Abstract
BMS-790052 is the most potent hepatitis C virus (HCV) inhibitor reported to date, with 50% effective concentrations (EC50s) of ≤50 pM against genotype 1 replicons. This exceptional potency translated to rapid viral load declines in a phase I clinical study. By targeting NS5A, BMS-790052 is distinct from most HCV inhibitors in clinical evaluation. As an initial step toward correlating in vitro and in vivo resistances, multiple cell lines and selective pressures were used to identify BMS-790052-resistant variants in genotype 1 replicons. Similarities and differences were observed between genotypes 1a and 1b. For genotype 1b, L31F/V, P32L, and Y93H/N were identified as primary resistance mutations. L23F, R30Q, and P58S acted as secondary resistance substitutions, enhancing the resistance of primary mutations but themselves not conferring resistance. For genotype 1a, more sites of resistance were identified, and substitutions at these sites (M28T, Q30E/H/R, L31M/V, P32L, and Y93C/H/N) conferred higher levels of resistance. For both subtypes, combining two resistance mutations markedly decreased inhibitor susceptibility. Selection studies with a 1b/1a hybrid replicon highlighted the importance of the NS5A N-terminal region in determining genotype-specific inhibitor responses. As single mutations, Q30E and Y93N in genotype 1a conferred the highest levels of resistance. For genotype 1b, BMS-790052 retained subnanomolar potency against all variants with single amino acid substitutions, suggesting that multiple mutations will likely be required for significant in vivo resistance in this genetic background. Importantly, BMS-790052-resistant variants remained fully sensitive to alpha interferon and small-molecule inhibitors of HCV protease and polymerase.
Hepatitis C virus (HCV) is a major cause of chronic liver disease, affecting up to 180 million people worldwide (24, 32). HCV is an enveloped, positive-strand RNA virus and is the sole member of the Hepacivirus genus of the Flaviviridae family (17). HCV is classified into six major genotypes, each with multiple subtypes, based on sequence diversity (17, 33). The current standard of care for HCV infection includes therapy with a combination of pegylated alpha interferon (pegIFN-α) and ribavirin. This treatment is often associated with limiting side effects, and effectiveness is highly genotype dependent (3, 30, 38). For genotypes 1a and 1b, accounting for ∼60% of global infections, long-term efficacy or a sustained virological response is achieved in only ∼50% of chronically infected individuals (3, 24). An increasing number of small-molecule inhibitors targeting specific viral proteins are entering into clinical evaluation (1, 25, 27, 30, 38). Collectively, these inhibitors are often referred to as direct-acting antiviral agents (DAA). The most advanced of these inhibitors target enzymatic activities of the HCV nonstructural proteins NS3 (serine protease) and NS5B (RNA-dependent RNA polymerase) (1, 27). We recently described a potent inhibitor of HCV RNA replication, BMS-790052, that targets HCV NS5A (6). In a phase I clinical study, BMS-790052 was well tolerated and exhibited impressive antiviral activity, achieving mean reductions in HCV RNA of >3 log10 IU/ml 24 h following single doses of 10 or 100 mg (6).
NS5A, an essential component of the HCV replication complex (RC), does not possess any known enzymatic activities, and its role(s) in viral replication remains ill defined (reviewed in references 11 and 23). NS5A possesses an RNA binding activity and is comprised of three distinct structural domains, as well as an amphipathic α-helix at its N terminus (amino acids [aa] 5 to 25) that functions in membrane localization (10, 28, 34). Domain I (aa 37 to 213) is essential for viral RNA replication, possesses a zinc binding motif, and has been crystallized as a homodimer (22, 35). Domains II (aa 250 to 342) and III (aa 356 to 447) are less well characterized and are natively unfolded (7, 18).
The potential for viral resistance is a major concern in managing HCV infection through DAA strategies (14, 37). While clinical trials of compounds targeting NS3 and NS5B have demonstrated potent antiviral activities, these trials have also revealed the rapid emergence of resistant viral variants (reviewed in references 12 and 29). In this report, we provide a comprehensive analysis of the in vitro resistance profile of BMS-790052 against genotypes 1a and 1b. Our analysis significantly extends previous findings (6) and identifies both similarities and differences between genotype 1a and 1b resistance patterns. We also show that replicons highly resistant to BMS-790052 remain fully sensitive to pegIFN-α and DAA inhibitors of HCV NS3 and NS5B, underscoring the potential utility of including BMS-790052 in combination approaches to HCV therapy.
MATERIALS AND METHODS
Cell culture and HCV inhibitors.
Human hepatoma (Huh-7) cells were maintained in Dulbecco's modified Eagle's medium supplemented with fetal bovine serum (10%), l-glutamine (2 mM), penicillin (100 units/ml), and streptomycin (100 μg/ml). Genotype 1a (H77c) and 1b (con1) replicon cell lines used in this study have been described previously (15, 16, 26). A genotype 1b replicon cell line with an adaptive mutation in NS5B (R2884G) was obtained from R. Bartenschlager (Huh-9-13) (21). For cells harboring HCV replicons, complete medium was supplemented with 0.5 mg/ml G418 (Geneticin; Invitrogen Corp., Carlsbad, CA). Replicon cell lines with specific NS5A amino acid substitutions were established by selection with G418 as described previously (16). A previously described cured Huh-7 replicon cell line which is highly permissive for HCV replicon replication was used for transient transfection assays (16). BMS-790052 has been described previously (6). HCV reference inhibitors were ITMN-191 (a protease inhibitor [PI]) (31), an analog of JTK-109 (NS5B-1; the desfluoro derivative of example 10h in reference 9), and HCV796 (NS5B-2) (13). pegIFN-α was purchased from Hoffmann-La Roche, Inc. (Nutley, NJ).
Resistance selection.
Replicon cells plated in 100- or 150-mm tissue culture dishes were maintained in medium supplemented with 0.5 mg/ml G418 and the desired concentration of BMS-790052 in dimethyl sulfoxide (DMSO) (Sigma-Aldrich Corp., St. Louis, MO). Control cells were maintained with an equivalent percent volume of DMSO (≤0.01%). Cells were split or fed with fresh medium containing inhibitor twice weekly to maintain a subconfluent monolayer. After approximately 4 to 5 weeks, pooled cells were expanded for phenotypic and genotypic analyses.
Genotypic analysis.
Total RNA was isolated from replicon cells by use of Trizol (Invitrogen Corporation, Carlsbad, CA) following the manufacturer's protocol. For reverse transcription-PCR (RT-PCR), first-strand cDNA synthesis was performed with random hexamers, using a Superscript III first-strand cDNA synthesis kit (Invitrogen Corporation, Carlsbad, CA). PCR was performed with Platinum Taq high-fidelity DNA polymerase (Invitrogen Corporation, Carlsbad, CA). For genotype 1b replicons, primers were 5′-AGCCCGCTCACCACCCAACATACC (forward) and 5′-TCCTCCGCAGCGCATGGCGTG (reverse). Genotype 1a primers were 5′-GGCTAATAGCCTTCGCCTCCCG (forward) and 5′-CGTCCTGGTAATGGCTGTCCAGAAC (reverse). Amplicons spanning NS5A were subjected directly to sequence analysis and were also used as inserts for cloning with a TOPO TA cloning kit (Invitrogen Corporation, Carlsbad, CA). Sequence analysis and alignments were performed with Lasergene DNAStar software (Madison, WI).
Phenotypic analysis.
