Abstract
We assessed the natural genotypic and phenotypic susceptibilities to enfuvirtide of 171 HIV group O (HIV-O) samples and 29 strains, respectively. The N42D resistance-associated mutation in the gp41 region was detected in 98% of cases. The phenotypic assay showed a wide range of baseline susceptibilities, with 50% inhibitory concentrations (IC50s) from 4 to 5,000 nM, a range similar to that reported for HIV-1 group M. Thus, despite the natural genotypic resistance conferred by the N42D signature mutation, HIV-O variants appear to be phenotypically susceptible. Enfuvirtide could therefore potentially be used in antiretroviral treatments for HIV-O-infected patients.
Human immunodeficiency virus (HIV) is characterized by high levels of genetic diversity within HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is subdivided into four groups: M (major), O (outlier), N (non-M, non-O), and P. HIV-1 group O (HIV-O) is endemic in Western Central Africa, including Cameroon in particular, where the prevalence of this group is estimated at about 1% of all HIV infections (about 10,000 patients) (2, 26). The prevalence of HIV-O remains low, with limited spread, outside Western Central Africa. In France, the first HIV-O-infected patient was described in 1992 (1), and the viral strain was characterized in 1994 (3). As part of our surveillance activities on divergent variants of HIV, we set up, in 2005, a network (RES-O) of physicians and biologists for the surveillance and monitoring of HIV-O infections in France. This network has identified 123 patients (6; also our unpublished data), most of whom were originally from Cameroon or had a partner originating from Cameroon.
HIV-O displays strong genetic divergence from HIV-1 group M and a high degree of intragroup diversity (22, 28). This divergence has clinical implications, complicating the processes of diagnosis (18) and virological monitoring (11, 19), and necessitating the use of adapted techniques and treatment strategies. Indeed, HIV-O is considered to be naturally resistant to nonnucleoside reverse transcriptase inhibitors (NNRTI), due to the presence of the Y181C mutation (6). Few studies have assessed the virological response to treatment regimens containing nucleoside reverse transcriptase inhibitors (NRTI) and protease inhibitors (PI) (5, 21), and this response has not been evaluated for the most recently developed drugs.
Enfuvirtide (T20), an HIV-1 fusion inhibitor, is a peptide based on the sequence of the heptad repeat 2 (HR2) region of the gp41 transmembrane glycoprotein derived from the subtype B HIV-1LAI sequence. It was not clear whether T20 would be active against HIV-O, because this peptide was designed to work against HIV-1 group M. Furthermore, the reported natural presence in HIV-O of an N42D mutation (4, 20), associated with resistance in HIV-1 group M according to the interpretation of the algorithm defined by the ANRS (http://hivfrenchresistance.org) and REGA (http://www.rega.kuleuven.be/cev/), whether in isolation or associated with other mutations, would be expected to compromise the activity of T20 in HIV-O variants (14, 15, 16, 23). However, preliminary data showed that T20 was active against six HIV-O primary isolates in vitro (4) and in one HIV-O-infected patient in vivo (20). This apparent discordance with the expected findings and the limited number of cases of HIV-O infection evaluated in these studies precluded the development of a reliable representation of the high level of intragroup diversity.
In this study, we assessed the genotypic susceptibility to T20 of a large panel of HIV-O samples (n, 171) representative of the diversity of group O. Phenotypic susceptibility to T20 was evaluated for 29 of these samples, and the results obtained were compared with the genotypic interpretation. None of the patients studied had ever been treated with T20, and all were identified by the French RES-O network (n, 91) and through collaboration with the Centre Pasteur du Cameroun in Yaoundé (n, 80).
