Abstract
Shiga toxin (Stx) in Escherichia coli O157:H7 is encoded as a late gene product by temperate bacteriophage integrated into the chromosome. Phage late genes, including stx, are silent in the lysogenic state. However, stress signals, including some induced by antibiotics, trigger the phage to enter the lytic cycle, and phage replication and Stx production occur concurrently. In addition to the Stx produced by O157:H7, phage produced by O157:H7 can infect harmless intestinal E. coli and recruit them to produce Shiga toxin. To understand how antibiotics influence Stx production, Stx lysogens were treated with different classes of antibiotics in the presence or absence of phage-sensitive E. coli, and Stx-mediated inhibition of protein synthesis was monitored using luciferase-expressing Vero cells. Growth-inhibitory levels of antibiotics suppressed Stx production. Subinhibitory levels of antibiotics that target DNA synthesis, including ciprofloxacin (CIP) and trimethoprim-sulfamethoxazole, increased Stx production, while antibiotics that target the cell wall, transcription, or translation did not. More Stx was produced when E. coli O157:H7 was incubated in the presence of phage-sensitive E. coli than when grown as a pure culture. Remarkably, very high levels of Stx were detected even when growth of O157:H7 was completely suppressed by CIP. In contrast, azithromycin significantly reduced Stx levels even when O157:H7 viability remained high.
Escherichia coli O157:H7 causes about 73,000 cases of disease annually within the United States (16). While intestinal colonization by E. coli O157:H7 leads to diarrhea, E. coli O157:H7 also produces Stx, which mediates systemic complications, including neurologic damage (30) and hemolytic-uremic syndrome (HUS) (12). Two antigenic forms of Stx—Stx1 and Stx2—share ca. 60% amino acid identity. Strains of O157:H7 can possess genes for Stx1, Stx2, or both. Epidemiological studies suggest that Stx2 is associated with systemic disease (2), and primates injected with purified Stx2 but not Stx1 developed HUS (29), suggesting that Stx2 is sufficient to produce HUS.
Stx is an AB5 toxin, with five identical B, or binding subunits, and an enzymatically active A subunit. The B-pentamer binds to a glycolipid receptor, globotriaosylceramide, to gain entry into the host cytoplasm. The A subunit is an RNA-glycohydrolase and removes an adenine residue from the rRNA, disrupting protein synthesis (3). The A and B subunits are secreted to the periplasm, where the toxin is assembled. AB5 toxin production has only been reported in Gram-negative bacteria, and the Gram-negative periplasmic compartment likely plays an important role in the biogenesis of AB5 toxins by ensuring that the concentration of toxin subunits is high enough to drive assembly.
While promoting toxin assembly, the outer membrane of the periplasm can act as a barrier to secretion. Cholera toxin is released from the periplasm by a type 2 secretion system (27), while pertussis toxin uses a type 4 secretion system (35). In contrast, Stx has a very unusual strategy for cellular release; it is released by the process of phage-mediated lysis. The genes for Stx are encoded within the late-gene region of temperate bacteriophages integrated in the bacterial chromosome (19, 26). The phage late genes encode proteins responsible for viral replication, assembly, and lysis of the host E. coli. The late genes are silent during lysogeny and only become expressed during the lytic cycle. Both Stx and new viral particles are released when the bacteria undergo lysis, thus eliminating the need for a periplasmic secretion system. The switch from lysogeny to the lytic cycle is controlled by the bacterial SOS stress response (33), which is induced by certain antibiotics. Especially effective are antibiotics that interfere with DNA replication, and these include the quinolone antibiotic ciprofloxacin (CIP), which inhibits DNA gyrase, and trimethoprim-sulfamethoxazole (SXT), which blocks nucleotide synthesis (9, 34). These antibiotics induce Stx production in vitro and in animal models of disease in vivo (7, 40). Furthermore, treatment of patients with CIP or SXT is epidemiologically associated with the development of Stx-mediated systemic complications (36).
The ability of other antibiotics to induce Stx production in vivo is less clear. Some studies report antibiotic treatment of patients with STEC infection to be beneficial and others do not (reviewed in reference 23). Similarly, studies examining the ability of antibiotics to promote Stx production in vitro have given mixed results (9, 13, 18, 21, 25, 37). In several studies Stx expression was assessed by enzyme-linked immunosorbent assay (ELISA), Western blotting, or mRNA levels, and these indirect assays may not accurately reflect levels of biologically active Stx, since not all subunits may be part of assembled functional AB5 complexes.
In addition to the concern regarding the ability of antibiotics to induce Stx production, little experimental attention has been given to the potential for Stx-encoding phage released during the lytic cycle to infect other E. coli and elevate Stx production. Usually, when an antibiotic kills toxin-producing bacteria, the potential for toxin production is destroyed. However, Stx-phage released from dead O157:H7 can infect phage-sensitive E. coli. Lysogeny is a rare event and, typically, newly infected bacteria will undergo lytic infection, resulting in more phage and Shiga toxin production. In vitro, E. coli O157:H7 incubated with Stx-phage-susceptible E. coli produced up to 1,000-fold more Stx than O157:H7 grown in pure culture (8). Similarly, mice infected with O157:H7 produced significantly higher levels of Stx if their intestinal flora contained Stx-phage-sensitive E. coli rather than Stx-phage-resistant E. coli (7, 8). About 10% of human fecal isolates are sensitive to Stx-phage (5, 6, 8); the composition of the intestinal flora could influence the amount of Stx produced during human infection and ultimately the course of disease.
In the present study, we examined the influence of antibiotics on the production of biologically active Stx by E. coli O157:H7 grown in pure culture or in the presence of Stx-phage-sensitive E. coli. We confirmed that antibiotics, such as CIP and SXT, that alter DNA metabolism increase Stx production; however, translation inhibitors, especially azithromycin (AZM), did not induce Stx production and prevented subsequent phage-mediated toxin production by phage-sensitive E. coli.
MATERIALS AND METHODS
Bacterial strains.
