and a reporter construct (PL880) to monitor transcriptional activity of BMAL1 have been described (26) and kindly provided by C. Bradfield, University of Wisconsin, Madison. To generate pTARGETΔ, PL832 was digested with SmaI and XhoI, treated with T4 DNA polymerase, and religated. pTA-ICP0 was constructed by cloning α0 cDNA (a kind gift from S. Silvertein, Columbia University, New York) into pTARGETΔ to yield pTA-ICP0. pME-BMAL1(F) was described (27). pEgr-PolyA in UL3–4 was constructed by inserting into blunt-ended BamHI site of pRB422 (24), the blunt-ended SphI fragment of pRB5160 (13) containing the Egr-1-promoter region, multicloning sites, and the bidirectional polyadenylation signals of HSV-1 UL21 and UL22 genes. The full-length of BMAL1 cDNA was inserted into pEgr-PolyA in UL 3–4 to generate pBMAL1/UL 3–4.
Yeast Two-Hybrid Screen.
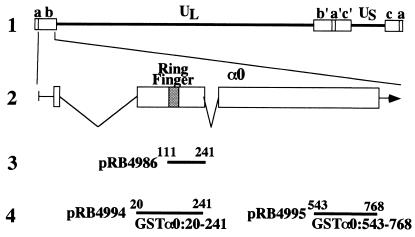
The ICP0 sequences encoded in pRB4986 (Fig. 1) served as bait. The yeasts were transformed with an Epstein–Barr virus-transformed human peripheral blood lymphocyte cDNA library (CLONTECH) as described (7, 13).
Figure 1.
(A) Schematic diagram of the sequence of the HSV-1 genome and the location of the α0 gene. Line 1, a linear representation of the HSV-1 genome. The unique sequences are represented as the regions. The terminal repeats flanking the unique long (UL) and unique short (US) sequences are shown as open rectangles with their designation letters shown above. Line 2, an expanded section of the domain encoding α0 gene. The transcribed and coding domains are shown for one copy of the α0 gene. An identical copy is located in the inverted repeat (b′) flanking UL. Line 3, the region used as “bait” in the yeast two-hybrid screen. Line 4, the domains of the α0 gene fused to GST to generate chimeric proteins.
Production and Purification of GST Fusion Protein.
GST fusion proteins were expressed in Escherichia coli BL21 transformed with either pRB4994, pRB4995, pBH1023, pBH1024, or pGEX4T-1, purified on glutathione-Sepharose beads (Amersham Pharmacia) and quantified as described (7).
Transient Expression Assays.
COS-7 cells were transfected with the expression vectors, the reporter construct PL880, or the βgalactosidase control plasmid pRSV-LacZ by the DEAE-dextran method (27) and harvested 2 days after transfection. Cell extraction, luciferase assays, and β-galactosidase assays were performed according to manufacturer's instructions (Promega).
Antibodies and Immunoblotting.
Polyclonal antibody to BMAL1 was made in rabbits immunized with purified GST-BMAL1467–627 by standard protocols at Josman Laboratories, Napa, CA (13). The serum used in this report here was collected 97 days after first immunization. The mouse mAb to ICP0 (H1083) and Flag epitope (M2) have been described (7, 27).
Cell Fractionation.
Nuclear and cytoplasmic fractions of SK-N-SH cells were obtained as described (7).
Construction of the Recombinant Virus YK101.
Recombinant virus YK101 was constructed by cotransfection of rabbit skin cells with intact R7205 viral DNA and pBMAL1/UL3–4 as described (13).
Results
ICP0 Interacts with a Transcription Factor BMAL1 in a Yeast Two-Hybrid System.
