Abstract
Paratyphoid fever is considered an emerging systemic intracellular infection caused by Salmonella enterica serotypes Paratyphi A, B, and C. We performed in vitro time-kill studies on three clinical isolates of nalidixic acid-resistant Salmonella serotype Paratyphi (NARSP) with different concentrations of ciprofloxacin and cefotaxime to identify combinations of antibiotics with synergistic activity against paratyphoid fever. Furthermore, we identify the frequency of mutations to ciprofloxacin, cefotaxime, and rifampin resistance and also sequenced the gyrA, gyrB, parC, and parE genes to identify the cause of resistance in NARSP. When the activity of ciprofloxacin at 0.75× MIC (0.012 to 0.38 μg/ml) with cefotaxime at the MIC (0.125 to 0.25 μg/ml) against all three NARSP isolates was investigated, synergy was observed at 24 h, and the bacterial counts were reduced by >3 log10 CFU/ml. This synergy was elongated for up to 72 h in two out of three isolates. When ciprofloxacin at 0.75× MIC (0.012 to 0.38 μg/ml) was combined with cefotaxime at 2× MIC (0.25 to 0.50 μg/ml), synergy was prolonged for up to 72 h in all three isolates. Both Salmonella serotype Paratyphi A isolates carried single mutations in codon 83 of the gyrA gene and codon 84 of the parC gene that were responsible for their reduced susceptibility to ciprofloxacin, while no mutations were found in the gyrB or parE gene. The ciprofloxacin-plus-cefotaxime regimen was very effective in reducing the bacterial counts at 24 h for all three isolates, and this combination therapy may be helpful in reducing the chance of the emergence of fluoroquinolone-resistant mutants in patients with severe paratyphoid fever.
Enteric fever (typhoid fever and paratyphoid fever) is a systemic intracellular infection caused by Salmonella enterica serotype Typhi or Salmonella enterica serotype Paratyphi A, B, and C. Paratyphoid fever caused by S. Paratyphi is considered an emerging disease, as its incidence has increased alarmingly in recent years (29), causing more asymptomatic infections than S. Typhi. Now enteric fever is a major public health problem in many countries. It is endemic in the Indian subcontinent, Southeast Asia, and Africa, causing at least 22 million cases of illness each year and 200,000 deaths (21), and more than 5 million of these cases were due to S. Paratyphi A (19). According to the findings of a 12-month survey by Ochiai et al. (20), 14% of the cases of enteric fever in Indonesia were caused by S. Paratyphi; these rates were 15% in Pakistan, 24% in India, and 64% in China. Shirakawa et al. (28) reported that in Kathmandu, Nepal, enteric fever caused by S. Paratyphi A is more prevalent than that caused by S. Typhi. The outbreak of enteric fever due to S. Paratyphi in Guangxi Province in southeast China and the increasing rate of isolation of serotype Paratyphi in other areas may signal that S. Paratyphi A is an emerging pathogen in Asia (20).
Since 1997, cases of infection with nalidixic acid-resistant Salmonella serotype Typhi (NARST) with decreased susceptibility to ciprofloxacin have been reported in many parts of the world and are now common in the Indian subcontinent and Southeast Asia (23). In Europe and Wales, high-level fluoroquinolone resistance in salmonellae is associated with foreign travel, especially to Southeast and South Asia (11). Some patients with typhoid fever caused by NARST that is susceptible to fluoroquinolones in vitro, according to current Clinical and Laboratory Standards Institute (CLSI) interpretive criteria, can show a delayed response to ciprofloxacin or treatment failure or can even be refractory to treatment both clinically and bacteriologically (26). The failure of treatment with fluoroquinolones in the cases of S. Typhi and S. Paratyphi A in the Indian subcontinent and Southeast and Central Asia due to decreased susceptibility to ciprofloxacin is now a serious concern (30). Fluoroquinolone-resistant S. Paratyphi A has already been reported in Japan (1), as has the relapse of an S. Paratyphi B infection in a child after treatment with ciprofloxacin (7). Fluoroquinolone resistance in most bacterial isolates is due to mutations in the genes encoding DNA gyrase (gyrA, gyrB) or DNA topoisomerase IV (parC, parE) (10). Additionally, increased efflux activity is also a major mechanism of resistance in many Gram-negative bacteria, including Salmonella (9). Currently, however, there is no treatment of choice for patients infected with multidrug-resistant (MDR) and nalidixic acid-resistant Salmonella enterica serotype Paratyphi (NARSP) (24). Indeed, clinicians recommend either the longer-duration use of high-dose fluoroquinolone or the expanded-spectrum drugs cephalosporin or azithromycin, or the combination of these regimens, for the treatment of NARST and NARSP infections. To date, however, no time-kill studies have demonstrated the efficacy of such combination therapy for NARSP infections.
We conducted time-kill studies on three clinical isolates, including an isolate from a patient at Chosun University Hospital infected with S. Paratyphi A and one isolate each of S. Paratyphi A and B obtained from the South Korea Center for Disease Control and Prevention (KCDC). In addition, we investigated the association of quinolone resistance with mutations in the genes coding for gyrase A and topoisomerase IV in these isolates.
