Abstract
A multidrug resistance gene, cfr, and a phenicol resistance gene, fexA, were detected in a Bacillus strain, BS-01, isolated from swine feces. The cfr gene was carried on a novel 16.5-kb plasmid, designated pBS-01. A complete Tn917 structure, which harbors the macrolide-lincosamide-streptogramin B resistance gene erm(B), was located downstream of the cfr gene. The fexA gene was discovered in the chromosomal DNA of the BS-01 strain and identified in a Tn558 variant.
The gene cfr was initially discovered on the 17.1-kb multiresistance plasmid pSCFS1 in a bovine Staphylococcus sciuri isolate (12). This gene was recently shown to encode Cfr, an RNA methyltransferase that affects the binding of at least five chemically unrelated antimicrobial classes: phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics, ultimately leading to a multidrug-resistant phenotype (PhLOPSA) (6). The gene cfr has been detected in staphylococcal isolates of animal and human origin, located on different plasmid types (2, 3, 7, 12), and also in the chromosomal DNA (14). Recently, the first human outbreak of cfr-carrying Staphylococcus aureus was reported in Spain (8). To date, no cfr-carrying bacteria other than staphylococcal species have been reported.
During a surveillance study for bacterial susceptibility to phenicols on animal farms in Shandong province, China, in 2008, a Bacillus strain, BS-01, which was isolated from swine feces, exhibited elevated MICs for florfenicol and chloramphenicol. Bacterial culture and morphological observation showed that BS-01 cells are aerobic, Gram-positive, spore-forming motile rods. The API 50 CH system, associated with the API 20 E system (bioMérieux, France), identified strain BS-01 as a Bacillus species without a specific species name. Analysis of the 16S rRNA sequence of this strain using the universal prokaryotic primers fD1 and rD1 (15) showed a maximum identity of 96% with the 16S rRNA sequence of Bacillus sphaericus and formed a distinct phylogenetic line within the genus Bacillus. Therefore, it is suggested that the BS-01 strain belongs to a novel Bacillus species. Further identification of this Bacillus strain is in progress.
BS-01 was screened for genes cfr and fexA (4) that contribute to florfenicol resistance using previously described primers (3). Positive PCR results were obtained and then confirmed by sequencing. In order to localize these two genes, genomic DNA was isolated from the BS-01 strain using a commercial kit (TianGen, Beijing, China) and following the manufacturer's instructions, and plasmid DNA was extracted following the maxiprep alkaline lysis procedure (10). Only a single plasmid was detected; it was subsequently transformed into the S. aureus recipient strain RN4220 by electroporation as described previously (11). This plasmid was designated pBS-01. Comparison of the restriction pattern of the plasmid from BS-01 and from its transformant did not show any differences. Southern blotting of the HindIII-digested pBS-01 plasmid DNA showed the cfr gene hybridized to an approximately 6-kb fragment. The transformant S. aureus RN4220::pBS-01 exhibited elevated MICs (at least 4-fold) for phenicol, lincosamide, oxazolidinone, and pleuromutilin antibiotics (Table 1), which confirmed the functional activity of the gene cfr.
TABLE 1.
