Abstract
Artemisinin-based combination therapies (ACTs) are highly effective for the treatment of Plasmodium falciparum malaria, yet their sustained efficacy is threatened by the potential spread of parasite resistance. Recent studies have provided evidence that artemisinins can inhibit the function of PfATP6, the P. falciparum ortholog of the ER calcium pump SERCA, when expressed in Xenopus laevis oocytes. Inhibition was significantly reduced in an L263E variant, which introduced the mammalian residue into a putative drug-binding pocket. To test the hypothesis that this single mutation could decrease P. falciparum susceptibility to artemisinins, we implemented an allelic-exchange strategy to replace the wild-type pfatp6 allele by a variant allele encoding L263E. Transfected P. falciparum clones were screened by PCR analysis for disruption of the endogenous locus and introduction of the mutant L263E allele under the transcriptional control of a calmodulin promoter. Expression of the mutant allele was demonstrated by reverse transcriptase (RT) PCR and verified by sequence analysis. Parasite clones expressing wild-type or L263E variant PfATP6 showed no significant difference in 50% inhibitory concentrations (IC50s) for artemisinin or its derivatives dihydroartemisinin and artesunate. Nonetheless, hierarchical clustering analysis revealed a trend toward reduced susceptibility that neared significance (artemisinin, P ≈ 0.1; dihydroartemisinin, P = 0.053 and P = 0.085; and artesunate, P = 0.082 and P = 0.162 for the D10 and 7G8 lines, respectively). Notable differences in the distribution of normalized IC50s provided evidence of decreased responsiveness to artemisinin and dihydroartemisinin (P = 0.02 for the D10 and 7G8 lines), but not to artesunate in parasites expressing mutant PfATP6.
Artemisinin (ART)-based combination therapies (ACTs) are endorsed by the World Health Organization as first-line treatment for multidrug-resistant Plasmodium falciparum malaria in most regions of endemicity in the world, including almost all of sub-Saharan Africa (14, 28; http://www.who.int/malaria/publications/atoz/9789241563901/en/index.html). However, emerging resistance to ART has recently begun to compromise the treatment of patients suffering from P. falciparum infections in western Cambodia (13, 37). ART resistance, if it spreads, would have enormous public health consequences, as there are no suitably efficacious alternatives. Understanding the mechanisms of action of ARTs has therefore gained urgency, because this knowledge may contribute to methods used for monitoring emerging resistance, as well as to the design of newer ARTs (22) that, for example, could be aimed at treating parasites that have become resistant to the first-generation derivatives.
ARTs are sesquiterpenes containing an endoperoxide bridge that is critical to their antimalarial activity. How this endoperoxide bridge kills parasites is a controversial subject (6, 18, 19, 23, 29, 55). One theory holds that an interaction of ART with Fe2+-heme in the parasite might generate toxic carbon-centered free radicals that damage parasite biomolecules, possibly via alkylation, resulting in cell death (21, 38). Evidence supporting a possible effect on the heme detoxification pathway includes reports of interactions between ARTs and heme both in vitro and in an in vivo rodent malaria model (12, 40, 43). Nonetheless, a recent study reported that exposure of Fe2+-heme to carbon monoxide does not alter the activity of ARTs, whereas it should if interaction with Fe2+-heme were important (8). Other proposed mechanisms of ART action have included interference with protein export pathways of the malarial tubulovesicular network, inhibition of parasite endocytic pathways, interference with mitochondrial membrane potential, and interactions with the calcium-binding P. falciparum translationally controlled tumor protein ortholog (1, 2, 4, 24, 31).
ARTs may also act by inhibiting the malarial sarcoplasmic-endoplasmic reticulum (ER) Ca2+-ATPase ortholog of SERCA, known as PfATP6 (15). When expressed in Xenopus laevis oocytes, PfATP6 can be specifically inhibited by ART, which had no action against several other transporters, including PfATP4 (a P. falciparum non-SERCA Ca2+ ATPase). ART inhibition of PfATP6 was antagonized by thapsigargin, a specific inhibitor of mammalian SERCA (15). Based on these data and related studies, ART compounds were proposed to bind to and inhibit PfATP6 in the P. falciparum ER, leading to parasite death.