Parental genotype 1a and 1b replicon clones have been described previously (15, 16). Amino acid substitutions in the NS5A coding region were introduced into replicon clones by recombinant PCR and standard cloning methods (8). Cloning sites were Eco47III (in NS4B) and EcoRI (in NS5A) for the genotype 1b replicon and BstEII (in NS4B and NS5A) for the genotype 1a replicon. Desired changes were confirmed by sequence analysis. Parental replicon clones carried a Renilla luciferase reporter gene and had one (1b; S2204I) or two (1a; P1496L and S2204I) cell culture-adaptive mutations. Replicon clones were linearized with ScaI and transcribed in vitro with a T7 MEGAScript kit (Ambion Inc., Austin, TX). For transient replicon transfection assays, equal amounts of RNA (10 to 20 μg) were transfected into Huh-7 cells (5,000,000 in 100-mm dishes) by use of DMRIE-C reagent (Invitrogen Corporation, Carlsbad, CA) following the manufacturer's protocol. After 16 h, cells were transferred to 96-well assay plates (10,000 cells/well in 200 μl) (Corning Inc., Corning, NY), and dilutions of inhibitors in DMSO were added (1 μl/well). After an additional 72 h, plates were washed (200 μl phosphate-buffered saline [PBS]/well) and assayed with a Renilla luciferase assay system as described by the manufacturer (Promega Corporation, Madison, WI). The background, determined for wells containing a high concentration of an HCV NS3 PI (inhibiting ≥95% of replication), was subtracted from all wells (26). Luminescence values averaged for 16 DMSO-treated wells were used to estimate the relative replication capacities of replicon variants. These values are reported relative to the value obtained for the parental replicon clone (100%), which was included as a control in each transient transfection assay. The 50% effective concentration (EC50) was calculated as the concentration of inhibitor required for a 50% reduction in luciferase activity. Some differences in EC50s derived from transient transfection assays and replicon cell lines were observed (compare values in Tables 1, 3, and 5), possibly reflecting differences in replicon replication between the two assays.
TABLE 1.
Potency and selection concentration of BMS-790052 on replicon cell lines
| Genotype | Cell culture-adaptive mutation | EC50a (pM) (n) | Selection concn (pM, unless shown otherwise) |
|---|---|---|---|
| 1b | S2204I | 5.5 ± 4.3 (11) | 50, 100, 500 |
| ΔS2197 | 12.3 ± 8.5 (5) | 200 | |
| R2884G | 9.0 ± 3.2 (4) | 200 | |
| 1a | P1496L and S2204I | 49.4 ± 27.1 (5) | 200, 1 nM, 1 μM |
| 1b1a100b | S2204I | 25.4 ± 2.7 (2) | 200, 1 nM |
Mean ± standard deviation, determined with the parental cell line.
Hybrid 1b replicon with first 100 amino acids of NS5A from genotype 1a.
TABLE 3.
Effects of NS5A amino acid substitutions on BMS-790052 potency for genotype 1b
| Amino acid substitutiona | EC50 (pM)b | Fold resistance | Replication level (%) |
|---|---|---|---|
| None (parent) | 2.6 ± 0.3 | 1 | 100 |
| L23F | 1.5 ± 0.2 | 1 | 39 ± 9 |
| L28M | 4.1 ± 1.1 | 2 | 121 ± 54 |
| L28T | 52 ± 14 | 20 | 13 ± 5 |
| ΔR30 | NDd | NDd | None |
| R30E | 15.6 ± 8.1 | 6 | 11 ± 2 |
| R30Q | 1.5 ± 0.3 | 1 | 92 ± 2 |
| L31F | 12.6 ± 1.2 | 5 | 146 ± 44 |
| L31M | 8.4 ± 1.9 | 3 | 99 ± 23 |
| L31Vc | 61 ± 15 | 23 | 145 ± 44 |
| P32L | 44.7 ± 21 | 17 | 18 ± 6 |
| F37L | 1.7 ± 0.2 | 1 | 138 ± 28 |
| Q54H | 3.2 ± 0.5 | 1 | 83 ± 18 |
| P58H | 2.2 ± 0.5 | 1 | 103 ± 23 |
| P58S | 2.3 ± 0.4 | 1 | 121 ± 18 |
| Y93Hc | 49.2 ± 12.8 | 19 | 20 ± 7 |
| Y93N | 73.5 ± 5.5 | 28 | 21 ± 6 |
| L23F + L31F | 33.8 ± 5.1 | 13 | 65 ± 5 |
| L23F + Y93H | 208.3 ± 36.9 | 80 | 8 ± 1 |
| ΔR30 + P32L | >1,000,000 | >384,615 | 9 ± 9 |
| R30Q + L31F | 234.1 ± 98.7 | 90 | 224 ± 39 |
| L31F + Y93H | 14,874.3 ± 1,762.9 | 5,721 | 29 ± 8 |
| L31M + Y93H | 10,989.4 ± 122.9 | 4,227 | 36 ± 5 |
| L31V + P58S | 421.8 ± 69.8 | 162 | 82 ± 2 |
| L31V + Y93H | 21,674.5 ± 9,461.3 | 8,336 | 30 ± 10 |
| F37L + Y93H | 48.6 ± 6.3 | 19 | 34 ± 4 |
| Q54H + Y93H | 24.7 ± 6.4 | 10 | 22 ± 7 |
Substitutions identified in inhibitor-treated replicon cell lines are indicated in bold. Others are theoretical mutations. All replicons contain a S2204I adaptive mutation.
Mean ± standard deviation, determined in transient transfection assays (n ≥ 3).
Data were taken from the work of Gao et al. (6).
ND, not determined.
TABLE 5.
Effects of NS5A amino acid substitutions on BMS-790052 potency for genotype 1a
| Amino acid substitutiona | 1a replicon with P1496L and S2204I mutations |
1b1a100 replicon with S2204I mutation |
||||
|---|---|---|---|---|---|---|
| EC50 (nM)b | Fold resistance | Replication level (%) | EC50 (nM)b | Fold resistance | Replication level (%) | |
| None (parent) | 0.006 ± 0.004 | 1 | 100 | 0.007 ± 0.002 | 1 | 100 |
| M28Tc | 4.1 ± 0.4 | 683 | 31 ± 23 | 0.35 ± 0.02 | 50 | 8 ± 1 |
| ΔQ30 | NDd | ND | ND | >5,000 | >714,286 | 7 ± 2 |
| Q30E | 149.6 ± 89.1 | 24,933 | 130 ± 56 | ND | ND | ND |
| Q30K | 145.9 ± 70.5 | 24,317 | 18 ± 9 | ND | ND | ND |
| Q30Hc | 8.7 ± 1.9 | 1,450 | 75 ± 31 | 0.54 ± 0.09 | 77 | 99 ± 19 |
| Q30Rc | 7.3 ± 1.1 | 1,217 | 41 ± 16 | 1.0 ± 0.08 | 143 | 35 ± 7 |
| L31Mc | 2.1 ± 0.6 | 350 | 55 ± 15 | ND | ND | ND |
| L31Vc | 20.1 ± 6.0 | 3,350 | 117 ± 29 | 3.5 ± 0.8 | 500 | 121 ± 17 |
| P32L | 1.4 ± 0.2 | 233 | 18 ± 5 | ND | ND | 0.08 ± 0.01 |
| H54Y | ND | ND | ND | 0.004 ± 0.001 | 1 | 107 ± 20 |
| Y93Cc | 11.1 ± 4.0 | 1,850 | 11 ± 7 | 0.67 ± 0.16 | 96 | 34 ± 13 |
| Y93H | 32.2 ± 9.4 | 5,367 | 18 ± 11 | ND | ND | ND |
| Y93N | 282.1 ± 64.7 | 47,017 | 13 ± 8 | ND | ND | ND |
| M28T + Q30H | 622.6 ± 314.3 | 103,767 | 31 ± 22 | ND | ND | ND |
| Q30H + Y93H | 553.3 ± 207.4 | 92,217 | 20 ± 6 | ND | ND | ND |
| L31V + Y93H | >1,000 | >166,667 | 20 ± 2 | ND | ND | ND |
All mutations except for the Q30H-plus-Y93H mutation were identified in inhibitor-treated replicon cell lines.