Genotypic assays involved amplification of the whole ectodomain of gp41, residues 1 to 179, encompassing the HR1 and HR2 regions and the start of the transmembrane region. Viral RNA was extracted from 142 cell-free plasma samples and 29 cell-free viral supernatant samples (obtained by coculture [see below]) and was subjected to reverse transcription-PCR with the primers and cycling conditions described elsewhere (17, 26). We developed a phenotypic test adapted to the gp41 region, using a peripheral blood mononuclear cell (PBMC) assay, as previously described (8). Briefly, 29 strains were isolated by coculturing PBMCs from HIV-O-infected patients with PBMCs from healthy donors. We used 1 ml of the viral supernatant, corresponding to a viral density of 1 × 106 copies/ml and one 50% tissue culture infective dose (TCID50) per ml of sample, to infect cells in the presence of various concentrations of T20 (range, 4.5 nM to 12,000 nM). Viral replication was then evaluated by an HIV-O-specific real-time PCR assay (10). We interpreted 50% inhibitory concentrations (IC50) using the values obtained for two laboratory reference strains: (i) HIV-1BRU, which is susceptible to T20, and (ii) HIV-2ROD, which is considered to be naturally resistant to T20 (27).
We found that 168 of the 171 HIV-O samples (98%) carried the N42D mutation within the HR1 domain of gp41. One sample carried the N42S mutation; one carried the natural residue; and the remaining sample was found to correspond to an N42D N43K double mutant virus, detected in a patient followed up at the Centre Pasteur du Cameroun in Yaoundé.
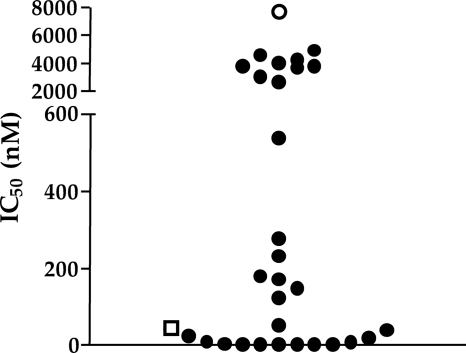
Phenotypic susceptibility was successfully determined for all 29 HIV-O viral supernatants tested. The IC50 obtained were widely scattered over a range extending from 4 nM to 5,000 nM, with a median value of 149 nM (Fig. 1). The sample characterized genotypically as a double mutant (N42D N43K) gave a low value (IC50, 6.4 nM), similar to that for the HIV-1 group M control. Even for the HIV-O samples with the highest IC50 in this series, the IC50 obtained were lower than those for HIV-2ROD.
FIG. 1.
Baseline susceptibilities to enfuvirtide (T20) of 29 HIV-1 group O viral strains. The 50% inhibitory concentrations (IC50) are expressed as nanomolar concentrations for each of the 29 HIV-1 group O viral supernatants. The results were obtained in two independent experiments, with quadruplicate determinations for each point. The IC50 of the reference laboratory virus HIV-1BRU is indicated by an open square, and that of the reference laboratory virus HIV-2ROD is indicated by an open circle.