Strains are described in Table 1. Bacteria were cultured at 37°C in Mueller-Hinton (MH) broth or Luria-Bertani (LB) broth or agar. Modified LB agar containing 10 mM CaCl2 was used in phage infection studies.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| C600::933W | Laboratory strain C600 lysogenized with Stx2-converting phage 933W | 20 |
| C600::H19B | Laboratory strain C600 lysogenized with Stx1-converting phage H19B | 20 |
| PT-32 | Clinical isolate of E. coli O157:H7; stx1stx2 | 8 |
| 185 | Strr mutant of PT-32 transformed with pBBR1MCS-2; Kanr | 8 |
| ECOR-4 | Human E. coli intestinal isolate, susceptible to Stx phage | 5, 6 |
| CMUC-170 | ECOR-4 transformed with pUW2138; Gentr | This study |
| CMUC-172 | E. coli intestinal isolate ECOR-4 susceptible to Stx phage, transformed with pBBR1MCS-3, Ampr Tetr | This study |
| Plasmids | ||
| pBBR1MCS-2 | Kanr | 15 |
| pBBR1MCS-3 | Ampr Tetr | 15 |
| pGL4.11[luc2p] | Luciferase gene tagged with PEST degradation sequence | Promega |
| pMig-R1 | Retroviral vector, multiple cloning site, IRES, GFP | 24 |
| pUW2138 | Gentr; OriT in pBluescript SK(+) | 4 |
Strr, streptomycin resistant; Gentr, gentamicin resistant; Ampr, ampicillin resistant; Tetr, tetracycline (doxycycline) resistant.
Luciferase assay for Stx.
The gene for destabilized luciferase, luc2p, was excised from pGL4.11[luc2p] (Promega) with HindIII and XbaI, blunted using Klenow, and cloned into the HpaI site of MigR1 (1). HEK GP2-293 retrovirus packaging strain (Clontech) was incubated with 3 μg of Luc2p-MigR1 DNA and 1 μg of pVSV-G envelope as previously described (28). Transfected Vero cells were sorted by flow cytometry; green fluorescent protein (GFP)-positive cells were propagated and sorted a second time to obtain cells expressing high levels of GFP. These cells have been deposited with the Biodefense and Emerging Infections Research Resources Repository (http://www.beiresources.org/).
Stx was quantified by determining the effective dose to inhibit protein synthesis by 50% (ED50). Culture supernatant was diluted 1:100 in phosphate-buffered saline (PBS), followed by half-log serial dilutions, and 25 μl of each dilution was added to white tissue culture-treated, Falcon 96-well microtiter plates (Becton Dickinson, Franklin Lakes, NJ). Then, 100 μl containing 104 luciferase-expressing Vero cells was added to each well, followed by incubation for 4 h at 37°C with 5% CO2. The cells were washed three times with PBS, 25 μl of Superlite luciferase substrate (Bioassay Systems, Hayward, CA) was added, and light was measured by using a Luminoskan Ascent (Thermo Labsystems, Helsinki, Finland). Culture supernatants from C600::933W containing 258 μg of Stx2/ml or from C600::H19B containing 108 μg of Stx1/ml were used as standards (5, 6). Luciferase-expressing Vero cells incubated without toxin were used as the negative control to determine the maximum light production. The ED50 was calculated using the two points above and below the midpoint. Values from the luciferase assay were comparable to those obtained using the standard 3-day Vero cell assay (8), where a loss of cells is observed after staining with Giemsa (data not shown).
Detection of antigenic Stx.
Western blots were performed essentially as described previously (8). Biotinylated receptor mimics specific for Stx1 and Stx2 were used as previously described (11).
Antibiotic susceptibility.
TMP and sulfamethoxazole (i.e., SXT) were combined at a ratio of 1:5. The MIC of the antibiotic was determined by the broth macrodilution assay (10). Briefly, E. coli specimens were grown overnight in MH broth and diluted to 1:1,000 and then to 1:200. To examine cells in logarithmic phase, overnight cultures were diluted to 1:1,000, grown to early logarithmic phase (optical density at 600 nm [OD600] of ∼0.04), and diluted as described above. Next, 1 ml was added to 1.0 ml of antibiotic in MH broth, and the cultures were incubated at 37°C overnight with rotary aeration. The MIC was visually determined and corresponded to ∼107 CFU/ml. Log ED50 values were analyzed for statistical significance. Experimental samples were compared to the no-antibiotic control by using an unpaired Student t test with Welch's correction for unequal variance (GraphPad Prism 5). In cases where toxin levels were below the LOD, the square root of the LOD was used as the estimated value for statistical analysis.
Coincubation of O157:H7 with Stx-phage-susceptible E. coli.
O157:H7 strain 185 and antibiotic-resistant derivatives of human intestinal strain ECOR-4, susceptible to Stx2 phage infection (5, 6), were incubated separately overnight in LB broth. Cells were washed by centrifugation to remove toxin and free phage particles, and the OD600 was adjusted to 1. Strain 185 and the ECOR-4 derivative were mixed at a ratio of 1:100 CFU (70 μl of strain 185 and 7 ml of the ECOR-4 derivative) in sterile test tubes containing 1 ml of LB medium with the appropriate antibiotic. Bacterial suspensions were overlaid onto modified LB agar in deep petri dishes, followed by incubation overnight at 37°C as a static culture. Stx-containing culture supernatants were filter sterilized and stored frozen. For coculture experiments, the growth-inhibitory concentration, or the concentration of antibiotic that resulted in <107 CFU/ml, was determined by plating serial dilutions on selective media. Samples with CFU/ml below the limit of detection (50 CFU/ml) were incorporated into the statistical analysis by using the square root of the limit of detection).
RESULTS
Luciferase assay to quantify Stx.