Mean ± standard deviation, determined in transient transfection assays (n ≥ 3).
Data were taken from the work of Gao et al. (6).
ND, not determined.
Replicon cell assays.
Inhibitor susceptibilities on replicon cell lines were determined with an HCV NS3 protease-based fluorescence resonance energy transfer (FRET) assay as previously described (16, 26). For cell lines harboring replicon clones with a Renilla luciferase reporter, assays were performed as described for transient transfection assays.
RESULTS
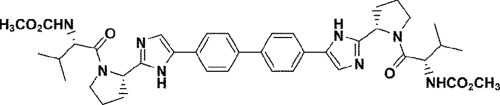
The discovery of BMS-790052 as a potent inhibitor of HCV RNA replication that targets NS5A was recently reported (6). The chemical structure of BMS-790052 is shown in Fig. 1. Preliminary characterization of the inhibitor revealed amino acid substitutions within the N-terminal region of NS5A that are associated with decreased inhibitor susceptibility (6). Identified mutations were L31V and Y93H for genotype 1b and M28T, Q30H/R, L31M/V, and Y93C for genotype 1a. In preparation for advancing BMS-790052 to clinical evaluation, and to further assess the potential for in vivo resistance to this unique inhibitor, we greatly expanded our characterization of the in vitro resistance profile of BMS-790052 by performing resistance selections on multiple replicon cell lines and with various selective pressures. In total, 10 independent selections were performed on five cell lines harboring genotype 1b, 1a, or 1b/1a hybrid replicons. Parental cell lines, EC50s for BMS-790052, and inhibitor concentrations used for selections are summarized in Table 1.
FIG. 1.
Chemical structure of BMS-790052.
Genotype 1b resistance.
For resistance selection, genotype 1b replicon cells were maintained in the presence of BMS-790052 at concentrations ranging from 50 to 500 pM (∼10 to 100 times the EC50) (Table 1). The effects of different cellular backgrounds were assessed by using replicon cell lines with three different primary cell culture-adaptive mutations (20) (Table 1). For all selections, cells experienced reduced growth rates and cell death followed by a return to normal growth, at which time cells were tested for susceptibility to BMS-790052. As shown in Fig. 2, the potency of BMS-790052, but not that of an HCV NS3 PI, was decreased in the inhibitor-treated cells relative to that in the DMSO-treated control cells. Although replicon RNA levels were not measured directly, uninhibited reporter signals from the control and inhibitor-selected cells were generally within a 2-fold difference of one another, suggesting that the cells had similar replicon replication capacities.
FIG. 2.
Resistance of replicon cells selected with BMS-790052. Pooled populations of replicon cells selected for resistance to BMS-790052 at the indicated concentrations were assayed for sensitivity to BMS-790052 and an HCV NS3 PI. Fold resistance was plotted as the EC50 of inhibitor on the resistant cells divided by the EC50 on DMSO-treated control cells. Error bars are standard deviations for three assays.
Sequence analysis revealed variations at NS5A residues L31 and Y93 in bulk cDNAs recovered from all of the inhibitor-treated cells (data not shown). Sequencing of individual NS5A cDNA clones confirmed the prevalence of mutations at these residues (Table 2). L31V and Y93H were the predominant mutations observed in all of the cell line selections. Other amino acid substitutions at these positions were observed less frequently (L31F and Y93C/N/S) (Table 2). In total, 69/79 cDNA clones isolated from BMS-790052-treated cells contained a mutation at either L31 or Y93; an additional 6 clones had substitutions at both of these positions (Table 2). Of the remaining four clones, one each had a P32L, P58S, or I63V substitution, and one did not contain any changes within NS5A. Amino acid substitutions at L31, P32, P58, I63, and Y93 were not detected in 53 NS5A cDNA clones isolated from DMSO-treated control cells (data not shown). Several of the cDNA clones that had an L31 or Y93 mutation contained one or more additional changes within NS5A compared to the con1 replicon reference sequence. The inhibitor susceptibility phenotypes induced by most of these additional changes were not determined, since these changes were also observed in cDNA clones from the control cells or were represented infrequently in the cDNA clones from the inhibitor-treated cells. In clones from both inhibitor-treated and control cells, apparently random amino acid substitutions were more prevalent in the C-terminal region of NS5A, a part of the protein largely dispensable for replicon replication (19, 36). Some secondary changes, however, were of potential interest because they were in close proximity to L31 or Y93 or occurred at residues that differed between the genotype 1a and 1b replicons (L23F, F37L, R30Q, and Q54H) (Table 2).
TABLE 2.
Frequencies of NS5A amino acid substitutions in cDNA clones isolated from BMS-790052-treated genotype 1b replicon cellsa
| Amino acid substitution | No. of clones at indicated BMS-790052 concn |
Total no. of clones | ||||
|---|---|---|---|---|---|---|
| S2204I cells |
ΔS2197 cells, 200 pM | R2884G cells, 200 pM | ||||
| 50 pM | 100 pM | 500 pM | ||||
| L31F | 2 | 1 | 1 | 4 | ||
| L31V | 1 | 5 | 10 | 5 | 21 | |
| P32L | 1 | 1 | ||||
| P58S | 1 | 1 | ||||
| I63V | 1 | 1 | ||||
| Y93H | 10 | 8 | 7 | 3 | 6 | 34 |
| Y93N | 1 | 1 | ||||
| Y93S | 1 | 1 | ||||
| L23F + L31F | 2 | 2 | ||||
| R30Q + L31F | 3 | 3 | ||||
| L31V + Y93C | 1 | 1 | ||||
| L31V + Y93H | 2 | 1 | 1 | 4 | ||
| L31V + Y93N | 1 | 1 | ||||
| F37L + Y93H | 1 | 1 | 2 | |||
| Q54H + Y93H | 1 | 1 | ||||
| None | 1 | 1 | ||||
| Total | 15 | 13 | 18 | 16 | 17 | 79 |
Replicon cells are described in Table 1.
To assess the contributions of specific mutations to inhibitor sensitivity, changes were introduced into a 1b replicon clone and analyzed in transient replicon transfection assays (Table 3). As individual amino acid substitutions, L31V, P32L, Y93H, and Y93N all conferred similar and modest levels of resistance to BMS-790052, with increases in EC50s ranging from 17- to 28-fold above the wild-type control level (Table 3). When L31V and Y93H mutations were combined in the same replicon, the effect on the inhibitor increased dramatically (8,336-fold resistance). Likewise, by itself, the L31F mutation had only a slight impact on BMS-790052 potency (5-fold), but when it was combined with the Y93H mutation, resistance was much greater (5,721-fold). The P58S mutation, identified in a single NS5A cDNA clone (Table 2), did not affect BMS-790052 potency as a single mutation. However, the combination of P58S and L31V mutations yielded 162-fold resistance, which was much greater than that for L31V mutation alone (23-fold).