Antimicrobial susceptibility profiles of strain BS-01, S. aureus RN4220, and the transformant S. aureus RN4220::pBS-01
| Antimicrobial agent | MIC (μg/ml) |
||
|---|---|---|---|
| BS-01 | S. aureus RN4220 | S. aureus RN4220::pBS-01 | |
| Chloramphenicol | 32 | 4 | 16 |
| Florfenicol | 32 | 4 | 16 |
| Linezolid | 2 | 1 | 4 |
| Clindamycin | >32 | <0.625 | >32 |
| Erythromycin | >64 | 0.25 | >64 |
| Tiamulin | 256 | <0.5 | 32 |
| Gentamicin | 8 | 0.5 | 0.25 |
| Ciprofloxacin | 8 | 0.5 | 0.5 |
| Tetracycline | 32 | 1 | 1 |
| Vancomycin | 0.5 | 1 | 1 |
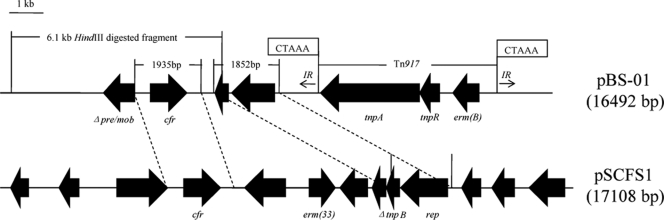
To investigate the genetic environment of the cfr gene, the 16,492-bp plasmid pBS-01 was sequenced by primer walking starting from the terminal parts of the amplified cfr gene. The plasmid pBS-01 contains two regions with homology to sequence in plasmid pSCFS1 from S. sciuri (Fig. 1). The first 1,935-bp fragment, with only 4 nucleotides that differ, comprises a potential promoter structure and the reading frame of the cfr gene (12). The second fragment comprises 1,852 bp with 90% similarity to the corresponding sequence of pSCFS1 and another newly reported multiresistance plasmid, pKKS825 (1), in which there are a truncated tnpB gene (coding for transposase B of Tn554) and a rep gene (coding for a putative protein possibly involved in the plasmid replication). Upstream from cfr, a 3′ truncated segment of the gene pre/mob is present. The product of pre/mob is considered essential for plasmid recombination and mobilization. This Δpre/mob nucleotide sequence revealed high similarities to those on plasmids of various staphylococci, including S. aureus (GenBank accession no. FM207105), Staphylococcus epidermidis (GenBank accession no. GQ900381), and Staphylococcus saprophyticus (GenBank accession no. AM159501), and to plasmid/chromosomal DNAs from other species, such as Enterococcus faecalis (GenBank accession no. AF503772) and Streptococcus suis (GenBank accession no. FM252032). Analysis of the region downstream of the cfr gene revealed the presence of a complete transposon structure of Tn917 from E. faecalis (13) in the opposite transcriptional direction to the cfr gene. It contains two transposase genes, tnpA and tnpR, and the resistance gene erm(B), which encodes an rRNA methyltransferase and confers resistance to macrolides, lincosamides, and streptogramin B (MLSB resistance) (13). The 5-bp direct repeats, 5′-CTAAA-3′, found at the left- and right-end junctions revealed the presumable integration site. The sequence of the pBS-01 plasmid did not show the presence of the IS21-558 element, which was considered to be very important in the mobility of the cfr gene in either animal isolates (2, 3) or clinical strains (7, 14).
FIG. 1.
Physical maps of the entire plasmids pBS-01, extracted from isolate BS-01, and pSCFS1, from a staphylococcal isolate. The positions and orientations of the genes are indicated by solid arrows, with the direction of transcription shown by the arrowhead. The 5-bp direct repeats at the junctions of transposon Tn917 are shown in boxes. The two regions of homology on plasmids pBS-01 and pSCFS1 are connected by dashed lines. The two 482-bp imperfect inverted repeat sequences (with 93% similarity) flanking Tn917 in pBS-01 are shown by arrows. The indicated 6,145-bp HindIII-digested fragment is the area where the cfr probe hybridized.