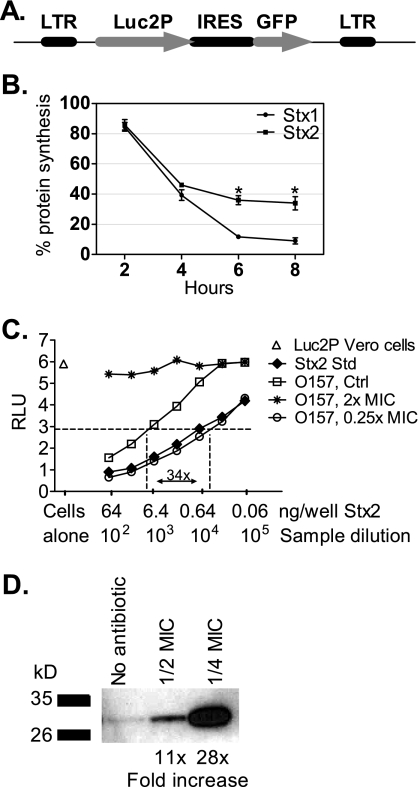
We developed an improved assay to assess Stx-mediated inhibition of protein synthesis based on the method of Zhao et al. (41). Vero cells were engineered to stably express the Luc2p variant of luciferase. Luc2p is tagged with a PEST sequence (rich in proline, glutamic acid, serine, and threonine), which promotes rapid turnover by targeting the protein to the proteasome for degradation. Therefore, luciferase activity reflects the current level of protein synthesis (41). Luc2p was cloned upstream of the GFP gene in the MigR1 retrovirus (Fig. 1 A). GFP and luciferase are transcriptionally coupled, and Vero cells transfected with MigR1-Luc2p were sorted for GFP expression. Luc2p Vero cells were incubated with 0.6 ng of Stx1 or Stx2 (Fig. 1B). The response to Stx1 and Stx2 was similar at 4 h but, after longer incubations, Stx1 caused a greater reduction in protein synthesis. Protein synthesis was evaluated after 4 h in all subsequent experiments.
FIG. 1.
Luciferase assay for Stx. (A) The Luc2p-MigR1 retroviral vector with the gene for Luc2p cloned upstream of the internal ribosomal entry site (IRES) of the gene for enhanced GFP. (B) Luciferase activity of Luc2p Vero cells incubated with 0.6 ng of Stx1 or Stx2, assessed as a function of time. The results are the average of three trials. The statistical significance between Stx1 and Stx2 is indicated by an asterisk (P < 0.05, Student t test). (C) Luciferase activity of Luc2p Vero cells. The Stx2 standard (Std) contains 258 μg of Stx2/ml. The average for six wells of untreated Luc2p Vero cells was determined to be 5.8 RLU, and the level of 50% inhibition or ED50 (2.9 RLU) is indicated by the horizontal dashed line. (D) E. coli O157:H7, strain PT-40, was grown in the presence or absence (control) of SXT as indicated. Equal toxin loads (64 ng) based on the Luc2p Vero cell assay were resolved by electrophoresis on an 8 to 16% Tris-glycine gel and blotted with polyclonal antibody to Stx2. Signal relative to the no-antibiotic control was determined by using ImageQuant.
Stx production was determined by using limiting dilution assays (Fig. 1C). In this example, untreated Luc2p Vero cells displayed ∼6 relative light units (RLU) of luciferase activity (Fig. 1C, open triangle). Incubation with the Stx2 standard caused a concentration-dependent decrease in luciferase expression (Fig. 1C, filled diamonds), and the ED50 for the Stx2 standard was determined to be 0.6 ng/well. Culture supernatant from the O157:H7 strain grown in the absence of antibiotics (Fig. 1C, unfilled squares) displayed less activity than the Stx2 standard. At growth-inhibitory levels of SXT (2× the MIC), no Stx was detected (Fig. 1C, stars). In contrast, incubation with subinhibitory levels of SXT (1/4 MIC) resulted in 34 times more Stx compared to untreated cultures (Fig. 1C, closed circles). This assay was used to quantify biologically active Stx in all subsequent experiments.
Antigenic versus active Stx.
Antigenic assays, including ELISA, have been used to quantify Stx production. However, the presence of unassembled, inactive toxin subunits could give false positive results. The presence of unassembled Stx2 was assessed by Western blotting (Fig. 1D). Culture supernatants from PT-40 incubated either without antibiotic or with SXT at 1/2 or 1/4 the MIC were loaded at 64 ng/well of Stx based on the Luc2p Vero cell assay. Signal was highly variable, likely due to the presence of unassembled toxin. Unlike cellular activities that are highly regulated, phage lysis is a highly variable process. Rapid bacterial lysis likely leads to large quantities of unassembled toxin, whereas slower lysis may promote subunit assembly and a higher percentage of functional, fully assembled toxin. These results suggest that antigenic assays cannot be used to assess levels of biologically active Stx.
Antibiotic treatment.
MIC values for various classes of antibiotics were determined (Table 2). All Shiga toxin-producing strains were antibiotic susceptible, and the MIC values were comparable to those reported previously. Since bacteria are more likely to be actively growing during infection, we also tested bacteria in the logarithmic phase. MICs for logarithmic-phase cultures were similar to stationary-phase cultures (Table 2).
TABLE 2.