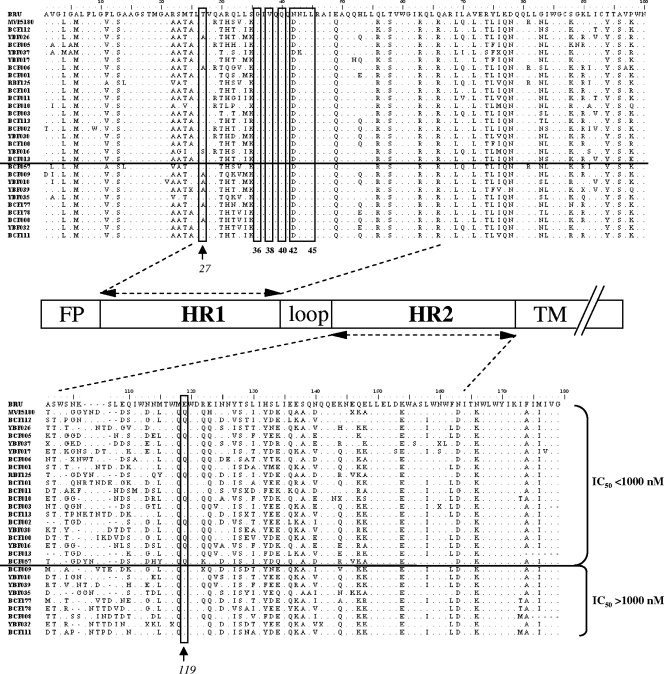
We then analyzed the sequences of the HR1 and HR2 regions of these 29 HIV-O strains in order to identify possible determinants linked to high or low IC50. A comparison between the HIV-O sequences and the sequences of the HIV-1 group M reference strain HIV-1BRU showed a high level of polymorphism in HIV-O viruses, associated with a high level of intragroup diversity, particularly for the HR2 region (Fig. 2). This high level of polymorphism was confirmed for the 142 sequences obtained from plasma samples (data not shown). Furthermore, comparisons of sequences of viruses for which the IC50 exceeded 1,000 nM with sequences of viruses for which the IC50 were below 1,000 nM (Fig. 2) showed two positions (residues 27 and 119) to be significantly different between these two groups of sequences (alpha, 5% by Pearson's chi-square test). The T27A mutation was more frequent in the group of HIV-O sequences with IC50 of >1,000 nM (54% versus 6% [P, 0.008]), whereas the E119Q mutation was less frequent among HIV-O sequences with IC50 of >1,000 nM (0% versus 44% [P, 0.026]).
FIG. 2.
Alignment of the sequences of the 29 HIV-1 group O strains used in the phenotypic analysis. The sequences correspond to the ectodomain of gp41; HIV-O sequences are compared with HIV-1BRU, used as a reference, and are ranked in ascending order of associated IC50. The IC50 threshold of 1,000 nM is indicated by a horizontal line. Resistance-associated positions are boxed, and the residues at positions 27 and 119 are boxed and indicated by arrows. A schematic linear structure of the gp41 ectodomain is shown in the center. FP, fusion peptide; HR1 and HR2, heptad repeats 1 and 2; TM, transmembrane region. The positions of the HR1 and HR2 domains were defined according to reference 9.
Finally, we assessed the possible impact of genetic diversity on phenotypic susceptibility, since we had previously demonstrated the clade dependence of the Y181C mutation associated with resistance to NNRTI (7). A phylogenetic tree including all 29 HIV-O gp41 sequences used for the phenotypic analysis was generated. No relationship was found between IC50 and a specific genetic cluster or a specific clade (data not shown).
We have reported the genotypic susceptibilities of 171 HIV-O viruses to T20, an HIV-1 fusion inhibitor, and the phenotypic susceptibilities of 29 of these viruses. In our study, 98% of HIV-O viruses naturally harbored the N42D mutation in the gp41 region, associated with a wide range of baseline levels of phenotypic susceptibility to T20. The lowest IC50 in our study were similar to those previously reported (4, 20), probably due to the limited number of samples tested. Our results, obtained on a larger panel, indicate that IC50 are more scattered than previously thought, with a range potentially spanning 3 log orders of magnitude, as reported for HIV-1 group M viruses, which had IC50 from 1 to 480 ng/ml in the study by Sista et al. (23), 3 to 1,002 ng/ml in the study by Labrosse et al. (12), and 4 to 8,192 ng/ml in the study by Melby et al. (13). The similar results obtained for HIV-1 groups M and O led us to conclude that this specific antiretroviral drug displayed a broad baseline susceptibility. In our study, the highest IC50, approaching that for HIV-2ROD, which is considered naturally resistant to T20 (27), require careful interpretation. One important consequence of this wide range of values is that it is difficult to state phenotypic resistance based solely on the finding of a high IC50. Thus, for a more accurate definition of resistance, a comparison of IC50 is required, with the definition of a significant fold change between the IC50 for the baseline sample and those for samples obtained during treatment in the context of virological failure with a T20-containing regimen, as described for HIV-1 group M.