MATERIALS AND METHODS
Bacterial isolates.
One clinical isolate (S. Paratyphi A CUH 61675) and two clinical NARSP isolates (S. Paratyphi A KCDC 1733 and S. Paratyphi B KCDC 3631) obtained from the KCDC (Table 1) were used in our in vitro studies. CUH 61675 was isolated from a blood sample obtained from a patient when that patient was admitted to Chosun University Hospital and was stored at −80°C for MIC and time-kill studies. Similarly, a nalidixic acid-susceptible S. Typhi strain (ATCC 9992) was also used for analysis of time-kill kinetics.
TABLE 1.
MICs and nucleotide changes in gyrA, gyrB, parC, and parE sequences of nalidixic acid-resistant and -susceptible Salmonella enterica serovar Paratyphi A and B and Salmonella enterica serovar Typhi
| Strain | MIC (μg/ml)a |
Nucleotide change at the indicated amino acid inb: |
||||
|---|---|---|---|---|---|---|
|
gyrA |
parC, 84GAA (Glu) | |||||
| NAL | CIP | CTX | 83TCC (Ser) | 87GAC (Asp) | ||
| S. Paratyphi A CUH 61675 | 256 | 0.50 | 0.25 | TAC (Tyr) | None | AAA (Lys) |
| S. Paratyphi A KCDC 1733 | 256 | 0.25 | 0.125 | TTC (Phe) | None | AAA (Lys) |
| S. Paratyphi B KCDC 3631 | 128 | 0.016 | 0.125 | None | AAC (Asn) | None |
| S. Typhi ATCC 9992 | 4 | 0.016 | 0.063 | None | None | None |
NAL, nalidixic acid; CIP, ciprofloxacin; CTX, cefotaxime.
No nucleotide changes were detected in gyrB or parE.
Media and antimicrobial agents.
Mueller-Hinton broth (MHB; Difco Laboratories, Detroit, MI) supplemented with 25 μg/ml calcium and 12.5 μg/ml magnesium was utilized for all broth microdilution susceptibility testing and time-kill analyses. Mueller-Hinton agar (Difco Laboratories) plates were used for growth of the isolates and time-kill experiments. Standard powders of the antimicrobials ciprofloxacin and cefotaxime were obtained from Bayer Health Care, South Korea, and Handok-Aventis, South Korea, respectively.
Amplification of QRDR sequences of the gyrA, gyrB, parC, and parE genes.
The quinolone resistance-determining regions (QRDRs) of the gyrA, gyrB, parC, and parE genes were amplified by PCR on a PCR express Hyaid cycler using as the template total extracted DNA, as described by Ausubel et al. (2). The oligonucleotide primers used for PCR assays were those described by Kariuki et al. (13): for gyrA the forward primer was 5′-ATGAGCGACCTTGCGAGAGAAATTACACCG-3′ and the reverse primer was 5′-CTTCTGTAGTCGTAACTTCCCGACTACCTT-3′, for gyrB the forward primer was 5′-AAGCGCGATGGCAAAGAAG-3′ and the reverse primer was 5′-AACGGTCTGCTCATCAGAAAGG-3′, for parC the forward primer was 5′-ATGAGCGATATGGCAGAGCG-3′ and the reverse primer was 5′-TGACCGAGTTCGCTTAACAG-3′, and for parE the forward primer was 5′-GACCGAGCTGTTCCTTGTGG-3′ and the reverse primer was 5′-GCGTAACTGCATCGGGTTCA-3′. The NCBI BLAST program was used to align the amplified sequences with the genome sequence of serovar Typhi strain CT 18 (GenBank accession number AL513382). The sequences of the QRDRs from amino acid residues 54 to 71 of gyrA, 397 to 520 of gyrB, 12 to 130 of parC, and 421 to 524 of parE were determined as described by Kim et al. (15).
MICs.
The MICs of each antimicrobial agent tested, nalidixic acid, ciprofloxacin, and cefotaxime, were determined in duplicate on the basis of the CLSI guidelines for the broth microdilution procedure (4). MIC testing was performed in duplicate with inocula of 5 × 105 CFU/ml. The MIC was read after 18 h of incubation at 35 ± 2°C in ambient air in an incubator shaking at 200 rpm. Ciprofloxacin-susceptible and -resistant strains were defined as isolates with MICs of ≤1 and ≥4 μg/ml, respectively. The MIC breakpoint value for reduced ciprofloxacin susceptibility was set equal to ≥0.125 μg/ml, on the basis of findings described in earlier publications (18). Additionally, to identify the genetic mutations of the isolates, MICs for each of ciprofloxacin and cefotaxime were also determined for the isolates at 24 h and 72 h of incubation.
Mutation frequencies.