MIC with cells starting in the stationary or logarithmic phase
| Antibiotic | MIC range (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|
| C600::933W (Stx2) |
C600::H19B (Stx1) |
PT-32 (Stx1 and Stx2) |
PT-40 (Stx2) |
|||||
| Stationary | Log | Stationary | Log | Stationary | Log | Stationary | Log | |
| CIP | 0.01-0.02 | 0.01-0.03 | 0.01 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 |
| SXT | 0.03-0.06 | 0.01-0.03 | 0.03 | 0.004-0.02 | 0.13 | 0.13-0.25 | 0.13 | 0.13-0.25 |
| AMP | 8-16 | 2-8 | 8-16 | 4-8 | 4-8 | 2-8 | 8-16 | 2-4 |
| CRO | 0.03-0.06 | 0.03-0.06 | 0.06-0.13 | 0.03 | 0.03-0.06 | 0.03-0.06 | 0.06 | 0.06 |
| FOS | 2 | 2-4 | 0.5-2 | 2 | 16 | 16-64 | 16-32 | 16-32 |
| RIF | 8-32 | 8-32 | 16-32 | 4-8 | 2-8 | 4-8 | 16 | 4-16 |
| GEN | 0.5-1 | 0.25-1 | 0.5-1 | 0.5 | 0.25-0.5 | 0.5 | 0.5-1 | 0.25-1 |
| DOX | 1 | 0.5-1 | 0.5-1 | 0.25-0.5 | 0.5-1 | 1-2 | 1-2 | 1-2 |
| AZM | 2-4 | 2-8 | 2-16 | 2-8 | 2.6 | 2.6-4 | 2.6-5.2 | 4 |
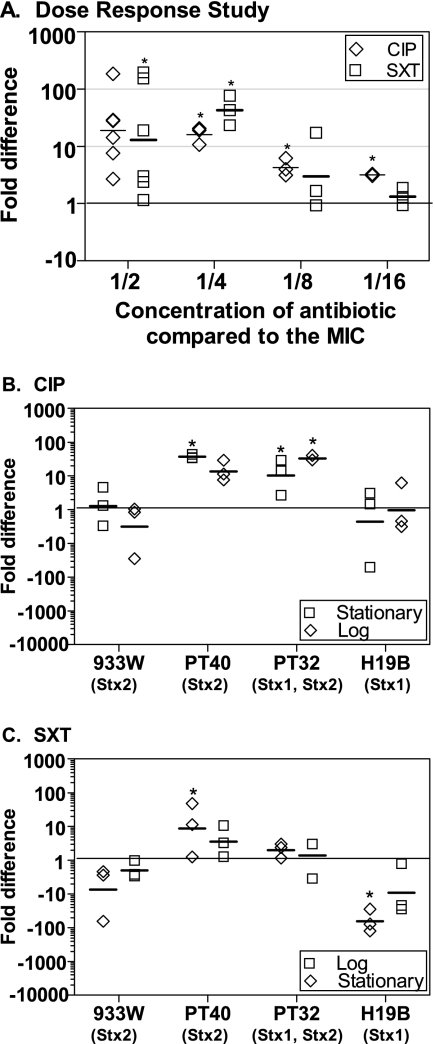
Subinhibitory levels of CIP and SXT increase Stx expression.
To verify previous studies showing that agents that inhibit DNA synthesis induce Stx production, O157:H7 strain PT-32 was incubated with serial dilutions of CIP or SXT. As expected, no Stx was detected when bacterial growth was inhibited by antibiotic at or above the MIC (data not shown). Subinhibitory concentrations of antibiotic increased Stx expression compared to the no-antibiotic control, and the most Stx was produced by bacteria grown in highest concentration of antibiotic that still supported bacterial growth (Fig. 2 A).
FIG. 2.
Effect of CIP and SXT on the production of Stx. The fold difference in toxin production compared to the no-antibiotic control was determined by using the luciferase assay. Each symbol represents one of three to six independent trials, and the horizontal dash denotes the geometric mean. Statistical significance (P < 0.05, Student unpaired t test with Welch's correction for unequal variance) compared to the no-antibiotic control is indicated by an asterisk. (A) Dose-response study. The O157:H7 strain PT-32 was incubated with subinhibitory concentrations of CIP or SXT. (B and C) Response of other strains to CIP (B) and SXT (C). E. coli strains inoculated in the stationary phase or the logarithmic phase were incubated overnight with CIP or SXT at 1/2 the MIC. In the absence of antibiotic, C600::933W, PT-32, and PT-40 produced between 8 and 27 μg of Stx/ml, whereas C600::H19B produced between 32 and 58 μg of Stx/ml.
The production of Stx by different Stx-encoding phage is very diverse (9, 32), and Stx production by other strains, as well as bacteria in stationary or logarithmic phase was assessed (Fig. 2B and C). Two clinical isolates of O157:H7, PT-40 (encoding Stx2), and PT-32 (encoding both Stx1 and Stx2), expressed significantly higher (P < 0.05, Student t test) levels of Stx in the presence of CIP or SXT (Fig. 2B and C), although the growth state of the bacteria appeared to influence Stx production (Fig. 2C). Increased expression of Stx was not observed for laboratory isolate C600 lysogenized with 933W (Stx2) or H19B (Stx1). These results are likely due to the different E. coli backgrounds, and it is important to perform such studies with clinical isolates.
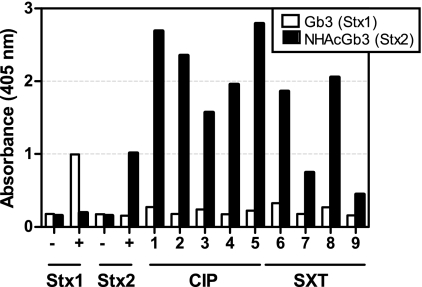
Stx2 is preferentially expressed.
Strains of O157:H7 can be lysogenized with multiple copies of Stx-encoding phage; however, not all stx genes are expressed (14, 17). PT-32 has the genes for both Stx1 and Stx2. ELISAs using receptor mimics (11) that preferentially capture Stx1 (Gb3) or Stx2 (NHAcGb3) were used to assess which forms of Stx are produced by strain PT-32. Signal was detected for Stx1 and Stx2 standards added at 1 μg/well (Fig. 3). Stx1-specific signal was at or near background levels for all of the cultures (Fig. 3, white bars); however, Stx2 was detected for cultures incubated with antibiotics (Fig. 3, black bars). These results suggest that PT-32 preferentially expresses Stx2 in the presence of CIP or SXT.
FIG. 3.
PT-32(stx1 stx2) produces Stx2. E. coli O157:H7 strain PT-32 was grown in the presence of CIP (samples 1 to 5) or SXT (samples 6 to 9) at 1/2 the MIC, and binding to receptor-specific analogues for Stx1 (Gb3) or Stx2 (NHAcGb3) was assessed. Stx1 or Stx2 added at 1 μg was used as a positive control (+), and wells incubated without Stx but incubated with primary and secondary antibody (−) served as a negative control. Nearly identical results were obtained when this assay was repeated (data not shown).