The evidence that ICP0 interacted with BMAL1 in the yeast two-hybrid studies was as follows. Of 4.6 × 106 transformants, 31 displayed strong expression of β-galactosidase activity. Partial sequence analyses and restriction endonuclease analyses of plasmids obtained from the positive yeast revealed that two plasmids (pBH1010) contained partial cDNAs encoding BMAL1, a member of basic helix–loop–helix–PER-ARNT-SIM (bHLH-PAS) transcription factors (28, 29). The positive plasmids retransformed with various plasmids into yeast yielded evidence of positive interactions of the cDNA products with ICP0 and negative interaction with human lamin, murine p53, or the GAL4 DNA-binding domain alone.
ICP0 Specifically Forms Complexes with BMAL1 in Reciprocal GST-Pull-Down Experiments.
To verify and extend the observed interaction between ICP0 and BMAL1 in yeast, we expressed various GST fusion proteins in E. coli and performed two series of GST pull-down experiments.
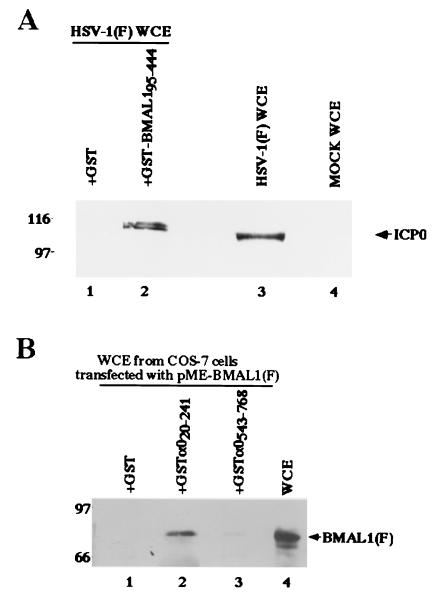
In the first series of experiments, we examined whether GST-BMAL195–450 pulls down ICP0 in HSV-1(F)-infected cells. Preliminary experiments indicated that the BMAL1 domain codons 95–450 interacted with ICP0 in the yeast two-hybrid system (data not shown). This portion of BMAL1 encodes a part of bHLH and PAS domains that are reported to be involved in dimerization with other proteins (30–32). The GST-BMAL195–450 or GST protein bound to glutathione-Sepharose beads was reacted with an extract from HSV-1(F)-infected HEp-2 cells and processed for detection of ICP0 by immunoblot analysis. As shown in Fig. 2A, the GST-BMAL195–450 fusion protein was able to pull down in ICP0, whereas GST, alone, did not. The electrophoretic mobility of the ICP0 that complexes with GST-BMAL195–450 fusion protein was similar to that present in infected cells. These results indicate that BMAL1 can interact with native, full-length ICP0 and a part of bHLH, and PAS domains of BMAL1 mediate this interaction.
Figure 2.
(A) Photographic image of an immunoblot of infected cell proteins bound to GST or chimeric GST-BMAL1 fusion protein, electrophoretically separated in a denaturing gel and reacted with a mouse mAb to ICP0 (H1083). Lysates of infected HEp-2 were reacted with GST or GST-BMAL1 chimeric protein immobilized on glutathione-agarose beads. The beads were pelleted, rinsed extensively, subjected to electrophoresis on a denaturing gel, transferred to a nitrocellulose sheet, and reacted with the ICP0 antibody. Lines 1 and 2, whole-cell extracts (WCE) from HSV-1(F)-infected HEp-2 cells reacted with GST or GST-BMAL1, respectively; lanes 3 and 4, whole-cell extracts from HSV-1(F)-infected or mock-infected HEp-2 cells, respectively. Molecular weights (×1,000) are shown on the left. (B) Photographic image of an immunoblot of proteins extracted from transfected cells and bound to GCT, GST-α020–241, or GST-α0543–768, electrophoretically separated in a denaturing gel, and reacted with a mouse mAb to the Flag epitope (M2). Lysates of COS-7 cells transfected with pME-BMAL1(F) were reacted with GST or GST-ICP0 chimeric protein immobilized on glutathione-Sepharose beads. The beads were pelleted, rinsed extensively, subjected to electrophoresis on a denaturing gel, transferred to a nitrocellulose sheet, and reacted with the anti-Flag epitope antibody. Lanes 1, 2, and 3, whole-cell extracts from COS-7 cells transfected with pME-BMAL1(F) bound to GST, GST-α020–241, and GST-α0543–768, respectively; lane 4, whole-cell extracts from COS-7 cells transfected with pME-BMAL1(F). Molecular weights (103) are shown on the left.