L23F and R30Q mutations were both found in cDNA clones linked to the L31F mutation (Table 2). By themselves, these mutations did not affect BMS-790052 potency, but they did enhance the resistance phenotype of L31F mutants (Table 3). In contrast, F37L and Q54H mutations, identified in clones linked to the Y93H mutation, had no effect on inhibitor potency either as individual mutations or in combination with Y93H mutation. The resistance phenotypes induced by the I63V, Y93C, and Y93S mutations, which were each observed in a single cDNA clone, have not yet been examined.
In addition to determining susceptibility to BMS-790052, an estimate of the replication fitness of the resistant variants was assessed (Table 3). Changes at L31 (to F, M, or V) were well tolerated, allowing replication levels similar to the wild-type control level. In contrast, replication of the Y93H/N and P32L variants was partially impaired (∼20% of the wild-type level). Values for all of the variants are provided in Table 3.
Genotype 1a resistance.
For genotype 1a resistance selection, replicon cells were maintained in the presence of 200 pM or 1 nM BMS-790052 (∼4- to 20-fold over the EC50) (Table 1). Testing of selected cells for inhibitor sensitivity revealed that the increase in resistance was greater for genotype 1a cells than for genotype 1b cells selected at the same inhibitor concentration (compare fold resistances observed for genotype 1a and 1b cells selected at 200 pM BMS-790052 in Fig. 2). Sequencing of bulk cDNAs from inhibitor-treated cells also revealed a more complicated pattern of drug-induced mutations in the genotype 1a cells, including apparent substitutions at NS5A residues 28, 30, 31, and 93 (data not shown). To better identify potential resistant variants, sequence analysis was performed on individual cDNA clones (Table 4). Of 49 clones isolated from the inhibitor-treated cells, all but 3 had an amino acid substitution at residue 28 (M28T), 30 (Q30E/H/R), 31 (L31M/V), 32 (P32L), or 93 (Y93C/H/N). No changes at any of these positions were identified in 30 clones obtained from DMSO-treated cells (data not shown). Mutations at residue 30 were the most common, with 27 clones bearing an amino acid substitution at this residue. Of the three remaining clones, one had the M28T mutation together with the Y93C mutation, one had the H54Y mutation, and one was wild type at all of these positions (Table 4).
TABLE 4.
Frequencies of NS5A amino acid substitutions in cDNA clones isolated from BMS-790052-treated genotype 1a and 1b1a hybrid replicon cells
| Amino acid substitution | No. of clones at indicated BMS-790052 concn |
||||||
|---|---|---|---|---|---|---|---|
| 1a P1496L/S2204I cells |
1b1a100 S2204I cells |
||||||
| 200 pM | 1 nM | 1 μMa | Total | 200 pM | 1 nM | Total | |
| M28T | 5 | 4 | 9 | ||||
| ΔQ30 | 9 | 9 | |||||
| Q30E | 1 | 1 | 2 | 3 | 3 | ||
| Q30K | 1 | 1 | 2 | ||||
| Q30H | 4 | 5 | 9 | 3 | 3 | 6 | |
| Q30R | 5 | 4 | 9 | 3 | 3 | ||
| L31M | 3 | 3 | 3 | 5 | 8 | ||
| L31V | 1 | 1 | 2 | ||||
| P32L | 3 | 1 | 4 | ||||
| H54Y | 1 | 1 | |||||
| Y93C | 3 | 3 | 6 | 2 | 3 | 5 | |
| Y93H | 1 | 1 | 2 | 1 | 1 | 2 | |
| Y93N | 1 | 2 | 3 | ||||
| M28T + Q30H | 9 | 9 | |||||
| M28T + Q30L | 1 | 1 | |||||
| M28T + P32L | 3 | 3 | |||||
| M28T + Y93C | 1 | 1 | |||||
| M28T + Y93H | 3 | 3 | |||||
| M28T + Y93N | 1 | 1 | |||||
| Q30H + P32L | 3 | 3 | |||||
| Q30E + Y93C | 1 | 1 | |||||
| Q30E + Y93H | 1 | 1 | |||||
| Q30R + L31M | 2 | 2 | |||||
| Q30R + L31V | 2 | 2 | |||||
| Q30R + P32L | 1 | 1 | |||||
| Q30R + Y93H | 1 | 1 | |||||
| L31M + Y93C | 1 | 1 | |||||
| L31V + Y93H | 1 | 1 | |||||
| None | 1 | 1 | |||||
| Total | 29 | 20 | 42 | 91 | 16 | 13 | 29 |
Cells selected with 200 pM BMS-790052 were subsequently selected with 1 μM BMS-790052.
In transient replicon transfection assays, the M28T, Q30H/R, L31M/V, P32L, and Y93C/H mutants showed reduced susceptibility to BMS-790052, by factors ranging from 233- to 5,367-fold compared to the 1a replicon control level (Table 5). The levels of resistance were substantially greater with the Q30E and Y93N variants (24,933- and 47,017-fold, respectively). Substitutions at residue 93 (Y93C/H/N) had the greatest impact on replication fitness, with variants replicating at 11 to 18% of the wild-type level. In contrast, the L31V and Q30E variants replicated at least as well as the parental replicon. The H54Y mutation was not tested in the 1a replicon, but it did not confer resistance to BMS-790052 in the context of a 1b/1a hybrid replicon (Table 5; see below).
Resistance selection with a 1b/1a hybrid replicon cell line.
Resistance studies identified both similarities (residues 31, 32, and 93) and differences (residues 28 and 30) in the patterns of resistance between genotypes 1a and 1b (Tables 2 and 4). BMS-790052 was ∼5 to 10 times less potent against genotype 1a than against genotype 1b replicon cells (Table 1). Other NS5A inhibitors have been identified that display greater genotype 1a and 1b potency differences, and these differences are mediated at least partly by sequences within the NS5A N terminus (15). To determine if the N-terminal region of NS5A is also sufficient to account for the differences in resistance selection observed with genotype 1a and 1b replicon cells, a cell line harboring a genotype 1b replicon clone with sequences encoding the first 100 amino acids of NS5A derived from genotype 1a (1b1a100) was used for resistance selection. The EC50 of BMS-790052 on the parental hybrid replicon cell line was 25.4 ± 2.7 pM; selections were carried out with BMS-790052 at 200 pM and 1 nM (Table 1). Similar to the results obtained with the authentic genotype 1a replicon cell line, sequence analysis of bulk cDNAs from the hybrid replicon cells revealed a complicated pattern of drug-induced mutations, with likely amino acid substitutions at residues 28, 30, 31, and 93 (data not shown). Sequence analysis of individual NS5A cDNA clones verified the presence of many of these substitutions (Table 4). Overall, drug-induced mutations identified in the 1b1a100 replicon cell line were similar to those observed in the authentic genotype 1a background. As with genotype 1a replicon cells, but distinct from the case for genotype 1b cells, substitutions at residue 30 were the most commonly identified drug-induced changes. The inhibitor susceptibility phenotypes induced by a subset of mutations introduced into the 1b1a100 hybrid replicon background were determined in transient transfection assays (Table 5). Qualitatively, the results were very similar to those obtained with the authentic 1a replicon. For example, in both replicon backgrounds, inhibitor susceptibilities were in the following order: M28T variants > Q30H/R variants > L31V variants. However, the fold resistance conferred by any given variant was ∼7- to 20-fold higher in the genotype 1a replicon than in the hybrid replicon (Table 5).
Genotype 1a selection with high selective pressure.