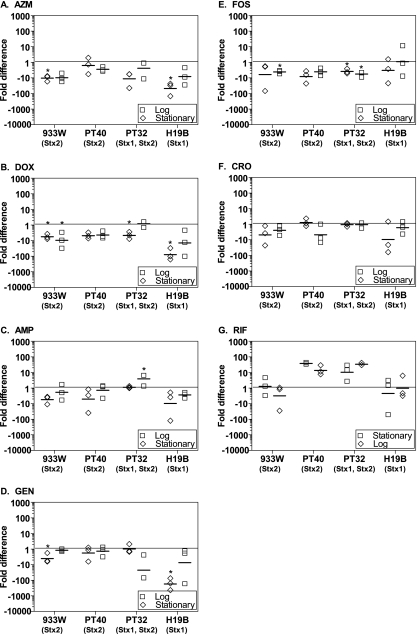
Other antibiotics do not induce Stx.
Stx production by cultures incubated with other antibiotics at 1/2 MIC was assessed. Under some conditions, AZM, doxycycline (DOX), fosfomicin (FOS), and gentamicin (GEN) caused a statistically significant decrease (P < 0.05, Student t test) in Stx production (Fig. 4 A to E). AMP caused a modest (<2-fold) but statistically significant increase in Stx2 production for PT-40 in logarithmic phase (Fig. 4E). Ceftriaxone (CRO) and rifampin (RIF) did not alter Stx production (Fig. 4F and G).
FIG. 4.
Influence of antibiotics on Stx production in pure culture. E. coli strains inoculated in the stationary phase (□) or the logarithmic phase (⋄) were incubated overnight with the following antibiotics at 1/2 the MIC: azithromycin (A), doxycycline (B), ampicillin (C), gentamicin (D), fosfomicin (E), ceftiaxone (F), or rifampin (G). The fold difference in toxin production compared to the no-antibiotic control is plotted. Each symbol represents one of three independent trials, and the horizontal dash denotes the geometric mean. The statistical significance (P < 0.05, Student t test unpaired t test with Welch's correction for unequal variance) compared to the no-antibiotic control is indicated by an asterisk.
Antibiotic susceptibility of O157:H7 in coculture with Stx-phage-susceptible E. coli.
To examine the influence of antibiotics on Shiga toxin production, we used a protocol published in previous studies which is designed to mimic the intestinal environment (7, 8). Briefly, overnight cultures of O157:H7 and the phage-sensitive strain were mixed at a ratio of 1:100. To mimic the static anaerobic environment of the intestine, 8 ml of the broth culture was layered onto modified L-agar plates, and the cultures were incubated overnight at 37°C without shaking. In coculture, the concentration of antibiotic that inhibits bacterial growth cannot be determined visually if the growth of one strain is not inhibited. To determine the growth-inhibitory concentration for O157:H7 in coculture, serial dilutions were plated on selective media. Since cultures containing fewer than 107 CFU/ml are not visibly turbid, the growth-inhibitory concentration for the O157:H7 strain was determined to be the lowest concentration of antibiotic resulting in <107 CFU/ml.
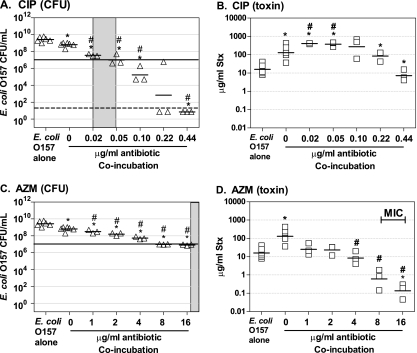
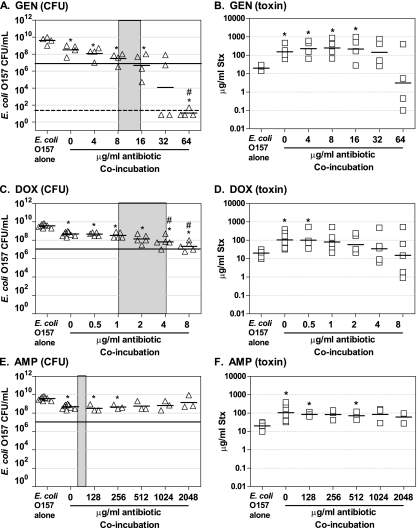
We assessed the antibiotic susceptibility of O157:H7 incubated in the presence of phage-susceptible, antibiotic-sensitive nontoxigenic E. coli (Fig. 5) or phage-susceptible, antibiotic-resistant nontoxigenic E. coli (Fig. 6). Recovery of O157:H7 is indicated by an open triangle with, the mean recovery indicated by a hatch and, for comparison, the MIC for O157:H7 in pure culture using the standard broth dilution method is indicated by the shaded area (Fig. 5A and C and Fig. 6A, C, and E). The growth-inhibitory concentration for O157:H7 incubated with CIP and AZM in coculture with antibiotic-sensitive strains was approximately the same as pure culture, as indicated by a mean CFU recovery of 107 occurring within the shaded region (Fig. 5A and C). Similarly, the growth-inhibitory concentration for O157:H7 incubated with GEN in the presence of a GEN-resistant strain was comparable to the MIC obtained with a pure culture (Fig. 6A). In contrast, growth of O157:H7 was not inhibited to below 107 when incubated with DOX or AMP in the presence of antibiotic-resistant strains (Fig. 6C and E). This was not due to transfer of the antibiotic resistance plasmid, since all O157:H7 recovered from coincubation were still sensitive to the DOX and AMP (data not shown). It is also important to note that the coincubation studies, unlike the standard broth dilution MIC method, were done with very high initial concentrations of bacteria. The ability of beta-lactamase-producing bacteria to inactivate the ampicillin in a culture is well described, suggesting the resistant strain could protect the antibiotic-sensitive O157:H7.
FIG. 5.
Influence of antibiotics on Stx production in coculture with antibiotic-sensitive, phage-susceptible E. coli. O157:H7 strain 185 was grown overnight in the presence of Stx-phage susceptible E. coli isolate (CMUC-170) susceptible to CIP (A and B) or AZM (C and D). Cultures were plated to determine the CFU (A and C), and Stx levels were determined by using the luciferase assay (B and D). Each symbol represents an independent trial, and the horizontal dash denotes the geometric mean. Controls (O157 alone and no-antibiotic coincubations) were repeated four to eight times, and all other coincubation experiments were repeated three to five times. Statistical significance (P < 0.05 Student unpaired t test with Welch's correction for unequal variance) compared to the pure culture of E. coli O157:H7 is denoted with an asterisk, and statistical significance compared to the no-antibiotic coculture control is denoted by a pound sign (#). The MIC for pure culture of E. coli O157:H7 is noted by a gray box. The horizontal line at 107 denotes the number of O157:H7 at the visual limit of detection. Samples with CFU/ml below the limit of detection (50 CFU/ml, which is represented by the dashed line) were incorporated into the statistical analysis using the square root of the limit of detection).