In the second series of experiments, the carboxyl-terminal domain (GST-α0543–768) or amino-terminal domain (GST-α020–241) of ICP0 fused to GST (Fig. 1) was expressed in E. coli and purified as described (7), reacted with cell extracts of COS-7 cells expressing full-length of BMAL1 tagged with Flag-epitope, and processed for detection of BMAL1 by immunoblot analysis. The results (Fig. 2B) were that GST-α020–241 was able to pull down BMAL1, whereas GST alone or GST-α0543–768 did not. The electrophoretic mobility of the BMAL1 that complexes with GST-α020–241 fusion protein was similar to that found in extracts of whole transfected cells. The faster migrating band of BMAL1 detected in Fig. 2B, lane 4, might be the degraded product because of overexpression of the protein in COS-7 cells. These results indicate that ICP0 can interact with full-length of BMAL1 and further affirm the interaction of ICP0 with BMAL1.
ICP0-BMAL1 Complex Drives Transcription in Mammalian Cells.
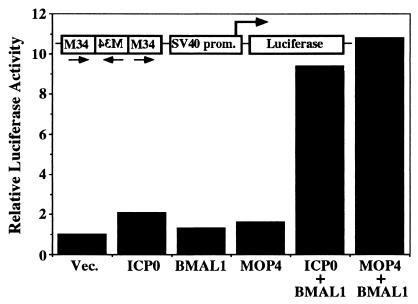
The bHLH PAS proteins interact with appropriate dimeric partners through their bHLH and PAS domains and function as transcription factors (30–32). The evidence that BMAL1 interacts with ICP0 through its bHLH and PAS domains suggested the possibility that the ICP0-BMAL1 complex functions as a transcription complex to enable transcription. To test this hypothesis, we carried out luciferase-reporter assays in COS-7 cells transfected with the expression plasmids for ICP0, BMAL1, and MOP4, and a reporter gene plasmid PL880 (26) containing three copies of the BMAL1-binding sites upstream of a minimal simian virus 40 promoter-luciferase reporter (Fig. 3). Luciferase activity was assayed at 48 h after transfection. As reported elsewhere (26), coexpression of BMAL1 and MOP4 resulted in approximately 10-fold activation of reporter gene, whereas neither BMAL1 nor MOP4 alone were capable of driving the transcription of the reporter construct above control levels (Fig. 3). Similarly, ICP0 by itself had little effect, whereas ICP0 and BMAL1 significantly enhanced transcription of the reporter construct (Fig. 3). These results indicated that ICP0 forms transcriptionally active complex with BMAL1 in mammalian cells.
Figure 3.
ICP0-BMAL1 complex enhances transcription of mRNA from the reporter gene containing BMAL1 response elements. COS-7 cells were transfected with the M34 reporter plasmid and pRSV-LacZ in combination with expression plasmid as indicated. Luciferase activity was assayed in cells that were harvested 48 h after transfection as described in Materials and Methods. Luciferase activity were normalized with that of β-galactosidase. The results represent averages of three independent experiments.
Identification of Endogenous BMAL1 in SK-N-SH Cells.