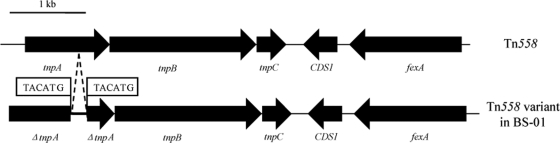
Repeated negative results were obtained for PCR amplification of fexA in the transformant S. aureus RN4220::pBS-01. Moreover, hybridizations of plasmid DNA extracted either from strain BS-01 or its transformant using the fexA fragment as the probe also showed negative results, but positive hybridization was observed when genomic DNA isolated from strain BS-01 was used as the template. All of the above-described evidence suggested a chromosomal location of the fexA gene in the BS-01 strain. Since Tn558 (5) was shown to be closely associated with the mobility of the fexA gene, previously described primers (3) were used to detect different parts in Tn558 and its intermediate circular form which precede the integration of the transposon into a new target sequence (5). The amplicons of the transposase genes tnpB and tnpC and of the tnpB-fexA segment showed the expected sizes, but the tnpA amplicons and the amplicon of the almost-complete Tn558 differed in their sizes from those discovered in staphylococci. No intermediate circular form was obtained. Sequence analysis of the tnpA amplicon identified a 1,212-bp fragment, with a 283-bp sequence inserted into the tnpA gene (Fig. 2). A direct repeat sequence, 5′-TACATG-3′, was detected immediately flanking the 283-bp sequence. Since TnpA and TnpB are essential for transposition (9), truncation of these genes resulted in the immobility of Tn558, as confirmed by the lack of detection of circular forms in the Tn558 variant of strain BS-01. Similar Tn558 variants were observed among staphylococcal isolates (3). The Tn558 variant detected in the BS-01 isolate suggests that Tn558 is important in the spread of the fexA gene among different bacterial species.
FIG. 2.
Organization of transposon Tn558 and its variant in strain BS-01. This Tn558 variant is shown in comparison with the original fexA-carrying transposon Tn558 located on plasmid pSCFS2 (GenBank accession no. AJ715531). The positions and orientations of the genes coding for transposition functions (tnpA, tnpB, and tnpC), resistance to florfenicol and chloramphenicol (fexA), and unknown functions (CDS1) are indicated by arrows, with the direction of transcription shown by the arrowhead. The 283-bp sequence inserted into the tnpA gene of the Tn558 variant is indicated between two dashed lines. The 6-bp direct repeats immediately flanking the 283-bp sequence in the Tn558 variant are shown in boxes.
In conclusion, this is the first time that the cfr and fexA genes have been found in a species other than the staphylococci. The potential dissemination of these resistance genes among different bacterial species, especially in common pathogens of human and animal origin, such as enterococci and streptococci, is worrisome and should be under surveillance.
Nucleotide sequence accession numbers.
The 6,678-bp sequence of the incomplete Tn558 variant of strain BS-01, as well as the 16,492-bp sequence of a cfr-carrying plasmid, pBS-01, have been deposited in the GenBank database under accession no. GU591496 and GU591497.
Acknowledgments
We thank Rocky Patil (Iowa State University) for revision of the manuscript.
This study was supported by a grant from the Program for Chang Jiang Scholars and the Innovative Research Team at the University of China (grant no. IRT0866).
Footnotes
Published ahead of print on 28 June 2010.
REFERENCES
- 1.Kadlec, K., and S. Schwarz. 2009. Novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 53:3589-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kehrenberg, C., F. M. Aarestrup, and S. Schwarz. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehrenberg, C., and S. Schwarz. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehrenberg, C., and S. Schwarz. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob. Agents Chemother. 48:615-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kehrenberg, C., and S. Schwarz. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob. Agents Chemother. 49:813-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendes, R. E., L. M. Deshpande, M. Castanheira, J. DiPersio, M. A. Saubolle, and R. N. Jones. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales, G., J. J. Picazo, E. Baos, F. J. Candel, A. Arribi, B. Pelaez, R. Andrade, M. A. de la Torre, J. Fereres, and M. Sanchez-Garcia. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50:821-825. [DOI] [PubMed] [Google Scholar]
- 9.Murphy, E., L. Huwyler, and C. de Freire Bastos Mdo. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 4:3357-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning; a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY.
- 11.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz, S., C. Werckenthin, and C. Kehrenberg. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toh, S. M., L. Xiong, C. A. Arias, M. V. Villegas, K. Lolans, J. Quinn, and A. S. Mankin. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]