FIG. 6.
Influence of antibiotics on Stx production in coculture with antibiotic-resistant, phage-susceptible E. coli. E. coli O157:H7 strain 185 was grown overnight in the presence of Stx-phage susceptible E. coli isolate resistant to GEN (CMUC-170) (A and B), DOX (CMUC-172) (C and D), or AMP (CMUC-172) (E and F). Cultures were plated to determine CFU (A, C, and E) and Stx levels were determined by using the luciferase assay (B, D, and F), as described in Fig. 5. Each symbol represents an independent trial, and the horizontal dash denotes the geometric mean, Controls (O157 alone and no-antibiotic coincubations) were repeated four to eight times, and all other coincubation experiments were repeated three to five times. Statistical significance (P < 0.05 Student unpaired t test with Welch's correction for unequal variance) compared to the pure culture of E. coli O157:H7 is denoted by an asterisk, and statistical significance compared to the no-antibiotic coculture control is denoted with a pound sign. The MIC for pure culture of E. coli O157:H7 is noted by the gray box. The horizontal line at 107 denotes the number of O157:H7 at the visual limit of detection. Samples with CFU/ml below the limit of detection (50 CFU/ml, which is represented by the dashed line) were incorporated into the statistical analysis using the square root of the limit of detection).
Stx production in coculture with Stx-phage-susceptible, antibiotic-sensitive E. coli.
Stx-encoding phage can infect Stx phage-susceptible E. coli, leading to lytic infection and additional production of Stx. Furthermore, previous studies have shown that Stx levels in coculture experiments can greatly exceed those produced by O157:H7 grown as a pure culture (8). We assessed Stx production by O157:H7 incubated in the presence of phage-susceptible, antibiotic-sensitive nontoxigenic E. coli (Fig. 5) or phage-susceptible, antibiotic-resistant nontoxigenic E. coli (Fig. 6).
Coculture in the absence of antibiotic (Fig. 5A and C, 0 μg/ml) resulted in recovery of ∼10-fold fewer O157:H7 compared to pure culture (Fig. 5A and C, E. coli O157:H7), likely due to nutrient limitation. However, 100-fold fewer O157:H7 organisms were in the initial inoculum, suggesting that the O157:H7 outcompeted the phage-susceptible strain. A likely explanation is that some phage-susceptible bacteria were killed by lytic infection with the Stx2 phage. Lytic phage infection is further supported by the observation that coculture produced a statistically significant (P < 0.05, Student t test) 10-fold increase in Stx production over pure culture (Fig. 5B and D), compare O157 alone and 0 μg of antibiotic/ml, even though 10-fold-fewer O157:H7 organisms were recovered compared to the pure culture.
Stx2 levels were also assessed for coculture in the presence of antibiotic. At concentrations of CIP below the growth-inhibitory concentration (0.05 μg/ml) for O157:H7, Stx production in coculture was greater than that observed in the absence of antibiotic (Fig. 5B). At concentrations of CIP above the growth-inhibitory concentration (>0.05 μg/ml), growth of O157:H7 was inhibited; however, Stx recovery was greater that of a pure culture of O157:H7 grown in the absence of antibiotic. Remarkably, this was true even at 0.44 μg of CIP/ml, when no O157:H7 were recovered from the coculture. This toxin was likely produced by the phage-sensitive strain after infection with Stx-phage produced before O157:H7 was killed. In contrast to CIP, AZM significantly reduced Stx production at subinhibitory levels of antibiotics (4 μg/ml) and at inhibitory levels (8 to 16 μg/ml), compared to coincubation in the absence of antibiotic (Fig. 5D, #).
Coculture with antibiotic-resistant strains.
We wanted to examine the most confounding clinical scenario, treatment of antibiotic-susceptible O157:H7 in the presence of antibiotic-resistant, Stx-phage-susceptible intestinal E. coli, where Stx phage produced by the dying O157:H7 would be able to replicate freely in the intestinal E. coli. Strain O157:H7 was completely resistant to AMP when incubated with AMP-resistant E. coli (Fig. 6E) and partially resistant to DOX (Fig. 6C) compared to the values determined by the standard MIC method. To test for transfer of the antibiotic resistance plasmid, serial dilutions were plated on kanamycin to select for O157:H7 and then replica plated onto AMP and DOX. All of the O157:H7 colonies were sensitive to AMP and DOX. The increased resistance was likely due to the high density of bacteria under the coculture conditions or due to the ability of β-lactamase to inactivate ampicillin in the periplasmic space, reducing availability of ampicillin.
As observed in Fig. 5, coculture produced a statistically significant (P < 0.05, Student t test) 10-fold increase in Stx production over pure culture (Fig. 6B, D, and F, compare O157 alone and 0 μg of antibiotic/ml). However, neither subinhibitory or inhibitory levels of antibiotic in mixed culture significantly reduced Stx production compared to mixed culture in the absence of antibiotic (Fig. 6B, D, and F).
DISCUSSION
Antibiotic therapy for E. coli O157:H7 is complicated. The growth of O157:H7 in the intestinal tract leads to diarrhea, and patients would presumably benefit if antibiotic treatment eliminated the bacteria. However, systemic dissemination of Stx2 produced by the bacteria in the intestinal tract can lead to life-threatening complications, including neurological damage and hemolytic-uremic syndrome. Antibiotic treatments that induce the phage lytic cycle, resulting in increased Stx production, could lead to more serious disease. Indeed, unlike most pathogens where elimination of the bacteria by antibiotics eliminates additional toxin production, the Stx-encoding phage released by O157:H7 can infect non-O157:H7 strains, recruiting them to produce Stx (5-8). Furthermore, lysogeny is rare; phage infection usually results in lytic infection, and E. coli infected with Stx-phage can produce very high levels of Stx (6-8). Although the influence of antibiotics on Stx expression has been studied in the past, the contribution of Stx produced by phage-sensitive bacteria during antibiotic treatment has received little attention. In the present study we compared the effects of antibiotics on Stx production by O157:H7 in pure culture and during coincubation with phage-sensitive non-O157:H7 strains.