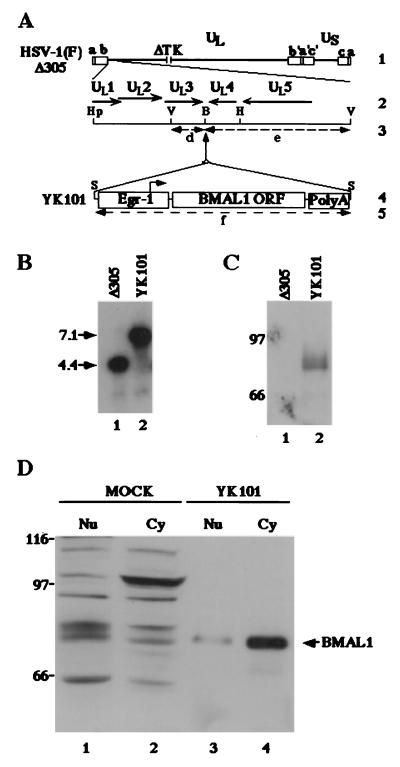
To facilitate identification of endogenous BMAL1, we constructed recombinant virus YK101, in which a BMAL1 expression cassette was inserted into the context of viral genome as described in Materials and Methods. Viral DNAs prepared from cells infected with HSV-1(F)Δ305 or the recombinant virus YK101 were analyzed for the presence of the BMAL1 expression cassette by Southern blotting. As could be predicted from the details of the construction of these viruses (Fig. 4A), the 32P-labeled probe hybridized to fragment d + e (4.4 kbp) in HSV-1(F)Δ305 (Fig. 4B, lane 1) and d + e + f (7.1 kbp) in YK101 (Fig. 4B, lane 2). To assay for expression of BMAL1, Vero cells were infected with 10 plaque-forming units (pfu) of HSV-1(F)Δ305 or YK101 per cell. The lysates of cells harvested at 24 h after infection were subjected to immunoblotting probed with the BMAL1 polyclonal antibody. As shown in Fig. 4C, the antibody reacted with a protein with an Mr of 80,000 in cells infected with YK101 whereas BMAL1 expression was not detected in HSV-1(F)Δ305-infected cells. We could not differentiate between YK101 or its control virus in which only the Egr-1 promoter and polyadenylation sequences were inserted between UL3 and UL4 with respect to virus yields from either HEp-2 or SK-N-SH cells infected at high or low multiplicities of infection (data not shown). With the aid of the polyclonal antibody to BMAL1 and the recombinant virus YK101, we tested various cell lines for expression of endogenous BMAL1 and found that a neuroblastoma cell line, SK-N-SH, clearly expresses BMAL1. Therefore, we used SK-N-SH cells for further experiments.
Figure 4.
(A) Schematic diagram of the sequence of HSV-1(F)Δ305 and the recombinant virus, YK101, derived from it. Line 1, sequence arrangement of the HSV-1(F)Δ305 DNA. Line 2, an enlarged portion of the domain of HSV-1 encoding UL1–UL5. The coding region and direction of the genes are indicated. Line 4, sequence of the recombinant virus, YK101, which was selected, as described in Materials and Methods, from among the progeny of transfection of rabbit skin cells with intact HSV-1(F)Δ305 DNA and plasmid pBMAL1 in UL3–4. The promoter sequence of Egr-1, the BMAL1 coding sequence, and the sequence containing bidirectional poly(A) signals of UL21 and UL22 of HSV-1(F) are shown as open rectangles. The arrow indicates the site of transcription initiation and direction of transcription. Lines 3 and 5 show expected sizes of DNA fragments generated by cleavage of the DNA. The fragment designations shown here are identical to those described in the text and in B. Restriction sites: B, BamHI site; Hp, HpaI site; S, SphI site; and V, EcoRV. (B) Characterization of an HSV-1 recombinant virus expressing the BMAL1 gene (YK101). Autoradiographic image of electrophoretically separated EcoRV digests of HSV-1(F)Δ305 or YK101 DNAs, hybridized to the radiolabeled EcoRV-HindIII fragment of pRB442 that contained sequences of UL3 and UL4. (C) Lane 1, digest of DNA of HSV-1(F)Δ305. Lane 2, digest of DNA of recombinant virus YK101. Molecular weights (×1,000) are shown on the left. (D) Photographic image of an immunoblot of electrophoretically separated lysates of HSV-1(F)Δ305 (lane 1) or YK101 (lane 2) infected with Vero cells harvested 24 h after infection. The blot was probed with the anti-BMAL1 polyclonal antibody. Molecular weights (×1,000) are shown on the left. (D) Photographic image of an immunoblot of electrophoretically separated cell fractions of SK-N-SH cells mock-infected (lanes 1 and 2) or infected with YK101 (lanes 3 and 4). Nuclear (Nu; lanes 1 and 3) and cytoplasmic (Cy; lanes 2 and 4) fractions of infected SK-N-SH cells harvested 24 h after infection were prepared as described in Materials and Methods and reacted with the anti-BMAL1 polyclonal antibody. BMAL1-specific band is indicated on the right, and molecular weights (×1,000) are shown on the left.