Mutation frequencies were determined as previously described by Gall et al. (8). Briefly, a single colony of S. Paratyphi isolates was suspended in 10 ml of Luria-Bertani (LB; Difco Laboratories) broth and incubated at 37°C for 24 h. This preculture was inoculated at a dilution of 1:50 in 5 ml of fresh LB broth and then further incubated at 37°C for 4 h until the mid-log growth phase was reached. Bacterial cell suspensions (1.5 ml) of each cultured strain were taken and centrifuged (8,000 × g, 3 min), the supernatants were discarded, and the pellets were resuspended in 0.9% saline. The cells were then adjusted to an optical density at 600 nm of 0.6 with saline solution in cuvettes with a 1-cm path length. The number of CFU was determined by spread plating of 0.1 ml of the bacterial suspension onto duplicate LB agar plate, one with rifampin (100 μg/ml; Sigma-Aldrich, Inc., St. Louis, MO) and one without rifampin. The results were related to the mean value obtained from three independent cultures of 108 CFU/ml spread on three different agar plates containing or not containing antibiotics. Along with these assays, all three isolates were also subjected to a serial passage experiment with ciprofloxacin or cefotaxime, as described earlier by Kim et al. (14). Briefly, the overnight growth of Salmonella isolates on Mueller-Hinton agar was swabbed onto fresh agar plates containing one-half of the MIC of ciprofloxacin or cefotaxime or on agar plates not containing antibiotics. At 24 h, the surface growth was picked and placed onto a fresh agar plate containing the same antibiotic at twice the concentration on the previous agar plate or on agar plates without antibiotics. This process was repeated serially up to 4-fold the MICs of both ciprofloxacin and cefotaxime for the respective isolates, even though the MICs in agar which may be different from the MICs in broth. Mutation frequencies are reported as the proportion of ciprofloxacin-, cefotaxime-, or rifampin-resistant colonies to the total viable counts.
Time-kill studies.
Time-kill studies were performed to evaluate synergistic activity against all three NARSP isolates and one S. Typhi isolate (Table 1), as described previously (17). Bacteria were diluted to a standardized cell suspension of about 5.0 × 105 CFU/ml in 50 ml fresh cation-adjusted MHB in a 250-ml conical flask and exposed to antibiotics at different concentrations. A growth control was assessed in an extra conical flask with 50 ml of the MHB described above but without antibiotic. Each flask was incubated at 37°C in an incubator shaking at 200 rpm. At 0, 2, 4, 8, 12, and 24 h of incubation, aliquots of 100 μl of the bacterial cultures were obtained, spun down by centrifugation at 3,200 × g for 15 min, resuspended in fresh MHB to minimize the drug carryover effect, serially diluted 10-fold in MHB, and cultured on duplicate drug-free Mueller-Hinton agar plates (5). Additionally, to evaluate the synergistic activity after 24 h, 4 ml freshly prepared MHB was incubated with the same concentrations of drugs described above and a bacterial cell suspension of 5.0 × 105 CFU/ml. To account for the potential breakdown of antibiotics in broth, bacterial cells grown in each tube were spun down and resuspended in fresh drug-containing broth at 24 and 48 h. Additionally, at 36, 48, and 72 h of incubation, culture aliquots of 100 μl were taken from each tube and the drug carryover was minimized by dilution in MHB, as described above. Bacterial counts were determined by plating 100 μl of the appropriate dilutions on duplicate Mueller-Hinton agar (MHA) plates and overnight incubation under ambient conditions at 37°C to enumerate the numbers of CFU/ml (16, 31). Time-kill curves were constructed by plotting the mean colony counts on a logarithmic scale versus the time with a lower limit detection of 2 log10 CFU/ml. According to the 2009 guidelines of Antimicrobial Agents and Chemotherapy, synergy was defined as a ≥2-log10-unit decrease (100-fold drop) in the numbers of CFU per ml achieved with the combination and with the most efficient agent used alone at 24 h. Indifference and antagonism were defined as ±1- to <2-log10-unit killing at 24 h compared to the activity of the most efficient agent alone and >1 log10 unit of growth at 24 h compared with the activity of the least active agent, respectively (31). The bactericidal activities of the drugs alone and the drug combinations were defined as a reduction of ≥3 log10 CFU/ml (99.9%) compared to the starting inoculum and to the most active antimicrobial agent at 24 h, respectively (15). To confirm the results, all the experiments were performed at least in duplicate.
RESULTS
Sequence analysis of gyrA and parC.
Sequencing of the S. Paratyphi B strain confirmed the presence of a single mutation in codon 87 of gyrA, in which a G → A transversion leads to the replacement of aspartate by asparagine (Asp87 → Asn). However, in the two isolates of S. Paratyphi A, mutations were present in both gyrA and parC. In clinical strain CUH 61675, the gyrA mutation is at codon 83, in which a C → A tranversion replaces serine by tyrosine (Ser83 → Tyr), while in the KCDC strain the codon 83 mutation (C → T transversion) results in Ser83 → Phe. Additionally, both isolates have a G → A transversion at codon 84 of parC, leading to the replacement of glutamine by lysine (Glu84 → Lys). The presence of this parC mutation, in addition to the gyrA mutations, causes these isolates to be more resistant than the S. Paratyphi B isolate. However, no mutations were reported in the gyrB or the parE genes of any of the three isolates of S. Paratyphi A or S. Paratyphi B.