Studies using O157:H7 in pure culture confirmed that subinhibitory levels of antibiotics that target DNA synthesis (CIP and SXT) can induce production of Stx (Fig. 2 and 5). However, subinhibitory levels of antibiotics that target the cell wall, ribosome, or RNA polymerase had little ability to induce Stx production and, in some cases, decreased Stx production compared to untreated cells (Fig. 4). These results support previous studies which demonstrated that antibiotics that inhibited protein synthesis decrease toxin production in vitro (25, 38, 39). Furthermore, two studies have shown that AZM affords protection from Stx-mediated disease in vivo. In an elegant study by Zhang, et al., neonatal pigs treated with AZM displayed increased survival and significantly reduced brain lesions compared to untreated controls (39). In another study (22), none of the 10 immature mice inoculated with O157:H7 survived, whereas all 10 mice in the AZM-treated group survived. However, normal intestinal flora was absent in both models; the gnotobiotic pig is a germfree model, and the immature mice were treated with streptomycin to promote colonization by O157:H7.
Our studies suggest that normal flora can have a profound impact on Stx production. Remarkably, coincubation with CIP at a concentration that completely killed the O157:H7 resulted in Stx levels equivalent to those produced by a pure culture of O157:H7 (Fig. 5). In contrast, AZM significantly reduced Stx levels, even when relatively high levels of O157:H7 were recovered. Stx production was also examined under conditions that mimic one of the most confounding clinical scenarios, treatment of antibiotic-susceptible O157:H7 in the presence of antibiotic-resistant, Stx-phage-susceptible intestinal E. coli. In direct contrast to the results obtained with pure cultures, where these antibiotics did not increase toxin production, under these coculture conditions elevated levels of Stx were recovered even when growth of O157:H7 was inhibited; this was likely due to Shiga toxin production by the phage-susceptible strain (Fig. 6). These results demonstrate that antibiotic efficacy, as judged by the elimination of either Stx production or the E. coli O157:H7 organisms in pure cultures grown in vitro cannot be used to predict elimination of Stx production in the complex intestinal environment.
While it is premature to advocate antibiotic treatment for individuals infected with O157:H7, antibiotic treatment should be reevaluated, with special consideration of the role of normal intestinal flora. The results presented here support the conclusion that CIP and SXT are not appropriate for treatment of O157:H7. With regard to other classes of antibiotics, antibiotics capable of producing rapid bactericidal activity would be preferable to slower-acting or bacteriostatic antibiotics. In addition, protein synthesis inhibitors that cut off the pipeline to new Stx production would be preferable to antibiotics that allow synthesis of toxin to continue prior to bacterial death. AZM reduced Stx production even in mixed culture, and when O157:H7 CFU remained high. Our studies also suggest that the normal intestinal flora of patients with O157:H7 should be evaluated for the presence of Stx-phage-sensitive bacterial strains. These patients may be at greater risk for the development of more severe disease. Interestingly, ca. 10% of human intestinal isolates are susceptible to Stx-encoding phage (5, 6, 8), and ca. 10% of people infected with O157:H7 develop HUS (31). It is possible that the presence of Stx-phage susceptible flora is a previously unrecognized risk factor for developing HUS. Our studies suggest that little can be done to prevent Stx production by phage-susceptible nontoxigenic strains, and it is possible that these individuals could have been misinterpreted as treatment failures in previous clinical studies.
Acknowledgments
This study was supported by grants from the National Institute of Allergy and Infectious Disease (RO1AI064893 and U01AI075498) to A.A.W.
Footnotes
Published ahead of print on 28 June 2010.
REFERENCES
- 1.Boerlin, P., S. Chen, J. K. Colbourne, R. Johnson, S. De Grandis, and C. Gyles. 1998. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect. Immun. 66:2553-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endo, Y., K. Tsurugi, T. Yutsudo, Y. Takeda, T. Ogasawara, and K. Igarashi. 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes: RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171:45-50. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez, R. C., and A. A. Weiss. 1998. Serum resistance in bvg-regulated mutants of Bordetella pertussis. FEMS Microbiol. Lett. 163:57-63. [DOI] [PubMed] [Google Scholar]
- 5.Gamage, S. D., C. M. McGannon, and A. A. Weiss. 2004. Escherichia coli serogroup O107/O117 lipopolysaccharide binds and neutralizes Shiga toxin 2. J. Bacteriol. 186:5506-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamage, S. D., A. K. Patton, J. F. Hanson, and A. A. Weiss. 2004. Diversity and host range of Shiga toxin-encoding phage. Infect. Immun. 72:7131-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamage, S. D., A. K. Patton, J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2006. Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect. Immun. 74:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamage, S. D., J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2003. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 71:3107-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grif, K., M. P. Dierich, H. Karch, and F. Allerberger. 1998. Strain-specific differences in the amount of Shiga toxin released from enterohemorrhagic Escherichia coli O157 following exposure to subinhibitory concentrations of antimicrobial agents. Eur. J. Clin. Microbiol. Infect. Dis. 17:761-766. [DOI] [PubMed] [Google Scholar]
- 10.Jorgensen, J. H., J. D. Turnridge, and J. A. Washington. 1999. Antibacterial susceptibility tests: dilution and disk diffusion methods, p. 1526-1543. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. K. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 11.Kale, R. R., C. M. McGannon, C. Fuller-Schaefer, D. M. Hatch, M. J. Flagler, S. D. Gamage, A. A. Weiss, and S. S. Iyer. 2008. Differentiation between structurally homologous Shiga 1 and Shiga 2 toxins by using synthetic glycoconjugates. Angew. Chem. Int. Ed. 47:1265-1268. [DOI] [PubMed] [Google Scholar]