Modeling studies of the PfATP6 sequence and docking simulations suggested that ARTs might interact directly with PfATP6, possibly via hydrophobic interactions (27, 35, 53). Further studies in oocytes showed that amino acid variants at residue 263 (L263, 263S, and 263A), predicted to be located at the apex of the predicted ART-binding pocket, could modulate the susceptibility of PfATP6 to inhibition by ART. Differences in susceptibility paralleled the rank order of the ART 50% inhibitory concentrations (IC50s) observed with P. falciparum, Plasmodium berghei, and Plasmodium vivax, respectively, from which these variants derive (53). An L263E substitution introduced the residue found in the position analogous to that of mammalian SERCA, which was resistant to ART when assayed in enriched rabbit muscle preparations. This resulted in loss of inhibition by ARTs.
These observations led to the hypothesis that mutations at this residue might change the conformation of the putative drug-binding site and potentially cause decreased susceptibility to ART. Additional support for PfATP6 as a possible target of ART compounds in P. falciparum emerged from a field study that reported an association between a S769N substitution in PfATP6 and elevated in vitro artemether IC50s in isolates from French Guiana, suggesting a potential role for polymorphisms in pfatp6 as candidate markers for emerging ART resistance (26).
In this study, we aimed to test whether a single amino acid change at position 263 of PfATP6 could modulate parasite resistance to this critical class of antimalarials. Utilizing allelic-exchange techniques, the wild-type pfatp6 allele was replaced via single-crossover recombination by a 263E variant allele in two distinct genetic backgrounds, 7G8 (Brazil) and D10 (Papua New Guinea). This article reports the generation of these recombinant parasites and an analysis of their susceptibilities to ARTs.
MATERIALS AND METHODS
Parasites and transfection.
The P. falciparum parasite lines 7G8 and D10 were obtained from MR4 (http://www.malaria.mr4.org; ATCC, Manassas, VA). They were propagated in leukocyte-free human erythrocytes at 3% hematocrit in complete medium (RPMI 1640 with l-glutamine, 50 mg/liter hypoxanthine, 10 mg/liter gentamicin, 25 mM HEPES, 0.225% NaHCO3, and 0.5% Albumax I [Life Technologies]) and cultured at 37°C under 5% CO2, 5% O2, and 90% N2 gas (17). The parasites were electroporated with purified plasmid DNA, and episomally transfected lines were selected with 2.0 μg/ml blasticidin HCl (Invitrogen), as described previously (45). Plasmid integration into the pfatp6 locus was detected by PCR and confirmed by Southern blot analysis (see below). Recombinant parasites were cloned on day 120 by limiting dilution and identified by their expression of parasite lactate dehydrogenase, as described elsewhere (20, 46).
DNA constructs.
A 2.5-kb pfatp6 coding-sequence fragment (nucleotides 1 to 2503 of the 3.7-kb pfatp6 gene, contained within exon 1 of this 3-exon gene; PlasmoDB gene ID, PFA0310c [http://www.plasmodb.org]) was amplified from Xenopus expression vectors that had either the L263 or the mutant 263E pfatp6 coding sequence cloned into pcDNA3.1(+) (15). Sequences were amplified with the primers 5′-AAACCTAGGATGGAAGAGGTTATTAAGAATGCTC (the AvrII site is underlined) and 5′-AAAGTCGACCTTTAGCTATAGCTCTGGCCG (the SalI site is underlined) and subcloned into the pLN_CAM plasmid digested with AvrII and SalI. This transfection plasmid expresses the blasticidin-S-deaminase (bsd)-selectable marker (32), under the control of a 0.6-kb Plasmodium chabaudi dihydrofolate reductase-thymidylate synthase (PcDT) 5′ untranslated region (UTR) and a 0.6-kb P. falciparum hrp2 3′ UTR. The pfatp6 insert was placed under the control of a functional 1.0-kb P. falciparum 5′ UTR from the calmodulin gene (PlasmoDB gene ID PF14_0323 [10]). The resulting 8.0-kb plasmids were designated pLN_L263 and pLN_E263. The pLN_E263 plasmid has three nucleotide mutations: T787G and T788A (which confer the L263E amino acid substitution) and T792C (a silent mutation that creates an EcoRI site) (53).
DNA analysis.
P. falciparum DNA was purified from saponin-released trophozoite pellets with DNeasy Tissue Kits (Qiagen). PCR-based detection of integration used the pfatp6-specific primers p1 (5′-CTTATTATATCTTTGTCATTCGTG) and p5 (5′-GAAAAATCTTCAAATTCTCTTCCA), the pLN-specific primer p2 (5′-ACACTATAGAATACTCAAGCTTGG), and the calmodulin 5′ UTR-specific primer p3 (5′-CCTAATAGAAATATATCACCTAGG). Gel-purified PCR products (Gel Extraction Kit; Qiagen) were sequenced with the pfatp6 internal coding sequence primer 5′-AAATATTTTATTTTCATCTACCGC.