We found no association between IC50 and a specific HIV-O phylogenetic cluster, as might have been expected following our recent description of the clade A dependence of the Y181C mutation responsible for NNRTI resistance (7). Interestingly, significant changes to residues 27 and 119 of gp41 have been associated with changes in IC50 but not with susceptibility, since all samples were nonetheless considered to be susceptible to T20. It is difficult to determine the individual roles of these specific mutations, because they have not been described in previous studies, and the accurate assessment of their roles is likely to be difficult, because the considerable intragroup genetic diversity within HIV-O viruses must be taken into account. These two mutations do not affect residues forming the leucine zipper, an essential part of the final structure of gp41, but they may help to stabilize the helix or may act as accessory mutations in the restoration of fitness, as suggested by previous studies (24, 25).
The apparent discrepancy between genotypic and phenotypic results raises questions about the real impact of the N42D mutation on the mechanism of resistance of HIV-O variants to enfuvirtide. This mutation may not be the only determinant of resistance. Compensatory mutations may also be present, though difficult to identify, due to the high degree of polymorphism of these variants. This discordance between phenotypic results and the interpretation of genotypic resistance on the basis of HIV-1 group M algorithms highlights the need to optimize current interpretations for HIV-O. Finally, our findings demonstrate that T20 may potentially be useful for the treatment of HIV-O-infected patients whose treatment options are already limited. The prevalence of HIV-O is low outside the Western Central Africa region, and the short-term use of T20 in Cameroon seems to be compromised by logistical obstacles. Nevertheless, this study shows that T20 was active in vitro against a large panel of diverse HIV-O viruses, increasing the number of antiretroviral drugs that may be considered active against this particular group of HIV-1 viruses.
Nucleotide sequence accession numbers.
The HIV-O sequences determined in this study have been submitted to GenBank under accession numbers GU935937 to GU936106 and HM021797.
Acknowledgments
We thank the Institut de Veille Sanitaire (InVS), the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales (ANRS), and Rouen University Hospital for financial support.
We thank all the physicians and biologists involved in the RES-O network for the surveillance of HIV-O infections in France.
Footnotes
Published ahead of print on 4 June 2010.
REFERENCES
- 1.Agut, H., D. Candotti, B. Rabanel, J. M. Huraux, G. Remy, D. Ingrand, T. Tabary, C. Chippaux, S. Chamaret, D. Guetard, and L. Montagnier. 1992. Isolation of atypical HIV-1-related retrovirus from an AIDS patient. Lancet 340:681-682. [DOI] [PubMed] [Google Scholar]
- 2.Ayouba, A., P. Mauclère, P. M. Martin, P. Cunin, J. Mfoupouendoun, B. Njinku, S. Souquières, and F. Simon. 2001. HIV-1 group O infection in Cameroon, 1986 to 1998. Emerg. Infect. Dis. 7:466-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charneau, P., A. M. Borman, C. Quillent, D. Guétard, S. Chamaret, J. Cohen, G. Rémy, L. Montagnier, and F. Clavel. 1994. Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology 205:247-253. [DOI] [PubMed] [Google Scholar]