MICs and mutation frequencies.
The MICs for nalidixic acid, ciprofloxacin, and cefotaxime against all three S. Paratyphi isolates tested are shown in Table 1. According to current CLSI breakpoints, all three isolates were highly resistant to nalidixic acid but remained susceptible in vitro to ciprofloxacin and cefotaxime. However, using a cutoff MIC value of ≥0.125 μg/ml, both S. Paratyphi A isolates showed reduced susceptibility to ciprofloxacin. Additionally, the MICs of the isolates at 24 h and 72 h remained stable. The distribution of the rifampin resistance mutation frequencies (f) of the S. Paratyphi isolates was determined by using the phenotypic categories described earlier by Gall et al. (8): hypomutable (f ≤ 8 × 10−9), normomutable (8 × 10−9 < f < 4 ×10−8), weakly hypermutable (4 × 10−8 ≤ f < 4 × 10−7), and strongly hypermutable (f ≥ 4 × 10−7). In all three isolates, ciprofloxacin exhibited 2- to 4-fold increased MICs and cefotaxime showed only 2-fold increased MICs in reference to the MICs of the colonies displaying rifampin resistance on 100-mg/liter rifampin plates. The ciprofloxacin resistance mutation frequencies for the CUH 61675, KCDC 738, and KCDC 3697 isolates were 5.76 × 10−5, 1.48 × 10−5, and 8.32 × 10−6, respectively. Similarly, the cefotaxime resistance mutation frequencies for the CUH 61675, KCDC 738, and KCDC 3697 isolates were 8.03 × 10−6, 4.17 ×10−5, and 6.0 × 10−6, respectively. The rifampin resistance mutation frequencies for the CUH 61675 and KCDC 738 isolates were 5.4 ×10−8 and 5.8 ×10−8 (both were weakly hypermutable), respectively, while the KCDC 3631 isolates were normomutable, with the resistance frequencies being 3.6 ×10−8. Hence, the mutation frequencies for all isolates indicated that it was extremely likely that the total initial inoculum of 5 × 105 CFU/ml in 50 ml broth contained preexisting resistant mutants.
Time-kill studies.
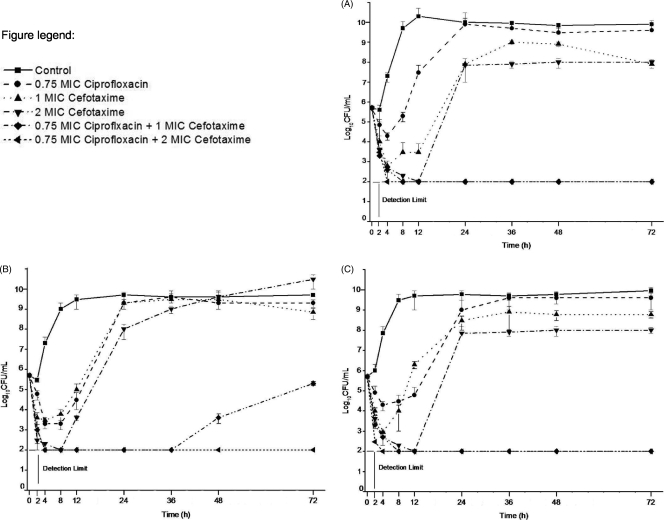
The time-kill study of clinical isolate CUH 61675 showed that ciprofloxacin at 0.38 μg/ml (0.75× MIC), cefotaxime at 0.25 μg/ml (MIC), and cefotaxime at (0.5 μg/ml [2× MIC]) had growth-inhibitory effects for up to 4, 8, and 12 h, respectively. Thereafter, the bacteria began to regrow and proliferated to an extent approaching the maximum level. In the combination regimens with ciprofloxacin at 0.75× MIC plus cefotaxime at the MIC and with ciprofloxacin at 0.75× MIC plus cefotaxime at 2× MIC, the reduction in the bacterial counts from the starting inoculum was more pronounced (≥3 log10 CFU/ml) at 24 h and the drugs in the combination regimens showed synergy (Fig. 1A).
FIG. 1.
Time-kill curves for nalidixic acid-resistant Salmonella enterica serovar Paratyphi A CUH 61675 (A) and KCDC 1733 (B) and Salmonella enterica serovar Paratyphi B KCDC 3631 (C). The detection limit was 2 log10 CFU/ml.
In the time-kill study of NARSP clinical isolate S. Paratyphi A KCDC 1733, the combination regimens of ciprofloxacin at 0.75× MIC plus cefotaxime at the MIC and ciprofloxacin at 0.75× MIC plus cefotaxime at 2× MIC gave a more pronounced reduction in bacterial counts (≥3 log10 CFU/ml) from the starting inoculum (in CFU/ml) at 24 h and had synergistic effects (Fig. 1B). Similarly, in the time-kill study of S. Paratyphi B KCDC 3631, for both combination regimens, ciprofloxacin at 0.75× MIC plus cefotaxime at the MIC and ciprofloxacin at 0.75× MIC plus cefotaxime at 2× MIC, the reduction in the bacterial counts from the starting inoculum was more pronounced (≥3 log10 CFU/ml) at 24 h and the drugs in the combinations had pronounced synergistic effects (Fig. 1C).