For Southern blot analysis, 1 μg of DNA was digested with XbaI and NcoI, electrophoresed, and transferred onto nylon membranes. Hybridizations were performed with a hexamer-primed 32P-labeled probe that was prepared from a 0.7-kb fragment of the pfatp6 coding sequence (nucleotides 373 to 1090), released by HindIII/KpnI digestion of the transfection plasmid pLN_E263.
RNA preparation and RT-PCR assays.
RNA was prepared from sorbitol-synchronized trophozoite stage parasites using Trizol (Invitrogen) and treated with DNase I (New England Biolabs). cDNA was prepared from purified RNA by reverse transcription with oligo(dT) primers. Reverse transcriptase (RT) PCR was performed with the pfatp6-specific primers p4 (5′-TGGAAGAGGTTATTAAGAATGCTC) and p5, as well as with the calmodulin 5′ UTR-specific primers p3 plus p5. RT-PCR products were gel purified with gel extraction kits (Qiagen), and the products were sequenced using the pfatp6 internal coding sequence primer 5′-AAATATTTTATTTTCATCTACCGC (nucleotides 633 to 656; located 130 bp upstream of the variant nucleotide at position 789).
In vitro antimalarial drug assays.
Artemisinin, dihydroartemisinin (DHA), and artesunate (AS) were kindly provided by William Ellis, Division of Experimental Therapeutics, Walter Reed Army Institute of Research, Washington, DC. Parasite susceptibilities to antimalarial drugs were assayed in vitro using [3H]hypoxanthine incorporation, as previously described (17). Briefly, cultures composed predominately of ring stage parasites were seeded in duplicate in 96-well plates at 0.4% parasitemia and 1.6% hematocrit; 1.5-fold serial dilutions were made to final-concentration ranges of 2.1 to 53.1 nM for ART for 7G8, 5.5 to 141.7 nM for ART for D10, 0.8 to 21.1 nM for DHA for both strains, and 2.8 to 104.0 nM for AS, also for both strains. After 48 h, [3H]hypoxanthine (0.5 μCi/well) was added, and cells were harvested at 72 h. IC50s were determined by linear regression as described previously (17). Each assay was performed in duplicate and repeated 4 to 9 independent times. Assays 3 to 7 and 8 to 11 (see Tables S1 and S2 in the supplemental material) were run as two groups 6 to 8 months apart and were thus treated as two separate data sets for the purpose of establishing normalized IC50s for the L263 controls (i.e., mean values were calculated for assays 3 to 7 and separately for assays 8 to 11 and were used to derive normalized IC50s for individual lines). Assays 1 and 2 were not considered, as they were dose-finding experiments to optimize the range of dilutions employed in these assays.
Heat map analyses and IC50 normalizations.
Heat maps (see Fig. 4) were generated from the six individual data sets created for each combination of drug (ART, DHA, and AS) and strain (7G8 and D10). Further heat maps that incorporated two or more drugs and/or that combined both 7G8 and D10 parasites were generated following normalization of the IC50s (as described above) in order to compare across drugs or strains irrespective of their baseline IC50s. For each drug, this produced a normalized mean IC50 of 1.0 for the control L263 lines (see Fig. 5B; see Table S2 in the supplemental material). After normalization, the data sets were combined by taking the set of concentrations that had a smaller range and imputing those values into the other sets.
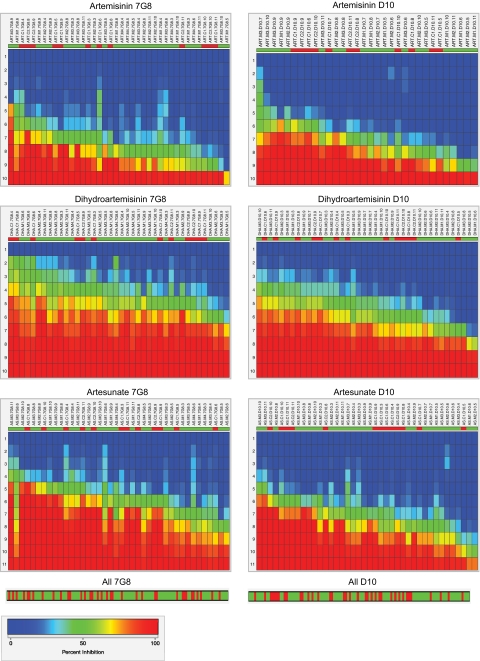
FIG. 4.