- 4.Chinnadurai, R., J. Münch, M. T. Dittmar, and F. Kirchhoff. 2005. Inhibition of HIV-1 group M and O isolates by fusion inhibitors. AIDS 19:1919-1922. [DOI] [PubMed] [Google Scholar]
- 5.de Baar, M. P., W. Janssens, A. de Ronde, K. Fransen, R. Colebunders, L. Kestens, G. van der Groen, and J. Goudsmit. 2000. Natural residues versus antiretroviral drug-selected mutations in HIV type 1 group O reverse transcriptase and protease related to virological drug failure in vivo. AIDS Res. Hum. Retroviruses 16:1385-1394. [DOI] [PubMed] [Google Scholar]
- 6.Depatureaux, A., M. Leoz, F. De Oliveira, M. Gueudin, F. Damond, D. Descamps, F. Brun-Vézinet, V. Lemée, F. Simon, F. Barin, and J. C. Plantier. 18 June 2010, posting date. Diagnostic spécifique et prise en charge des infections par un VIH-1 groupe O: données de RES-O. Méd. Malad. Infect. doi: 10.1016/j.medmal.2010.04.011. [DOI] [PubMed]
- 7.Depatureaux, A., M. Leoz, A. Vessière, D. Rousset, F. Damond, F. Simon, and J. C. Plantier. 2009. Natural presence of resistance mutation Y181C in HIV-1 group O variants is clade-dependent. Antivir. Ther. 14(Suppl. 1):A93. [Google Scholar]
- 8.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damond, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vézinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 71:8893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg, M. L., and N. Cammack. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54:333-340. [DOI] [PubMed] [Google Scholar]
- 10.Gueudin, M., V. Lemée, V. Ferre, A. Beby-Defaux, J. P. Pathé, O. Guist'hau, J. Braun, F. Simon, and J. C. Plantier. 2004. Virologic diagnosis and follow-up of children born to mothers infected by HIV-1 group O. J. Acquir. Immune Defic. Syndr. 36:639-641. [DOI] [PubMed] [Google Scholar]
- 11.Gueudin, M., J. C. Plantier, V. Lemée, M. P. Schmitt, L. Chartier, T. Bourlet, A. Ruffault, F. Damond, M. Vray, and F. Simon. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500-505. [DOI] [PubMed] [Google Scholar]
- 12.Labrosse, B., J. L. Labernardière, E. Dam, V. Trouplin, K. Skrabal, F. Clavel, and F. Mammano. 2003. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J. Virol. 77:1610-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melby, T., P. Sista, R. DeMasi, T. Kirkland, N. Roberts, M. Salgo, G. Heilek-Snyder, N. Cammack, T. J. Matthews, and M. L. Greenberg. 2006. Characterization of envelope glycoprotein gp41 genotype and phenotypic susceptibility to enfuvirtide at baseline and on treatment in the phase III clinical trials TORO-1 and TORO-2. AIDS Res. Hum. Retroviruses 22:375-385. [DOI] [PubMed] [Google Scholar]
- 14.Menzo, S., A. Castagna, A. Monachetti, H. Hasson, A. Danise, E. Carini, P. Bagnarelli, A. Lazzarin, and M. Clementi. 2004. Resistance and replicative capacity of HIV-1 strains selected in vivo by long-term enfuvirtide treatment. New Microbiol. 27:51-61. [PubMed] [Google Scholar]
- 15.Mink, M., M. Greenberg, S. Mosier, S. Janumpalli, D. Davison, L. Jin, T. Melby, P. Sista, D. Lambert, N. Cammack, M. Salgo, and T. Matthews. 2002. Impact of HIV-1 gp41 amino acid substitutions (positions 36-45) on susceptibility to T-20 (enfuvirtide) in vitro: analysis of primary virus isolates recovered from patients during chronic enfuvirtide treatment and site-directed mutants in NL4-3. Antivir. Ther. 7:S17. [Google Scholar]