In the combination of 0.75× MIC (0.012 to 0.38 μg/ml) of ciprofloxacin and 0.75× MIC (0.09 to 0.19 μg/ml) of cefotaxime, the reduction in bacterial counts was pronounced only by comparison with the activity of the most active single agent but not by comparison with the starting inoculum in both S. Paratyphi A isolates (mean decreases in counts from that of the most active single agent and the starting inoculum, 2.42 ± 0.18 and 0.51, respectively, in CUH 61675, and 4.11 ± 0.42 and 0.35, respectively, in KCDC 1733) at 24 h. However, in S. Paratyphi B strain KCDC 3631, the decrease in bacterial counts was pronounced, with the decreases being 5.6 ± 0.56 CFU/ml (mean ± standard deviation) and 2.36 from those achieved with the most active single agent and the starting inoculum, respectively (data not shown). Additionally, for the other combination regimens, ciprofloxacin at 0.25× MIC (0.004 to 0.13 μg/ml) plus cefotaxime at 0.5× MIC (0.06 to 0.13 μg/ml) and ciprofloxacin at 0.5× MIC (0.008 to 0.25 μg/ml) plus cefotaxime at 0.5× MIC (0.06 to 0.13 μg/ml), the reduction in the bacterial counts at 24 h was less than 2 log10 CFU/ml in all three isolates and the combinations did not show synergistic effects.
DISCUSSION
The main causes of resistance to quinolones in Gram-negative bacteria are mutations in the genes coding for DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) (6). Target protection mediated by the Qnr protein and reduced intracellular accumulation of antibiotic due to a lower outer membrane permeability have also been implicated in resistance (12). Additionally, increased efflux activity is also a general mechanism responsible for bacterial resistance, especially in Gram-negative bacteria, owing to the sophisticated cell envelope architecture. Giraud et al. (9) postulated that the enhanced active efflux and early overproduction of the AcrAB pump in isolates with the gyrA mutation could be responsible for the decrease in susceptibility to fluoroquinolones. Furthermore, in S. enterica, mutations associated with quinolone resistance predominantly occur in the QRDRs of the gyrA and parC genes (10). The changes in the topoisomerase gene are insufficient to account for the level of resistance to quinolones, and mutations in codons 83 and 87 of gyrA are frequently found in clinical isolates of S. enterica with decreased susceptibility to fluoroquinolones (28). The emergence of full fluoroquinolone resistance is due to additional mutations in patients with NARST infections, and S. Paratyphi A isolates with high-level ciprofloxacin resistance (MICs ≥ 128 μg/ml) have already been reported in Japan (1). Eaves et al. and others (6, 13) showed that mutations in gyrB and the parC and parE topoisomerase IV genes were rare in salmonellae and that isolates with decreased susceptibility to ciprofloxacin harbored a single mutation in gyrA, whereas resistant isolates contained two or more mutations in gyrA, gyrB, parC, or parE. In Salmonella, high-level quinolone resistance due to mutations in both gyrA and parC can arise in the long-term carrier state for S. Paratyphi A under the selection pressure of frequent quinolone administration, and multiple point mutations are probably responsible for the acquisition of full resistance (1). Furthermore, the failure of treatment of an infection caused by a resistant strain with a fluoroquinolone is likely to be because of a high mutation rate. In our isolates, both S. Paratyphi A isolates showed a mutation frequency classified as weakly hypermutable, while S. Paratyphi B had a normomutable mutation frequency and might be responsible for the reduced susceptibility to ciprofloxacin.
Sequencing of amplified DNA fragments of the QRDR of gyrA in our hospital isolate of S. Paratyphi A revealed mutations in gyrA codon 83 (Ser83 → Tyr) and parC codon 84 (Glu84 → Lys). Similarly, in KCDC 1733, mutations were found in codon 83 of parC (Ser83 → Phe) and codon 84 of parC (Glu84 → Lys). However, in S. Paratyphi B strain KCDC 3631, a single mutation was present in codon 87 of gyrA (Asp87 → Asn). A double mutation in gyrA and a single mutation in parC are evidently required for high-level ciprofloxacin resistance (MICs > 4 μg/ml); the second mutation in gyrA is essential for high-level resistance, whereas the role of the parC mutation is less clear. In our study, both clinical isolates of S. Paratyphi A had single mutations in codon 83 of gyrA and codon 84 (Glu84 → Lys) of parC responsible for reduced susceptibility to ciprofloxacin (MICs = 0.125 to 0.5 μg/ml). Moreover, the Asp87 → Asn change in amino acid residue 87 of gyrA apparently confers resistance only to nalidixic acid in our clinical strain of S. Paratyphi B.