Heat maps of dose-response data for pfatp6-modified parasite lines, grouped by host strain and compound. The rows represent the range of concentrations renumbered 1 to 10 or 1 to 11, with 1 being zero drug. Each column represents a separate line tested in a particular assay, with percent inhibition going from 0 (dark blue) to 50 (green) to 100 (red) as the compound concentration increased. This plot allows the visualization of both the IC50 level of a sample and the percent inhibition at all concentrations. The dose-response data are hierarchically clustered from left to right based on IC50s, with the left being the most sensitive response and the right being the least sensitive response (i.e., clustered from lowest IC50 on the left to highest IC50 on the right). The top row reflects the type of line; 263E lines are green, and L263 lines are red. Reviewing the top row from left to right illustrates the distribution of these lines from most susceptible to least susceptible, respectively. The heat maps for individual strains and drugs utilized raw IC50 values and dose-response data. At the bottom is shown the hierarchical clustering of the normalized IC50s obtained for all drugs for the 7G8 or D10 background, producing P values of 0.108 and 0.008, respectively (Table 2). 7G8 and D10 heat maps showing the combined data for all drugs are presented in Fig. S1 in the supplemental material.
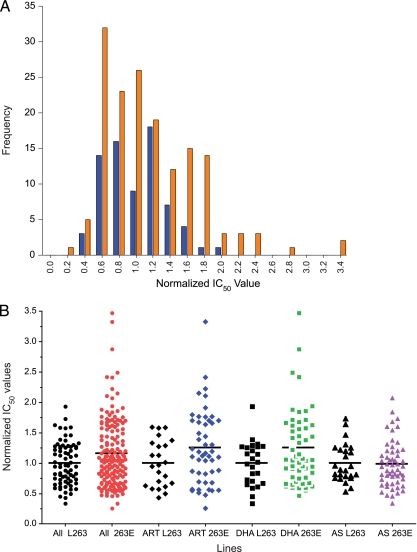
FIG. 5.
In vitro response of pfatp6-modified recombinant clones to ART, DHA, and AS. (A) Distributional plot of normalized (1 = 100%) IC50s for ART, DHA, and AS for control (L263, blue) and mutant (E263, orange) congenic cell lines. (B) Individual normalized IC50s for control (L263, black) and mutant (E263, red, blue, green, or purple) congenic lines. Aggregated data sets (ART plus DHA plus AS) include results for these drugs pooled for both 7G8 and D10 parasites. Individual assays are shown for each drug. The horizontal lines illustrate the normalized mean IC50s for each group.
Statistical analyses.
Data were compiled in Excel. One-way analysis of variance (ANOVA) tests were used to compare mean IC50s between control and mutant lines for a given strain and compound (Table 1). Following hierarchical clustering, hypergeometric distribution tests were used to analyze the extent to which the IC50s of the mutant samples were skewed to the left or right of the IC50s of the controls (Table 2; see Fig. 4 below and Fig. S1 in the supplemental material).
TABLE 1.
Summary of absolute and normalized IC50 values for mutant and control lines
| Drug | Measurement | Value (mean ±SEM) |
|||
|---|---|---|---|---|---|
| 7G8a |
D10b |
||||
| L263 | 263E | L263 | 263E | ||
| Artemisinin | Absolute IC50 (nM) | 20.7 ± 2.9 | 22.3 ± 1.7 | 42.8 ± 5.0 | 51.6 ± 5.5 |
| Normalized IC50 | 1.00 ± 0.11 | 1.30 ± 0.12 | 1.00 ± 0.12 | 1.20 ± 0.13 | |
| No. of assays | 12 | 28 | 11 | 21 | |
| Dihydroartemisinin | Absolute IC50 (nM) | 2.26 ± 0.29 | 2.37 ± 0.18 | 3.41 ± 0.36 | 3.96 ± 0.35 |
| Normalized IC50 | 1.00 ± 0.10 | 1.33 ± 0.13 | 1.00 ± 0.11 | 1.18 ± 0.10 | |
| No. of assays | 12 | 28 | 13 | 27 | |
| Artesunate | Absolute IC50 (nM) | 13.5 ± 1.7 | 14.9 ± 1.4 | 23.6 ± 3.1 | 26.2 ± 2.8 |
| Normalized IC50 | 1.00 ± 0.09 | 0.97 ± 0.07 | 1.00 ± 0.09 | 0.99 ± 0.08 | |
| No. of assays | 12 | 28 | 13 | 27 | |
7G8 L263 refers to C17G8 and C27G8; 7G8 263E refers to M17G8, M27G8, M37G8, and M47G8.