- 16.Peuchant, O., S. Capdepont, J. M. Ragnaud, V. Aurillac-Lavignolle, R. Thiébaut, H. Fleury, and B. Masquelier; ANRS CO3 Aquitaine Cohort. 2007. Primary resistance to enfuvirtide (T20) in recently HIV-1 infected, antiretroviral-naive patients from the ANRS Aquitaine Cohort. Antivir. Ther. 12:559-562. [DOI] [PubMed] [Google Scholar]
- 17.Plantier, J. C., R. Dachraoui, V. Lemée, M. Gueudin, F. Borsa-Lebas, F. Caron, and F. Simon. 2005. HIV-1 resistance genotyping on dried serum spots. AIDS 19:391-397. [DOI] [PubMed] [Google Scholar]
- 18.Plantier, J. C., M. Djemai, V. Lemée, A. Reggiani, M. Leoz, L. Burc, A. Vessière, D. Rousset, J. D. Poveda, C. Henquell, A. Gautheret-Dejean, and F. Barin. 2009. Census and analysis of persistent false-negative results in serological diagnosis of human immunodeficiency virus type 1 group O infections. J. Clin. Microbiol. 47:2906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plantier, J. C., M. Gueudin, F. Damond, J. Braun, P. Mauclère, and F. Simon. 2003. Plasma RNA quantification and HIV-1 divergent strains. J. Acquir. Immune Defic. Syndr. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Poveda, E., P. Barreiro, B. Rodés, and V. Soriano. 2005. Enfuvirtide is active against HIV type 1 group O. AIDS Res. Hum. Retroviruses 21:583-585. [DOI] [PubMed] [Google Scholar]
- 21.Rodes, B., C. De Mendoza, M. Rodgers, A. Newell, V. Jimenez, R. M. Lopez-Brugada, and V. Soriano. 2005. Treatment response and drug resistance in patients infected with HIV type 1 group O viruses. AIDS Res. Hum. Retroviruses 21:602-607. [DOI] [PubMed] [Google Scholar]
- 22.Roques, P., D. L. Robertson, S. Souquière, F. Damond, A. Ayouba, I. Farfara, C. Depienne, E. Nerrienet, D. Dormont, F. Brun-Vézinet, F. Simon, and P. Mauclère. 2002. Phylogenetic analysis of 49 newly derived HIV-1 group O strains: high viral diversity but no group M-like subtype structure. Virology 302:259-273. [DOI] [PubMed] [Google Scholar]
- 23.Sista, P. R., T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. L. Nelson, R. DeMasi, N. Cammack, M. P. Salgo, T. J. Matthews, and M. L. Greenberg. 2004. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 18:1787-1794. [DOI] [PubMed] [Google Scholar]
- 24.Su, C., T. Melby, R. DeMasi, P. Ravindran, and G. Heilek-Snyder. 2006. Genotypic changes in human immunodeficiency virus type 1 envelope glycoproteins on treatment with the fusion inhibitor enfuvirtide and their influence on changes in drug susceptibility in vitro. J. Clin. Virol. 36:249-257. [DOI] [PubMed] [Google Scholar]
- 25.Teixeira, C., D. de Sá-Filho, W. Alkmim, L. M. Janini, R. S. Diaz, and S. Komninakis. 2010. High polymorphism rates in the HR1 and HR2 gp41 and presence of primary resistance-related mutations in HIV type 1 circulating in Brazil: possible impact on enfuvirtide efficacy. AIDS Res. Hum. Retroviruses 26:307-311. [DOI] [PubMed] [Google Scholar]
- 26.Vessière, A., D. Rousset, A. Kfutwah, M. Leoz, A. Depatureaux, F. Simon, and J. C. Plantier. 2010. Diagnosis and monitoring of HIV-1 group O-infected patients in Cameroon. J. Acquir. Immune Defic. Syndr. 53:107-110. [DOI] [PubMed] [Google Scholar]
- 27.Witvrouw, M., C. Pannecouque, W. M. Switzer, T. M. Folks, E. De Clercq, and W. Heneine. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir. Ther. 9:57-65. [PubMed] [Google Scholar]
- 28.Yamaguchi, J., A. S. Vallari, P. Swanson, P. Bodelle, L. Kaptué, C. Ngansop, L. Zekeng, L. G. Gürtler, S. G. Devare, and C. A. Brennan. 2002. Evaluation of HIV type 1 group O isolates: identification of five phylogenetic clusters. AIDS Res. Hum. Retroviruses 18:269-282. [DOI] [PubMed] [Google Scholar]