Selections performed with the genotype 1a replicon cells yielded predominantly variants with single amino acid substitutions (Table 4). To determine the effect of increasing selective pressure, replicon cells originally selected with 200 pM BMS-790052 (Table 1) were subsequently treated with 1 μM BMS-790052 until a resistant cell population emerged. Sequence analysis of NS5A cDNAs isolated from the pooled resistant cell population was difficult to interpret, displaying evidence of multiple substitutions at previously identified sites of resistance (data not shown). The complexity of the resistance pattern was verified by analysis of individual cDNA clones (Table 4). Thirty of 42 clones contained a combination of two amino acid substitutions, at residues 28, 30, 31, 32, and/or 93 (Table 4). The combination of the M28T and Q30H mutations was the most prevalent (9 clones), but several other combinations were also observed (Table 4). To determine the phenotypes of variants with linked mutations, replicons with representative combinations (M28T + Q30H, Q30H + Y93H, and L31V + Y93H) were tested in transient replication assays. These variants displayed very high levels of resistance to BMS-790052 (92,217- to >166,667-fold) but also exhibited impaired replication (20 to 30%) relative to that of the parental clone (Table 5). Among the 12 clones that did not contain a double mutation, 2 had a Y93N change and 1 had a Q30E change. Not surprisingly, these were the single amino acid substitution variants with the greatest resistances to BMS-790052 in transient transfection assays (Table 5). The remaining nine clones contained an in-frame deletion of residue 30 (ΔQ30). The effect of the ΔQ30 mutation on BMS-790052 potency was examined in transient transfection assays in the 1b1a100 hybrid replicon background. No inhibition of this variant was observed, even at 5 μM BMS-790052 (Table 5).
Effect of genotype 1a-specific mutations in the genotype 1b replicon.
To explore differences in the genotype 1a and 1b resistance profiles, some of the mutations identified only in genotype 1a selections were introduced into the genotype 1b replicon. An alignment of the first 100 amino acids of the NS5A proteins from the genotype 1a (H77c) and 1b (con1) replicon clones is shown in Fig. 3. Among the primary BMS-790052 resistance sites, the amino acid sequences are the same at residues 31, 32, and 93 but differ at residues 28 and 30 (Fig. 3). M28T was a common resistance mutation in genotype 1a, but no substitutions at this residue were observed in genotype 1b (Tables 2 and 4). Replacing leucine with methionine (L28M) in genotype 1b did not substantially alter BMS-790052 susceptibility (Table 3). However, introducing a threonine at this position (L28T) resulted in a 20-fold decrease in inhibitor susceptibility, a level of resistance similar to that observed with the L31V and Y93H mutations (Table 3). This result therefore identifies L28T as a potential resistance mutation in genotype 1b.
FIG. 3.
Alignment of N-terminal 100 amino acids of NS5A proteins from the con1 (1b) (top) and H77c (1a) (bottom) replicon clones. Amino acid identities are indicated with dots. Residues identified as primary sites of BMS-790052 resistance are indicated with solid boxes. Secondary resistance sites are indicated with hatched boxes. The most common drug-induced substitutions at these positions are shown above (1b) and below (1a) the boxed residues.
A second difference between genotype 1a and 1b replicon selections was observed with the L31M mutation, which was selected in genotype 1a but not genotype 1b (Tables 2 and 4). Concordantly, an L31M change introduced into the genotype 1b replicon yielded minimal resistance to BMS-790052 (3-fold) (Table 3). However, if the L31M mutation was combined with the Y93H mutation, the level of resistance was greatly enhanced (4,227-fold) (Table 3), indicating that this mutation can also impact inhibitor sensitivity in genotype 1b.
The most distinct difference in the genotype 1a and 1b resistance profiles was observed at residue 30. Residue 30 was a major site of resistance in genotype 1a, with Q30R, Q30H, Q30E, and ΔQ30 mutations all identified as resistance mutations (Table 4). For genotype 1b, a mutation at residue 30 was observed (R30Q), but only as a secondary mutation in combination with L31F mutation (Table 2). To assess the potential impact of mutations at this position in genotype 1b, a glutamic acid residue was introduced at position 30 in the 1b replicon (R30E). While the Q30E mutation in genotype 1a yielded a very high level of resistance (∼25,000-fold) (Table 5), the R30E genotype 1b variant was only slightly less sensitive to BMS-790052 than the wild-type replicon (6-fold resistance) (Table 3). Perhaps the most intriguing resistance mutation identified in the resistance studies was the deletion of residue 30 in genotype 1a under high selective pressure. When this change was introduced into the genotype 1b replicon (ΔR30), the resulting clone did not replicate in transient transfection assays (Table 3). However, a cell line harboring a genotype 1b replicon with this deletion was established by selection with G418 (21). Sequence analysis of NS5A cDNA recovered from this cell line confirmed the ΔR30 mutation and also revealed a consensus P32L substitution. Since the ΔR30 cell line was selected in the absence of inhibitor, we hypothesized that the P32L mutation might act as a compensatory mutation, enabling replication of the ΔR30 variant. In transient transfection assays, a ΔR30-plus-P32L replicon replicated at ∼9% of the level of the wild-type control, confirming the compensatory ability of the P32L mutation (Table 3). The EC50 of BMS-790052 was >1 μM against the ΔR30-plus-P32L replicon, suggesting that deletion of residue 30 severely impacts BMS-790052 potency in genotype 1b as well as genotype 1a.
Conservation of NS5A residues important for BMS-790052 resistance.
The variation in NS5A in regions important for resistance development (residues 28 to 32 and 93) was examined in sequences deposited in the European HCV database (4). The genotype 1a and 1b consensus residues at these positions match the sequences from the H77c and con1 replicon clones (Fig. 3; Table 6). The primary residues involved in genotype 1b resistance, L31 and Y93, were conserved in 93.7 and 91.6%, respectively, of the 1,796 database sequences (Table 6). The most frequently observed variation at residue 31 was the L31M mutation (3.8%), a change that by itself minimally affected BMS-790052 potency in the 1b replicon (Table 3). The L31F and L31V resistance mutations were present in 0 and 2.1% of the sequences, respectively. The most common variation at residue 93 was the Y93H mutation, which was observed in 8.3% of the sequences. The Y93H substitution yielded ∼19-fold reduced susceptibility to BMS-790052 in the 1b replicon clone (EC50 = 49.2 pM) (Table 3).
TABLE 6.
Prevalence of BMS-790052 resistance mutations in the European HCV database
| Genotype (n) | Residue or mutation at NS5A positiona: |
|||||
|---|---|---|---|---|---|---|
| 28 | 29 | 30 | 31 | 32 | 93 | |
| 1b (1,796) | L (99.0) | P (99.9) | R (89.8) | L (93.7) | P (99.9) | Y (91.6) |
| T (0) | Q (3.1) | F (0) | L (0) | C (0.1) | ||
| ΔR (0) | M (3.8) | H (8.3) | ||||
| V (2.1) | N (0) | |||||
| 1a (548) | M (98.0) | P (100) | Q (97.6) | L (99.8) | P (100) | Y (99.5) |
| T (0) | E (0) | M (0.2) | L (0) | C (0.2) | ||
| K (0) | V (0) | H (0.4) | ||||
| H (1.1) | N (0) | |||||
| R (1.3) | ||||||
| ΔQ (0) | ||||||
Values in parentheses are the percentages of database sequences with the indicated amino acids.
For genotype 1a, M28, Q30, L31, and P32 were conserved in >97% of the 548 database sequences (Table 6). The most commonly observed resistance mutations were Q30H and Q30R, which were present in 1.1 and 1.3% of the sequences, respectively. Among the positions examined, residues 29 and 32 were the most conserved, with prolines present in ≥99.9% of the database sequences from genotypes 1a and 1b, suggesting that little variation may be tolerated at these positions in vivo.