SK-N-SH cells mock-infected or infected with YK101 were harvested at 24 h. The nuclei and cytoplasms were separated as described (7), and each fraction was electrophoretically separated in a denaturing gel and reacted with the polyclonal antibody to BMAL1. Although the rabbit antiserum to BMAL1 detected several bands in both nuclear and cytoplasmic fraction of mock-infected SK-N-SH cells, the electrophoretic mobility of a protein with Mr of 80,000 was identical to that of a protein overexpressed and detected by the BMAL1 antibody in cells infected with YK101 (Fig. 4D).
ICP0 Is Involved in the Loss of Cytoplasmic BMAL1 Induced by HSV-1 Infection in Infected SK-N-SH Cells.
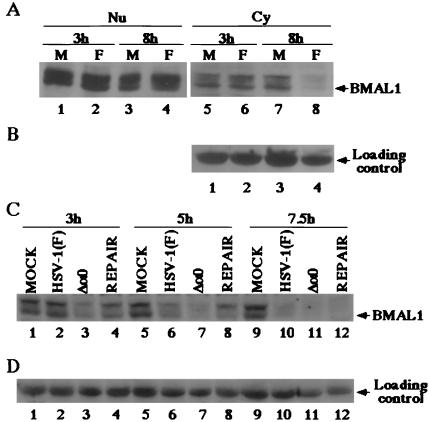
To monitor the level of BMAL1 expression in infected cells, SK-N-SH cells mock-infected or infected with 10 pfu of HSV-1(F) per cell were harvested 3 or 8 h after infection. The nuclear and cytoplasmic fractions were isolated as described (7), electrophoretically separated in a denaturing polyacrylamide gel, and reacted with the polyclonal antibody to BMAL1. The results (Fig. 5A) were as follows: BMAL1 levels remained relatively unchanged and similar in nuclei of mock-infected or infected cells at 3 and 8 h after infection. In the cytoplasmic fractions, the decrease in BMAL1 between 3 and 8 h was greater in infected cells that in mock-infected cells. In the same immunoblot shown in Fig. 5B, a cellular protein with Mr of approximately 55,000 that reacted with the BMAL1 polyclonal antibody served as a loading control in this experiment. These results indicate that HSV-1 infection induces specific loss of cytoplasmic BMAL1 between 3 and 8 h after infection.
Figure 5.
(A) Photographic images of electrophoretically separated cell fraction of SK-N-SH cells mock-infected (M) or infected with 10 pfu of HSV-1 (F) per cell and reacted with the rabbit polyclonal antibody to BMAL1. (B) Levels of expression of loading control in the cytoplasmic fractions. Fractions of the infected cells harvested at indicated times were prepared as described in Materials and Methods and reacted with the anti-BMAL1 antibody. (C) Photographic images of electrophoretically separated cytoplasmic fractions of SK-N-SH cells mock-infected or infected with 10 pfu of HSV-1 (F), R7910 (Δa0), or R7911 (REPAIR) per cells and reacted with the polyclonal antibody to BMAL1. (D) The levels of expression of loading control.