Hence, screening of S. Paratyphi isolates for susceptibility to nalidixic acid could identify those isolates that potentially carry the highest risk of becoming fluoroquinolone resistant, and use of an antibiotic to which resistance does not arise by mutation might be the preferred therapeutic option for the effective treatment of the patients. Both S. Paratyphi A isolates with reduced susceptibility to ciprofloxacin at MICs of ≥0.125 μg/ml were, however, susceptible to ceftriaxone, even though some patients had a slow clinical response, such as a delay in the fever clearance time, or failed treatment. The stable MICs at 24 h and 72 h showed that these genetic mutations remain stable. Patients infected with Salmonella have been reported to have fever clearance times exceeding 7 days and failure rates of more than 20% with ceftriaxone treatment (22). Additionally, the WHO recommends cefixime as the first-line treatment for typhoid fever caused by drug-resistant strains and for second-line therapy when the initial exposure to fluoroquinolone fails. However, the treatment failure rates for the cefixime-treated group were 4 to 23% after 14 days of treatment in a randomized trial in Patan Hospital, Kathmandu (21). Furthermore, isolates of S. Typhi with high-level resistance to ceftriaxone have been reported in Bangladesh (27), and S. Paratyphi A isolates with multidrug resistance and resistance to extended-spectrum β-lactamases (ESBLs) have also been reported in Nepal (25).
We selected therapy with the combination of ciprofloxacin (fluoroquinolone) and cefotaxime, expanded-spectrum cephalosporins having different targets, for comparison with monotherapy. Combining ciprofloxacin with another antibiotic may help to reduce the chances of the emergence of fluoroquinolone-resistant mutants. In our time-kill studies, none of the combinations of the two drugs at levels below the MICs (0.25× MIC [0.004 to 0.13 μg/ml] of ciprofloxacin plus 0.5× MIC [0.06 to 0.13 μg/ml] of cefotaxime and 0.5× MIC [0.008 to 0.25 μg/ml] of ciprofloxacin plus 0.5× MIC [0.06 to 0.13 μg/ml] of cefotaxime) did not reduce the bacterial counts by >3 log10 CFU/ml compared to the starting inoculum to form synergistic effects. However, when ciprofloxacin at 0.75× MIC was combined with cefotaxime at the MIC, the reductions in the bacterial counts were ≥3 log10 CFU/ml for all three NARSP isolates examined when the counts were compared with the starting inoculum, and the combination displayed synergy. The synergy was prolonged until 72 h in two out of the three isolates, isolates CUH 61675 and KCDC 3631 (Fig. 1A and C, respectively). However, in strain KCDC 1733, synergy was prolonged until 48 h, and thereafter, the bacteria began to regrow and the reduction of the bacterial counts at 72 h was less than 2 log10 CFU/ml compared with the starting inoculum when treatment was with 0.75× MIC of ciprofloxacin plus the MIC of cefotaxime (Fig. 1B). Hence, this regrowth of bacteria at a late time might be associated with cefotaxime because it slowed the regrowth of ciprofloxacin-resistant cells and synergy is likely to occur up to 48 h but not 72 h. Furthermore, when ciprofloxacin at 0.75× MIC was combined with 2× MIC of cefotaxime, the reductions in the bacterial counts of all three isolates were ≥3 log10 CFU/ml compared to the starting inoculum up to 72 h and the combination displayed synergy (Fig. 1A to C). Hence, our time-kill studies indicate that the combination of ciprofloxacin plus cefotaxime may be the treatment option for patients with paratyphoid fever due to S. Paratyphi A and B. Additionally, in our in vitro time-kill study with nalidixic acid-susceptible standard strain S. Typhi ATCC 9992, synergy was prolonged until 72 h when the combination of 0.75× MIC of ciprofloxacin plus the MIC of cefotaxime was used, with the reduction in the bacterial counts being ≥3 log10 CFU/ml compared to the starting inoculum (data not shown). To date, very little is known about the mechanism of the synergistic effect of these drugs against S. enterica infections. However, previous studies of Escherichia coli showed that quinolones interact with the outer membrane as chelating agents and thereby increase its permeability to β-lactam antibiotics by facilitating the diffusion of the β-lactam antibiotic into the cells and that the combination of ciprofloxacin plus cefotaxime had a synergistic effect (3). We could maybe apply this mechanism to S. Paratyphi. However, as the outer membranes of E. coli and Pseudomonas aeruginosa are very different with regard to their drug permeabilities, the mechanism of synergistic activity against S. Paratyphi might be different from that against E. coli. Another possible hypothesis for the mechanism of the synergy is suppression of resistant mutant selection. Basically, combination therapy with two antimicrobial agents having different targets might limit the risk of emergence of resistant mutant isolates compared to the risk resulting from monotherapy with ciprofloxacin. However, these hypotheses need to be confirmed with a further well-designed study.