Sensitivity of resistant variants to other HCV inhibitors.
Many of the resistant variants identified in this study were still inhibited by BMS-790052, with EC50s in the picomolar to low-nanomolar range (Tables 3 and 5). However, genotype 1a variants such as the Q30E and Y93N variants, as well as genotype 1a variants with linked resistance substitutions, exhibited much higher levels of resistance. We therefore tested a subset of the highly resistant variants for sensitivity to other HCV inhibitors. For this analysis, replicon cell lines with specific amino acid substitutions in NS5A (Q30E, Y93N, Q30H + Y93H, and M28T + Y93H) were established and tested for susceptibility to pegIFN-α, an HCV NS3 PI, and nonnucleoside inhibitors targeting the thumb (NS5B-1) and palm (NS5B-2) regions of NS5B. As reported in Table 7, these replicon cell lines, although highly resistant to BMS-790052, remained fully sensitive to the other HCV inhibitors.
TABLE 7.
Potencies of HCV inhibitors on BMS-790052-resistant replicon cell linesa
| Inhibitor | EC50 on cell line |
||||
|---|---|---|---|---|---|
| Parent | Q30E | Y93N | M28T + Q30H | Q30H + Y93H | |
| BMS-790052 (nM) | 0.038 | 217 | 399 | >1,000 | 471 |
| PI (nM) | 9.9 | 4.4 | 4.1 | 4.6 | 2.7 |
| NS5B-1 (μM) | 2.7 | 3.3 | 1.8 | 3.2 | 1.5 |
| NS5B-2 (nM) | 43.2 | 27.8 | 17.4 | 25.2 | 15.5 |
| pegIFN-α (ng/ml) | 1.7 | 2.3 | 1.7 | 2.9 | 3.0 |
Genotype 1a replicon cell lines with the indicated changes in NS5A were assayed for sensitivity to HCV replication inhibitors. Values are EC50s, with units as indicated in the first column, from a representative experiment. PI, ITMN-191; NS5B-1, analog of JTK-109; NS5B-2, HCV796 (see Materials and Methods for references).
DISCUSSION
BMS-790052 is a novel small-molecule inhibitor of HCV replication that targets NS5A and has shown impressive clinical efficacy in a phase 1, single-ascending-dose trial (6). With a target distinct from those of the many HCV protease and polymerase inhibitors in various stages of clinical development, BMS-790052 has the potential to be an important component in combination strategies for HCV therapy. As an initial step toward understanding the potential for in vivo resistance to this unique inhibitor, a comprehensive study of the resistance profile of BMS-790052 in genotype 1a and 1b replicon cells was undertaken.
For genotype 1b, resistance selection was performed on cells harboring replicons with three different cell culture-adaptive mutations (S2204I and ΔS2197 in NS5A and R2884G in NS5B). In each of these replicon cell backgrounds, the predominant mutations associated with BMS-790052 resistance were found at NS5A residues 31 (L31F/V) and 93 (Y93H/N). These results indicate that specific cell culture-adaptive mutations within the replicon did not substantially affect resistance selection outcome, thus providing confidence that the findings from these studies will also be relevant in vivo for viruses without adaptive mutations. In support of this conclusion, BMS-790052 was previously shown to be equally potent against genotype 2a replicons and viruses of the JFH-1 strain, which does not require adaptive mutations for efficient RNA replication (6).
In transient replicon transfection assays, individual changes at L31 and Y93 modestly affected BMS-790052 potency, and combinations of two of these changes had much greater effects. Substitutions at residues 28 and 32, although identified infrequently (P32L) or not at all (L28T) in inhibitor-selected cells, were also capable of acting as primary resistance mutations, moderately reducing BMS-790052 potency in transient transfection assays. In addition, L23F, R30Q, and P58S mutations were identified as secondary resistance substitutions, not affecting inhibitor potency themselves but markedly enhancing resistance when linked with a primary mutation.
In genotype 1a replicon cells, substitutions at NS5A residues 28 (M28T), 30 (Q30E/H/R), 31 (L31M/V), 32 (P32L), and 93 (Y93C/H/N) were all observed as drug-induced changes. In transient transfection assays, these individual substitutions conferred substantially higher levels of resistance than single amino acid changes in the genotype 1b replicon (Tables 3 and 5). The Q30E and Y93N substitutions, in particular, conferred very high levels of resistance to BMS-790052 in transient transfection assays (EC50s, ≥150 nM). When selective pressure on 1a replicon cells was increased sequentially, combinations of two of the resistance substitutions (e.g., M28T + Q30H) and a deletion of residue 30 (ΔQ30) were the predominant identified changes. Not surprisingly, replicon clones bearing these mutations exhibited very high levels of resistance to BMS-790052 in transient transfection assays (EC50s, ≥553 nM).
Collectively, the genotype 1a and 1b replicon selection studies identified NS5A residues 28 to 32 and 93 as key regions of the protein responsible for mediating inhibitor activity. As such, these residues likely delineate an inhibitor interaction site within the N terminus of NS5A, with binding of inhibitor disrupting one or more functions of NS5A required for RNA replication. While the precise role(s) of NS5A in RNA replication remains elusive, much has been learned about the structure and functions of the protein's N-terminal domains. NS5A amino acids 1 to 30, for example, function as a membrane anchor, directing NS5A or a heterologous protein to an endoplasmic reticulum (ER)-derived membrane compartment, likely the site of HCV replication (2, 5). Concordantly, NS5A amino acids 5 to 25 were demonstrated to form an amphipathic α-helix with a lipid bilayer-associated hydrophobic face and a solvent-exposed polar face (28). These findings suggest that NS5A is targeted to an ER-derived intracellular membrane in such a way that the hydrophobic surface of the amphipathic α-helix is embedded in-plane in a cytosolic leaflet of the membrane, while the polar surface of the helix remains exposed to the cytosol, potentially available for inter- or intramolecular interactions (28). Penin et al. (28) hypothesized that the cytosol-accessible polar surface of the α-helix could provide an essential platform for protein-protein interactions required for assembly of a functional HCV replication complex. Our resistance selection studies identified amino acid substitutions within (L23F) and very near (M28T) the N-terminal amphipathic α-helix of NS5A that alter BMS-790052 potency, thus suggesting that the structure or function of this domain could be impacted directly by inhibitor binding. Preliminary investigations suggest that BMS-790052-like NS5A inhibitors do not grossly affect NS5A membrane association, although more subtle effects on localization have not been excluded (D. Qiu, C. Wang, and R. Fridell, unpublished data). Even if they do not disrupt membrane association, BMS-790052 and related inhibitors could still potentially impair NS5A function by altering the structure or composition of a multiprotein complex proposed to assemble on the platform of the N-terminal amphipathic α-helix (28). Further investigations will be required to explore these possibilities.