Earlier publications reported that ICP0 controls stabilization of various cellular proteins (10, 11, 13). These results led us to examine whether ICP0 is involved in the loss of the cytoplasmic BMAL1 as well. SK-N-SH cells mock-infected or infected with 10 pfu of HSV-1(F), R7910, or R7911 per cell were harvested at 3, 5, or 7.5 h after infection. The cytoplasmic fractions were isolated as described in Materials and Methods, electrophoretically separated in a denaturing gel, and reacted with the anti-BMAL1 polyclonal antibody. The results (Fig. 5C) were as follows. At 3 and 5 h after infection the amount of cytoplasmic BMAL1 in cells infected with R7910 (α0-) mutant was significantly lower than those in cells infected with wild-type or the recombinant R7911 in which the ICP0 sequences were restored. At 7.5 h after infection, cytoplasmic BMAL1 was detected in mock-infected cells but not in any of the infected cell cultures. The cellular protein with Mr of approximately 55,000 reacted with the BMAL1 antibody and was present in equal amounts in all lanes (Fig. 5D). The levels of nuclear BMAL1 in ICP0-null mutant-infected cells was unchanged, compared to those in wild-type virus infected cells at 7.5 h after infection (data not shown). These results indicate that under conditions of cell maintenance to which mock-infected and infected cells were subjected caused a decrease of BMAL1 from the cytoplasm. ICP0 mediated a decrease in the rate of disappearance of ICP0 from the cytoplasm of these cells. The results suggest therefore that ICP0 mediates the stabilization of cytoplasmic BMAL1 in infected cells.
Discussion
In infected cells, the function of ICP0 is best described as that of a promiscuous transactivator. The studies done since its discovery have shed little light on the molecular basis of its apparent function in infected cells. Indeed, the data indicate that ICP0 is a multifunctional protein, that it regulates the stability of several cellular proteins through proteasomal interaction and it has both nuclear and cytoplasmic functions. This report presents evidence of physical and functional interaction with a transcription factor, BMAL1.
BMAL1 is a member of the bHLH-PAS family of transcriptional regulators. Members of this family are characterized by a bHLH motif contiguous with a conserved domain called PAS. They have been shown to bind to specific DNA sequences and form homo- or hetero-dimers through the bHLH and PAS with other bHLH-PAS transcription factors including MOP4, hypoxia-inducible factor (HIF)-1α, and HIF-2α, raising the possibility that BMAL1 is a general partner of a number of bHLH-PAS transcriptional factors (26). Members of this family play a role in development and adaptation to the environmental changes (30, 33–36). Although BMAL1 is expressed at highest levels in the brain, thymus, and muscle, it is present in most tissues (26, 28). Curiously, most of the literature on BMAL1 centers on the observations that BMAL1 forms a heterodimer with CLOCK and regulates circadian oscillation in mammalian clock system (37, 38).
In this report, we present two lines of evidence that ICP0 physically and functionally interacts with BMAL1. Specifically, (i) the domain encoded by exon II of ICP0 interacted with BMAL1 in the yeast two-hybrid system and GST-exon II pulled down Flag epitope-tagged BMAL1 transiently expressed in COS-7 cells. In a reciprocal experiment, GST-BMAL1 pulled down ICP0 from lysate of HEp-2 cell infected with HSV-1(F). (ii) ICP0 and BMAL1 together enhanced the expression of a chimeric gene consisting of multiple copies of the BMAL1-binding sites upstream of a minimal simian virus 40 promoter driving a luciferase reporter gene to a level significantly higher than either BMAL1 or ICP0 alone. These results are compelling evidence of a physical and functional interaction between ICP0 and BMAL1. Two caveats, however, should be noted. First, notwithstanding the string evidence of interaction between ICP0 and BMAL1 in yeast, in vitro and in mammalian cells, there is no direct evidence that ICP0 binds directly to BMAL1. HSV regulatory proteins sometimes associate with the other protein with the aid of bridging protein. Thus ICP0 complexes with another HSV-1 protein, ICP22, via a cellular protein designated p60 whose function is presently unknown (39). Second, although this study shows that ICP0 interacts with and stimulates the transcriptional activity of BMAL1, it remains to be proven that ICP0-BMAL1 complex acts as a transcriptional activator in the context of BMAL1 response elements native in infected cells. Transcription factors can act both as a transactivator and repressor and the phenotypes of the factor depend on the design of the experiments. Thus ICP4 acts both as a transactivator and a repressor depending on the nature and position of response elements in the viral genome (3). The possibility that ICP0 modulates BMAL1 negatively stems also from the evidence that BMAL1 is a transactivator that is active in stress responses that may be inimical to viral replication.