Some limitations of our experiments that can be pointed out are as follows. Only three isolates of S. Paratyphi were used in our experiments, and they had different levels of susceptibility to both antimicrobial agents, ciprofloxacin and cefotaxime; hence, this study merely provides preliminary data, and additional experiments would be helpful to draw a pronounced conclusion. S. Typhi and S. Paratyphi infections are intracellular (although obviously with both intracellular and extracellular components). Since β-lactams or aminoglycosides do not reach inhibitory levels within cells, such as macrophages, where the organism dwells, the in vitro synergy reported from a broth system may not be applicable to the clinical situation. Additionally, the initial inocula for our in vitro study were relatively low, and the duration of experiment was short compared to the length of therapy in patients. We emphasize that the synergistic activity compared with the activities of the single agents that we observed was against the initial inoculum, which most likely contained resistant cells. Analysis of our extended time-kill study data up to 72 h intuitively strongly supports the use of ciprofloxacin plus cefotaxime combination therapy against resistant isolates. Similarly, the ß-lactamase stability of the desacetyl metabolite of cefotaxime is higher than that of the parent compound, and this might be responsible for the synergistic effects of cefotaxime. Additionally, desacetyl-cefotaxime, which is occasionally used in in vitro combination studies of many of the members of the Enterobacteriaceae, was not included in the present study. Similarly, some reports pointed out the activity of cefotaxime. Hence, our data would therefore be of considerable interest in a long-term study with a fiber infection model. Indeed, studies with animal models of infection may be helpful, since an initial inoculum which contains resistant mutants may be chosen. We emphasize that these data would contribute valuable information before the combination therapy is analyzed in human clinical trials. Such clinical trials are, of course, ultimately important, but they can be designed much more rationally with these advanced in vitro and animal model data.
In conclusion, our in vitro time-kill studies demonstrated that the combination of ciprofloxacin plus cefotaxime showed synergistic effects against nalidixic acid-resistant S. Paratyphi A and B. This combination appears to be more effective than monotherapy and may help to reduce the chance that fluoroquinolone-resistant mutants will emerge in patients with severe typhoid fever.
Acknowledgments
No commercial interests or sponsorship provided poses a conflict of interest.
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea Government (MEST) through the Research Center for Resistant Cells (grant R13-2003-009).
Footnotes
Published ahead of print on 21 June 2010.
REFERENCES
- 1.Adachi, T., H. Sagara, K. Hirose, and H. Watanabe. 2005. Fluoroquinolone resistant Salmonella Paratyphi A. Emerg. Infect. Dis. 11:172-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, and R. E. Kingstone (ed.). 1996. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Chapman, J. S., and N. H. Georgopapadakou. 1988. Routes of quinolone permeation in Escherichia coli. Antimicrob. Agents Chemother. 32:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement, vol. 28, no 1. Document no. M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Drusano, G. L., W. Liu, D. L. Brown, L. B. Rice, and A. Louie. 2009. Impact of short course quinolone therapy on susceptible and resistant populations of Staphylococcus aureus. J. Infect. Dis. 199:219-226. [DOI] [PubMed] [Google Scholar]
- 6.Eaves, D. J., L. Randall, D. T. Gray, A. Buckley, M. J. Woodward, A. P. Wgite, and L. J. V. Piddock. 2004. Prevalence of mutations within the quinolone resistance determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone resistant Salmonella enterica. Antimicrob. Agents Chemother. 48:4012-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eltringham, I. J., N. Thalange, D. Ireland, H. Issler, and A. Teall. 1997. A relapse of paratyphoid fever after treatment with ciprofloxacin in a child with congential biliary atresia. Acta Paediatr. 86:547-548. [DOI] [PubMed] [Google Scholar]
- 8.Gall, S. L., L. Desbordes, P. Gracieux, S. Saffroy. L. Bousarghin, M. Bonnaure-Mallet, and A. Jolivet-Gougeon. 2009. Distribution of mutation frequencies among Salmonella enterica isolates from animal and human sources and genetic characterization of a Salmonella Heidelberg hypermutator. Vet. Microbiol. 137:306-312. [DOI] [PubMed] [Google Scholar]
- 9.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enteric serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirose, K., A. Hashimoto, K. Tamura, Y. Kawamura, T. Ezaki, H. Sagara, and H. Watanabe. 2002. DNA sequence analysis of DNA gyrase and DNA topoisomerase IV quinolone resistance-determining regions of Salmonella enterica serovar Typhi and serovar Paratyphi A. Antimicrob. Agents Chemother. 46:3249-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins, K. L., R. H. Davies, and E. J. Threlfall. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25:358-373. [DOI] [PubMed] [Google Scholar]