Crystal structures of NS5A domain I without the N-terminal amphipathic α-helix have been reported by Tellinghuisen et al. (35) and Love et al. (22). In both studies, NS5A was crystallized as a colinear homodimer. While the NS5A monomeric units are very similar in the two structures, the structures differ in having nonoverlapping dimer interfaces, suggesting the possibility that NS5A could assume two physiologically relevant dimer configurations, or potentially a multimeric structure (22). In the structure of Tellinghuisen et al. (35), the dimer interface is located near the N termini of the monomers, with the bodies of the monomers forming a “claw-like” structure encompassing a potential RNA binding groove. This RNA binding groove is absent in the dimer of Love et al. (22), perhaps reflecting different functions for the dimers in HCV replication (22). Both structures are compatible with membrane localization mediated by two N-terminal amphipathic α-helices extending from monomers at the same end of the dimer. Specific structural information on NS5A residues 28 to 32, which are important for BMS-790052 resistance, is lacking because these amino acids are not included (22) or are disordered (35) in the structures. These residues would necessarily link the membrane-associated N-terminal amphipathic α-helices with the monomer cores and could thus conceivably form an inhibitor binding pocket between the membrane and the core of the monomers, with residues from each monomer contributing inhibitor contact sites. Such a binding model is consistent with the symmetrical structure of BMS-790052 (Fig. 1). Tyrosine-93, another residue involved in BMS-790052 resistance, is located near the dimer interface in both structures and interacts with the side chain of phenylalanine-37 in the neighboring monomer in the structure of Tellinghuisen et al. (22, 35). The identification of Y93 as an important site of BMS-790052 resistance suggests the possibility that inhibitor binding could affect dimer formation. If transitioning between alternate dimer configurations is important for HCV replication, it is intriguing to speculate that BMS-790052 could influence this transition, for example, by binding across the dimer interface and stabilizing one conformation over the other. There is currently no empirical evidence to support such a model, however, and further work, including cocrystallization studies, will be required to better understand the binding mode and mechanism of inhibition of BMS-790052 and related inhibitors.
The results from our selection studies identified several differences in the genotype 1a and 1b resistance profiles. Among these was selection of drug-induced mutations at residues 28 and 30. For residue 28, the explanation for this difference is likely trivial. Although a threonine at residue 28 was found to confer resistance in both genotypes, this amino acid substitution was selected only in genotype 1a, likely because this change requires two nucleotide changes in the 1b replicon (L28T; CTC to ACC) but only one change in the 1a replicon (M28T; ATG to ACG). This finding suggests that resistance mutations not observed in the replicon cells due to codon or amino acid usage will likely surface in vivo, where the range of preexisting viral sequences is expected to be much greater. Differences observed in resistance mutations at residue 30 are less easily explained. Residue 30 was identified as a major site of resistance in genotype 1a (Q30H/R/E) but not in genotype 1b. Interestingly, the Q30R mutation, which replaces the wild-type genotype 1a residue with the genotype 1b residue, resulted in a >1,000-fold decrease in BMS-790052 potency. More dramatically, a glutamic acid residue at position 30 conferred a very high level of resistance in the genotype 1a replicon (∼25,000-fold) but had very little effect in the genotype 1b replicon (R30E; 6-fold resistance compared to wild type). A similar situation was also observed at residue 31, with the L31M substitution, which conferred substantial resistance in genotype 1a (350-fold) but very little resistance (3-fold) in genotype 1b. Thus, while the overall patterns of resistance between genotypes 1a and 1b were found to be similar, implying similar binding modes of BMS-790052, they also revealed important differences, including less flexibility for changes at some residues and an overall greater decrease in inhibitor susceptibility caused by most single amino acid substitutions in genotype 1a than in genotype 1b.
When selection was carried out with a genotype 1b hybrid replicon that had sequences encoding the first 100 amino acids of NS5A derived from genotype 1a (1b1a100), the pattern of resistance was very similar to that observed with the genotype 1a replicon, including a high prevalence of mutations at residue 30. This result is consistent with the N-terminal region of NS5A mediating the interaction with BMS-790052 and further implies that the sequence context of the first 100 amino acids of NS5A largely determines resistance selection. On the whole, the results also validate the use of hybrid replicons as tools to study genotype-specific NS5A inhibitor susceptibilities. One important difference, however, was observed between the authentic 1a replicon and the 1b1a100 hybrid replicon: the levels of resistance conferred by resistant variants in transient transfection assays were consistently greater in the authentic 1a replicon than in the 1b1a100 hybrid replicon. As an example, the M28T mutation conferred 683-fold resistance in the genotype 1a replicon but only 50-fold resistance in the hybrid replicon. The reason for this discrepancy is not clear, but it does suggest that the inhibitory activity of BMS-790052 is also affected by regions outside the first 100 amino acids of NS5A. One possibility is that interactions between the NS5A N-terminal region and other components of the HCV replication complex influence the impact of resistance mutations. To date, no amino acid substitutions that contribute to BMS-790052 resistance have been identified beyond the N-terminal 93 amino acids of NS5A. However, the range of resistance mutations identified and the identification of secondary substitutions that modulate the effects of primary mutations suggest that the current findings could underestimate the complexity of resistance to this inhibitor.
Across HCV genotypes, variation is observed at several of the residues identified as important sites of genotype 1 resistance. For example, for the majority of sequences in the European HCV database, residue 30 is a lysine in genotype 2a and an alanine in genotype 3a, residue 31 is a methionine in genotype 4a, and residue 93 is a threonine in genotype 5a (not shown). Despite this variation, BMS-790052 exhibited excellent potencies (EC50s, ≤146 pM) against replicon clones with NS5A sequences derived from these diverse genotypes (6). These results are consistent with the findings of the current report, which reveal that the sequence context of a particular resistance mutation is very important in determining its impact on BMS-790052 susceptibility.
Among the drug-induced mutations identified in this study, the deletion of residue 30, selected in genotype 1a at a very high selective pressure, conferred the highest level of resistance to BMS-790052. In transient replicon transfection assays, a replicon carrying this mutation replicated at ∼7% of the level of the parental replicon. In genotype 1b, replication of a replicon with a deletion of residue 30 required an additional compensatory change (P32L). Also, deletion of residue 30 was not observed in any of the genotype 1a or 1b sequences deposited in the European HCV database (n = 2,344) (Table 6), suggesting that this variant is unlikely to be encountered as a prevalent preexisting variant in vivo. Collectively, these findings suggest that this mutation may not figure prominently in clinical resistance to BMS-790052. In addition, the residue 30 deletion variants remained fully sensitive to other DAA (data not shown).
The number of mutations identified that confer resistance to BMS-790052 (e.g., M28T, Q30E/H/R, L31M/V, and Y93C/H/N mutations in genotype 1a) might at first glance suggest a relatively low resistance barrier for this inhibitor compared to other direct-acting antivirals. However, since BMS-790052 is such a potent inhibitor of wild-type genotype 1 replicons (EC50, <10 pM in transient transfection assays), most resistant variants with single amino acid substitutions were still inhibited by BMS-790052 at subnanomolar (genotype 1b) or low-nanomolar (genotype 1a) concentrations. Data from a single-ascending-dose clinical study indicated that plasma concentrations of BMS-790052 capable of suppressing most of the single-amino-acid variants with resistance should be readily achievable (6). In this case, only a few of the individual variants (e.g., Q30E and Y93N in genotype 1a) or double amino acid substitutions may ultimately prove clinically relevant.
Given the error rate of the HCV replicase and the high degree of genetic variability preexisting within infected individuals (14, 33), it is unlikely that BMS-790052 or any other DAA will be successful as a monotherapeutic agent for HCV infection. Importantly, NS5A variants exhibiting high levels of resistance to BMS-790052 remained fully sensitive to pegIFN-α as well as to small-molecule inhibitors of HCV protease and polymerase (Table 7), indicating that these variants would likely be suppressed in combination therapy approaches. With its extraordinary potency and nonoverlapping resistance profile, BMS-790052 has the potential to be an important component in future HCV therapeutic strategies.
Acknowledgments
We thank Mark Cockett and Nicholas Meanwell for leadership and support. We also thank Donald O'Boyle, Jin-Hua Sun, Peter Nower, Mengping Liu, Stacey Voss, Julie Lemm, Karen Rigat, and Susan Roberts for helpful discussions.