In this report, we also have demonstrated that BMAL1, like other transactivators, partitions in both nucleus and cytoplasm. It is thought that the inactive forms of BMAL1 accumulate in the cytoplasm and are transported to the nucleus on activation (28). Our data indicate that in wild-type virus-infected cells, BMAL1 accumulates in the cytoplasm for longer intervals of time than in cells infected with ICP0 null mutants. Hypotheses designed to explain the maintenance of BMAL1 in the cytoplasm of infected cells must take into account the observation that ICP0 is translocated from nuclei into the cytoplasm beginning ≈5 h after infection and reaching 85% completion by 9 h after infection (P. Lopez, C. Van Sant, and B.R., unpublished data). The decline of BMAL1 in the cytoplasm of cells infected with ICP0 null mutant was observed as early as 3 h after and in both mutant and wild-type virus-infected cells BMAL1 disappears by 7 h after infection. ICP0 appears to mediate the maintenance of cytoplasmic BMAL1 for several hours just before the time of transport of the bulk of ICP0 to the cytoplasm. Among the possible explanations of the data are the following: (i) ICP0 translocated into the cytoplasm early in this process binds BMAL1 and that these complexes perform a function in the cytoplasm of infected cells that is distinct from those normally performed by BMAL1. HSV-1 proteins have been shown to divert cellular proteins to perform novel functions. Thus the γ134.5 protein binds and diverts protein phosphatase 1 to dephosphorylate eIF-2α (40). Another example is that the interaction of ICP0 with cyclin D3 leads eventually to the translocation of ICP0 to the cytoplasm and not to activation of the S phase of the cell cycle (14, 16, 17). Once this function is completed maintenance of the complex is not longer essential. (ii) ICP0-BMAL1 function is restricted to the nucleus relatively early in infection. The extended maintenance of cytoplasmic BMAL1 reflects the alteration in the function of proteasomes that is mediated by ICP0. (iii) The extended maintenance of cytoplasmic BMAL1 reflects the need to maintain a reservoir of BMAL1 at least during the early stages of viral DNA synthesis. Less compelling is the hypothesis that ICP0 shuttles BMAL1 from the nucleus because at times when ICP0 is largely cytoplasmic the level of BMAL1 in the nucleus remains unchanged.
Overall, the status of cytoplasmic BMAL1 remains unclear and subject to a better understanding of the function of BMAL1-ICP0 complexes in infected cells. Further studies to unveil the direct significance of the interaction of ICP0 with BMAL1 in infected cells and in vivo are of interest.
Acknowledgments
We thank C. A. Bradfield for the plasmids (PL832, PL833, and PL881), K. Maruyama for pME18S, and C. Van Sant for invaluable discussions. The studies completed at Tokyo Medical and Dental University were supported in part by grants for Scientific Research (Y.K., K.H., and H.K.) from the ministry of Education, Science, Sports and Culture of Japan and Japan Science and Technology Cooperation. Y.K. was supported by a grant from the Inamori Foundation. The studies at the University of Chicago were aided by grants from the National Cancer Institute (CA47451, CA71933, and CA78766) and the United States Public Health Service.