- 12.Karmali, M. A., B. T. Steele, M. Petric, and C. Lim. 1983. Sporadic cases of haemolytic-uremic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619-620. [DOI] [PubMed] [Google Scholar]
- 13.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 1999. Induction of type 2 Shiga toxin synthesis in Escherichia coli O157 by 4-quinolones. Lancet 353:1588-1589. [DOI] [PubMed] [Google Scholar]
- 14.Koitabashi, T., V. Vuddhakul, S. Radu, T. Morigaki, N. Asai, Y. Nakaguchi, and M. Nishibuchi. 2006. Genetic characterization of Escherichia coli O157:H7/− strains carrying the stx2 gene but not producing Shiga toxin 2. Microbiol. Immunol. 50:135-148. [DOI] [PubMed] [Google Scholar]
- 15.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad host-range cloning vector pBR1MCS, carrying different antibiotic resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 16.Mead, P. S., and P. M. Griffin. 1998. Escherichia coli O157:H7. Lancet 352:1207-1212. [DOI] [PubMed] [Google Scholar]
- 17.Muniesa, M., M. De Simon, G. Prats, D. Ferrer, H. Paella, and J. Jofre. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71:4554-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami, J., K. Kishi, K. Hirai, K. Hiramatsu, T. Yamasaki, and M. Nasu. 2000. Macrolides and clindamycin suppress the release of Shiga-like toxins from Escherichia coli O157:H7 in vitro. Int. J. Antimicrob. Agents 15:103-109. [DOI] [PubMed] [Google Scholar]
- 19.Neely, M. N., and D. I. Friedman. 1998. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 223:105-113. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien, A. D., J. W. Newland, and S. F. Miller. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 21.Ochoa, T. J., J. Chen, C. M. Walker, E. Gonzales, and T. G. Cleary. 2007. Rifaximin does not induce toxin production or phage-mediated lysis of Shiga toxin-producing Escherichia coli. Antimicrob. Agents Chemother. 51:2837-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohara, T., S. Kojio, I. Taneike, S. Nakagawa, F. Gondaira, Y. Tamura, F. Gejyo, H. M. Zhang, and T. Yamamoto. 2002. Effects of azithromycin on Shiga toxin production by Escherichia coli and subsequent host inflammatory response. Antimicrob. Agents Chemother. 46:3478-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panos, G. Z., G. I. Betsi, and M. E. Falagas. 2006. Systematic review: are antibiotics detrimental or beneficial for the treatment of patients with Escherichia coli O157:H7 infection? Aliment. Pharmacol. Ther. 24:731-742. [DOI] [PubMed] [Google Scholar]
- 24.Pear, W. S., J. P. Miller, L. Xu, J. C. Pui, B. Soffer, R. C. Quackenbush, A. M. Pendergast, R. Bronson, J. C. Aster, M. L. Scott, and D. Baltimore. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/able-transduced bone marrow. Blood 92:3780-3792. [PubMed] [Google Scholar]
- 25.Pedersen, M. G., C. Hansen, E. Riise, S. Persson, and K. E. P. Olsen. 2008. Subtype-specific suppression of Shiga toxin 2 released from Escherichia coli upon exposure to protein synthesis inhibitors. J. Microbiol. 46:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plunkett, G., D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product? J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey Dirita, and V. J. Bagdasarian. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherrill, J. D., and W. E. Miller. 2006. G protein-coupled receptor (GPCR) kinase 2 regulates agonist-independent Gq/11 signaling from the mouse cytomegalovirus GPCR M33. J. Biol. Chem. 281:39796-39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegler, R. L., T. G. Obrig, T. J. Pysher, V. L. Tesh, N. D. Denkers, and F. B. Taylor. 2003. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic-uremic syndrome. Pediatr. Nephrol. 18:92-96. [DOI] [PubMed] [Google Scholar]
- 30.Steinborn, M., S. Leiz, K. Rüdisser, M. Griebel, T. Harder, and H. Hahn. 2004. CT and MRI in haemolytic-uraemic syndrome with central nervous system involvement: distribution of lesions and prognostic value of imaging findings. Pediatr. Radiol. 34:805-810. [DOI] [PubMed] [Google Scholar]
- 31.Tarr, P. I., C. A. Gordon, and W. L. Chandler. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073-1086. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, P. L., D. W. K. Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waldor, M. K., and D. I. Friedman. 2005. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8:459-465. [DOI] [PubMed] [Google Scholar]
- 34.Walterspiel, J. N., S. Ashkenazi, A. L. Morrow, and T. G. Cleary. 1992. Effect of subinhibitory concentrations of antibiotics on extracellular Shiga-like toxin I. Infection 20:25-29. [DOI] [PubMed] [Google Scholar]
- 35.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. U. S. A. 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoh, M., E. K. Frimpong, and T. Honda. 1997. Effect of antimicrobial agents, especially fosfomycin, on the production and release of Vero toxin by enterohaemorrhagic Escherichia coli O157:H7. FEMS Immunol. Med. Microbiol. 19:57-64. [DOI] [PubMed] [Google Scholar]
- 38.Yoh, M., E. K. Frimpong, S. P. Voravuthikunchai, and T. Honda. 1999. Effect of subinhibitory concentrations of antimicrobial agents (quinolones and macrolide) on the production of verotoxin by enterohemorrhagic Escherichia coli O157:H7. Can. J. Microbiol. 45:732-739. [PubMed] [Google Scholar]
- 39.Zhang, Q., A. Donohue-Rolfe, G. Krautz-Peterson, M. Sevo, N. Parry, C. Abeijon, and S. Tzipori. 2009. Gnotobiotic piglet infection model for evaluating the safe use of antibiotics against Escherichia coli O157:H7 infection. J. Infect. Dis. 199:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. K. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, L., and D. B. Haslam. 2005. A quantitative and highly sensitive luciferase-based assay for bacterial toxins that inhibit protein synthesis. J. Med. Microbiol. 54:1023-1030. [DOI] [PubMed] [Google Scholar]