- 12.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41:S120-S126. [DOI] [PubMed] [Google Scholar]
- 13.Kariuki, S., G. Revathi, J. Muyodi, J. Mwituria, A. Munyalo, S. Mirza, and C. A. Hart. 2004. Characterization of multidrug-resistant typhoid outbreaks in Kenya. J. Clin. Microbiol. 42:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, D. M., C. I. Kang. C. S. Lee, H. B. Kim, E. C. Kim, N. J. Kim, M. D. Oh, and K. W. Choe. 2006. Treatment failure due to emergence of resistance to carbapenem during therapy for Shewanella algae bacteremia. J. Clin. Microbiol. 44:1172-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, D. M., G. P. Neupane, S. J. Jang, S. H. Kim, and B. K. Lee. 2010. In vitro efficacy of the combination of ciprofloxacin and cefotaxime against nalidixic acid resistant Salmonella enterica serotype Typhi. Int. J. Antimicrob. Agents. 36:155-158. [DOI] [PubMed] [Google Scholar]
- 16.Kim, D. M., N. R. Yun, and J. H. Chung. 2008. Time kill studies of antibiotics against a nalidixic acid resistant Salmonella enterica serotype Typhi. Infect. Chemother. 40:207-211. [Google Scholar]
- 17.Kim, D. M., Y. Lym, S. J. Jang, H. Han, Y. G. Kim, C. H. Chung, and S. P. Hong. 2005. In vitro efficacy of the combination of ciprofloxacin and cefotaxime against Vibrio vulnificus. Antimicrob. Agents Chemother. 49:3489-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindgren, M. M., P. Kotilainen, P. Huovinen, S. Hurme, S. Lukinmaa, M. A. Webber, L. J. Piddock, A. Siitonen, and A. J. Hakanen. 2009. Reduced fluoroquinolone susceptibility in Salmonella enterica isolates from travelers, Finland. Emerg. Infect. Dis. 15:809-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair, S., M. Unnikrishnan, K. Turner, S. C. Parija, C. Churcher, J. Wain, and N. Harish. 2006. Molecular analysis of fluoroquinolone resistant Salmonella Paratyphi A isolate, India. Emerg. Infect. Dis. 12:489-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochiai, R. L., X. Y. Wang, L. V. Seidlein, J. Yang, Z. A. Bhutta, S. K. Bhattacharya, M. Agtini, J. L. Deen, J. Wain, D. R. Kim, M. Ali, C. J. Acosta, L. Jodar, and J. D. Clemens. 2005. Salmonella Paratyphi A rates, Asia. Emerg. Infect. Dis. 11:1764-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandit, A., A. Arjyal, J. N. Day, B. Paudyal, S. Dangol, M. D. Zimmerman, B. Yadav, K. Stepniewska, J. I. Campbell, C. Dolecek, J. J. Farrar, and B. Basnyat. 2007. An open randomized comparison of gatifloxacin versus cefixime for the treatment of uncomplicated enteric fever. PLoS One 2:e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry, C., J. Wain, N. T. Chinh, H. Vinh, and J. J. Farrar. 1998. Quinolone-resistant Salmonella Typhi in Vietnam. Lancet 351:1289. [DOI] [PubMed] [Google Scholar]
- 23.Parry, C. M., T. T. Hien, G. Dougan, N. J. White, and J. J. Farrar. 2002. Typhoid fever. N. Engl. J. Med. 347:1770-1782. [DOI] [PubMed] [Google Scholar]
- 24.Parry, C. M., V. A. Ho, T. Phuong le, P. V. Bay, M. N. Lanh, T. Tung le, N. T. Tham, J. Wain, T. T. Hien, and J. J. Farrar. 2007. Randomized controlled comparison of ofloxacin, azithromycin, and an ofloxacin-azithromycin combination for treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever. Antimicrob. Agents Chemother. 51:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokhrel, B. M., J. Koirala, R. K. Dahal, S. K. Mishra, P. K. Khadga, and N. R. Tuladhar. 2006. Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. Int. J. Infect. Dis. 10:434-438. [DOI] [PubMed] [Google Scholar]
- 26.Rupali, P., O. C. Abraham, M. V. Jesudason, T. J. John, A. Zachariah, S. Sivaram, and D. Mathai. 2004. Treatment failure in typhoid fever with ciprofloxacin susceptible Salmonella enterica serotype Typhi. Diagn. Microbiol. Infect. Dis. 49:1-3. [DOI] [PubMed] [Google Scholar]
- 27.Saha, S. K., S. Y. Talukder, M. Islam, and S. Saha. 1999. A highly ceftriaxone-resistant Salmonella Typhi in Bangladesh. Pediatr. Infect. Dis. J. 18:387. [DOI] [PubMed] [Google Scholar]
- 28.Shirakawa, T., B. Acharya, S. Kinoshita, S. Kumagai, A. Gotoh, and M. Kawabata. 2006. Decreased susceptibility to fluoroquinolones and gyrA gene mutation in the Salmonella enterica serovar Typhi and Paratyphi A isolated in Kathmandu, Nepal, in 2003. Diagn. Microbiol. Infect. Dis. 54:299-303. [DOI] [PubMed] [Google Scholar]
- 29.Thong, K. L., S. Nair, R. Chaudhry, P. Seth, A. Kapil, D. Kumar, H. Kapoor, S. Puthucheary, and T. Pang. 1998. Molecular analysis of Salmonella Paratyphi A from an outbreak in New Delhi, India. Emerg. Infect. Dis. 4:507-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Threlfall, E. J., and L. R. Ward. 2001. Decreased susceptibility to ciprofloxacin in Salmonella enterica serotype Typhi in the United Kingdom. Emerg. Infect. Dis. 7:448-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidaillac C., S. N. Leonard, H. S. Sader, R. N. Jones, and M. J. Rybak. 2009. In vitro activity of ceftaroline alone and in combination against clinical isolates of resistant Gram-negative pathogens, including β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:2360-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]