Abbreviations
- HSV-1
herpes simplex virus 1
- ICP0
infected cell protein no. 0
- EF-1δ
elongation factor-1δ
- GST
glutathione S-transferase
- pfu
plaque-forming unit
- bHLH
basic helix–loop–helix
- PAS
PER-ARNT-SIM homology domain
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041592598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041592598
References
- 1.Ogle W O, Ng T I, Carter K L, Roizman B. Virology. 1997;235:406–413. doi: 10.1006/viro.1997.8710. [DOI] [PubMed] [Google Scholar]
- 2.Purves F C, Roizman B. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roizman B, Sears A E. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J C, Monath J P, Roizman B, editors. Philadelphia: Lippincott; 1996. pp. 2231–2295. [Google Scholar]
- 4.Blaho J A, Mitchell C, Roizman B. J Virol. 1993;67:3891–3900. doi: 10.1128/jvi.67.7.3891-3900.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell C, Blaho J A, McCormick A L, Roizman B. J Biol Chem. 1997;272:25394–25400. doi: 10.1074/jbc.272.40.25394. [DOI] [PubMed] [Google Scholar]
- 6.Everett R D, Preston C M, Stow N D. Functional and Genetic Analysis of the Role of Vmw 110 in Herpes Simplex Virus Replication. Boston: CRC; 1991. [Google Scholar]
- 7.Kawaguchi Y, Bruni R, Roizman B. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maul G G, Guldner H H, Spivack J G. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 9.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett R D, Earnshaw W C, Findlay J, Lomonte P. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett R D. J Virol. 2000;74:9994–10005. doi: 10.1128/jvi.74.21.9994-10005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Sant, C., Lopez, P., Advani, S. & Roizman, B. (2000) J. Virol. in press. [DOI] [PMC free article] [PubMed]
- 15.Van Sant C, Kawaguchi Y, Roizman B. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Advani S J, Weichselbaum R R, Roizman B. Proc Natl Acad Sci USA. 2000;97:10996–11001. doi: 10.1073/pnas.200375297. . (First Published September 19, 2000; 10.1073/pnas.200375297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Advani S J, Weichselbaum R R, Roizman B. J Virol. 2000;74:7842–7850. doi: 10.1128/jvi.74.17.7842-7850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi Y, Matsumura T, Roizman B, Hirai K. J Virol. 1999;73:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter J D, Wasserman W J, Smith L D. Dev Biol. 1982;89:159–167. doi: 10.1016/0012-1606(82)90304-9. [DOI] [PubMed] [Google Scholar]
- 21.Wasserman W J, Richter J D, Smith L D. Dev Biol. 1982;89:152–158. doi: 10.1016/0012-1606(82)90303-7. [DOI] [PubMed] [Google Scholar]
- 22.Chou J, Roizman B. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ejercito P M, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 24.Baines J D, Roizman B. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Post L E, Mackem S, Roizman B. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 26.Hogenesch J B, Gu Y Z, Jain S, Bradfield C A. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi Y, Nakajima K, Igarashi M, Morita T, Tanaka M, Suzuki M, Yokoyama A, Matsuda G, Kato K, Kanamori M, Hirai K. J Virol. 2000;74:10104–10111. doi: 10.1128/jvi.74.21.10104-10111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda M, Nomura M. Biochem Biophys Res Commun. 1997;233:258–264. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 30.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 31.Matsushita N, Sogawa K, Ema M, Yoshida A, Fujii-Kuriyama Y. J Biol Chem. 1993;268:21002–21006. [PubMed] [Google Scholar]
- 32.Reyes H, Reisz-Porszasz S, Hankinson O. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 33.Burbach K M, Poland A, Bradfield C A. Proc Natl Acad Sci USA. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman E C, Reyes H, Chu F F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 35.Jackson F R, Bargiello T A, Yun S H, Young M W. Nature (London) 1986;320:185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- 36.Wang G L, Jiang B H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 38.King D P, Takahashi J S. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 39.Bruni R, Fineschi B, Ogle W O, Roizman B. J Virol. 1999;73:3810–3817. doi: 10.1128/jvi.73.5.3810-3817.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He B, Gross M, Roizman B. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]