Footnotes
Published ahead of print on 28 June 2010.
REFERENCES
- 1.Asselah, T., Y. Benhamou, and P. Marcellin. 2009. Protease and polymerase inhibitors for the treatment of hepatitis C. Liver Int. 29(Suppl. 1):57-67. [DOI] [PubMed] [Google Scholar]
- 2.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 3.Chevaliez, S., and J.-M. Pawlotsky. 2007. Hepatitis C virus: virology, diagnosis and management of antiviral therapy. World J. Gastroenterol. 13:2461-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Combet, C., N. Garnier, C. Charavay, D. Grando, D. Crisan, J. Lopez, A. Dehne-Garcia, C. Geourjon, E. Bettler, C. Hulo, P. L. Mercier, R. Bartenschlager, H. Diepolder, D. Moradpour, J. M. Pawlotsky, C. M. Rice, C. Trepo, F. Penin, and G. Deléage. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 35:D363-D366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elazar, M., P. Liu, C. M. Rice, and J. S. Glenn. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, M., R. E. Nettles, M. Belema, L. B. Snyder, V. N. Nguyen, R. A. Fridell, M. H. Serrano-Wu, D. R. Langley, J.-H. Sun, D. R. O'Boyle II, J. A. Lemm, C. Wang, J. O. Knipe, C. Chien, R. J. Colonno, D. M. Grasela, N. A. Meanwell, and L. G. Hamann. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical efficacy. Nature 465:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanoulle, X., D. Verdegem, A. Badillo, J. M. Wieruszeski, F. Penin, and G. Lippens. 2009. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 38:634-638. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. John, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 9.Hirashima, S., T. Suzuki, T. Ishida, S. Noji, S. Yata, I. Ando, M. Komatsu, S. Ikeda, and H. Hashimoto. 2006. Benzimidazole derivatives bearing substituted biphenyls as hepatitis C virus NS5B RNA-dependent RNA polymerase inhibitors: structure-activity relationship studies and identification of a potent and highly selective inhibitor JTK-109. J. Med. Chem. 49:4721-4736. [DOI] [PubMed] [Google Scholar]
- 10.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus non-structural protein 5A (NS5A) is an RNA binding protein. J. Biol. Chem. 280:36417-36428. [DOI] [PubMed] [Google Scholar]
- 11.Huang, Y., K. Staschke, R. De Francesco, and S. L. Tan. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Kieffer, T. L., A. D. Kwong, and G. R. Picchio. 2010. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs). J. Antimicrob. Chemother. 65:202-212. [DOI] [PubMed] [Google Scholar]
- 13.Kneteman, N. M., A. Y. Howe, T. Gao, J. Lewis, D. Pevear, G. Lund, D. Douglas, D. F. Mercer, D. L. Tyrrell, F. Immermann, I. Chaudhary, J. Speth, S. A. Villano, J. O'Connell, and M. Collett. 2009. HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology 49:745-752. [DOI] [PubMed] [Google Scholar]
- 14.Le Guillou-Guillemette, H., S. Vallet, C. Gaudy-Graffin, C. Payan, A. Pivert, A. Goudeau, and F. Lunel-Fabiani. 2007. Genetic diversity of the hepatitis C virus: impact and issues in the antiviral therapy. World J. Gastroenterol. 13:2416-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemm, J. A., D. R. O'Boyle II, M. Liu, P. T. Nower, R. Colonno, M. S. Deshpande, L. B. Snyder, S. W. Martin, D. R. St. Laurent, M. H. Serrano-Wu, J. L. Romine, N. A. Meanwell, and M. Gao. 2010. Identification of NS5A inhibitors. J. Virol. 84:482-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemm, J. A., M. Liu, R. E. Rose, R. Fridell, D. R. O'Boyle, R. Colonno, and M. Gao. 2005. Replication-competent chimeric hepatitis C virus subgenomic replicons. Intervirology 48:183-191. [DOI] [PubMed] [Google Scholar]
- 17.Lemon, S. M., C. Walker, M. J. Alter, and M. Yi. 2007. Hepatitis C virus, p. 1254-1304. In D. M. Knipe et al. (ed.), Fields virology, 5th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 18.Liang, Y., H. Ye, C. B. Kang, and H. S. Yoon. 2007. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46:11550-11558. [DOI] [PubMed] [Google Scholar]
- 19.Liu, S., I. H. Ansari, S. C. Das, and A. K. Pattnaik. 2006. Insertion and deletion analyses identify regions of non-structural protein 5A of hepatitis C virus that are dispensable for viral genome replication. J. Gen. Virol. 87:323-327. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 22.Love, R. A., O. Brodsky, M. J. Hickey, P. A. Wells, and C. N. Cronin. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83:4395-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macdonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485-2502. [DOI] [PubMed] [Google Scholar]
- 24.Magiorkinis, G., E. Magiorkinis, D. Paraskevis, S. Y. Ho, B. Shapiro, O. G. Pybus, J. P. Allain, and A. Hatzakis. 2009. The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med. 6:e1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moucari, R., and P. Marcellin. 2009. Hepatitis C: the role of new interferons in the era of STAT-C. Nat. Rev. Gastroenterol. Hepatol. 6:509-511. [DOI] [PubMed] [Google Scholar]
- 26.O'Boyle, D. R., P. T. Nower, J. L. Lemm, L. Valera, J.-H. Sun, K. Rigat, R. Colonno, and M. Gao. 2005. Development of a cell-based high-throughput specificity screen using a hepatitis C virus-bovine viral diarrhea virus dual replicon assay. Antimicrob. Agents Chemother. 49:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parfieniuk, A., J. Jaroszewicz, and R. Flisiak. 2007. Specifically targeted antiviral therapy for hepatitis C virus. World J. Gastroenterol. 13:5673-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279:40835-40843. [DOI] [PubMed] [Google Scholar]
- 29.Sarrazin, C., and S. Zeuzem. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447-462. [DOI] [PubMed] [Google Scholar]
- 30.Schinazi, R. F., L. Bassit, and C. Gavegnano. 2010. HCV drug discovery aimed at viral eradication. J. Viral Hepat. 17:77-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seiwert, S. D., S. W. Andrews, Y. Jiang, V. Serebryany, H. Tan, K. Kossen, P. T. Rajagopalan, S. Misialek, S. K. Stevens, A. Stoycheva, J. Hong, S. R. Lim, X. Qin, R. Rieger, K. R. Condroski, H. Zhang, M. G. Do, C. Lemieux, G. P. Hingorani, D. P. Hartley, J. A. Josey, L. Pan, L. Beigelman, and L. M. Blatt. 2008. Preclinical characteristics of the hepatitis C virus NS3/4A protease inhibitor ITMN-191 (R7227). Antimicrob. Agents Chemother. 52:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 33.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 34.Tellinghuisen, T. L., J. Marcotrigiano, A. E. Gorbalenya, and C. M. Rice. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576-48587. [DOI] [PubMed] [Google Scholar]
- 35.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tellinghuisen, T. L., K. L. Foss, J. C. Treadaway, and C. M. Rice. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, A. J., and J. G. McHutchison. 2009. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J. Viral Hepat. 16:377-387. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, A., K. Patel, H. Tillman, and J. G. McHutchison. 2009. Directly acting antivirals for the treatment of patients with hepatitis C infection: a clinical development update addressing key future challenges. J. Hepatol. 50:184-194. [DOI] [PubMed] [Google Scholar]