D10 L263 refers to C1D10 and C2D10; D10 263E refers to M1D10, M2D10, and M3D10.
TABLE 2.
Tests of significancea
| Drug | Line | P valueb |
|---|---|---|
| Artemisinin | 7G8 | 0.107 |
| Artemisinin | D10 | 0.101 |
| Artesunate | 7G8 | 0.162 |
| Artesunate | D10 | 0.082 |
| Dihydroartemisinin | 7G8 | 0.085 |
| Dihydroartemisinin | D10 | 0.053 |
| Artemisinin | 7G8 and D10 | 0.032 |
| Artesunate | 7G8 and D10 | 0.039 |
| Dihydroartemisinin | 7G8 and D10 | 0.057 |
| Artemisinin plus artesunate | 7G8 and D10 | 0.017 |
| Artemisinin plus dihydroartemisinin | 7G8 and D10 | 0.020 |
| Artesunate plus dihydroartemisinin | 7G8 and D10 | 0.009 |
| All three drugs | 7G8 | 0.108 |
| All three drugs | D10 | 0.008 |
| All three drugs | 7G8 and D10 | 0.026 |
Performed following hierarchical clustering analysis.
P values were calculated using a hypergeometric one-tailed test to compare the L263 and 263E lines. This test analyzed the extents to which the IC50 values of the 263E samples were skewed to the right (i.e., were higher) compared to the L263 controls.
Sample size calculations and normality assumptions for IC50s employed Shapiro-Wilks and Shapiro-Francia tests and were evaluated in Stata (v8.0 for Macintosh). Other statistical and graphical analyses were done in Prism (Graphpad v4.0a). Nonparametric Mann-Whitney U tests and parametric unpaired t tests (with Welch's correction for unequal variances) were used to compare control and mutant groups. Proportions were compared with chi-squared tests, and Fisher's exact values were calculated. Data values for each line are provided in Tables S1 and S2 in the supplemental material, which list the individual arithmetic and the group normalized means.
RESULTS
Generation of recombinant lines expressing the PfATP6 L263E mutation.
To investigate a potential role for the PfATP6 L263E mutation in mediating reduced in vitro susceptibility of P. falciparum to the ART class of antimalarials, we applied an allelic-exchange strategy to the P. falciparum ART-sensitive strains 7G8 and D10, as illustrated in Fig. 1.
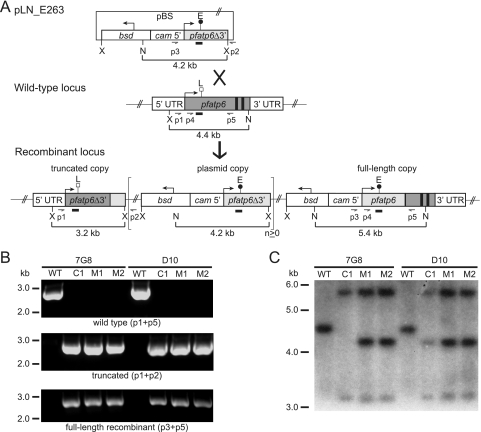
FIG. 1.
pfatp6 allelic-exchange strategy and molecular characterization of clones. (A) The transfection plasmid pLN_E263 contains the bsd selectable marker and a 2.5-kb pfatp6 truncated coding sequence fragment lacking the 3′ end of the gene (containing the putative transmembrane domains 6 to 10 and encoding the L263E mutation). The control plasmid pLN_L263 (not shown) encodes the wild-type L263 residue. Single-site crossover between the plasmid and the endogenous pfatp6 locus resulted in a recombinant locus in which the wild-type pfatp6 allele was replaced. The resulting upstream, truncated fragment of pfatp6 lacked the 3′ end of the gene (including exons 2 and 3) and a 3′ UTR. The downstream, functional, full-length allele was under the control of the calmodulin 5′ UTR and had the endogenous pfatp6 3′ UTR as its terminator element. The brackets delineate the plasmid sequence that in some lines integrated as tandem linear copies (total plasmid copy number, n ≥ 1). PCR primers (p1 to p5), XbaI (X) and NcoI (N) sites, and fragment sizes and the pfatp6 probe (narrow rectangular boxes) are indicated. pBS, pBluescript plasmid backbone. The large cross indicates a single crossover event, and the thick arrow points to the ensuing recombinant locus. The E above the black circle and the L above the white square refer to the position of the nucleotides encoding amino acids 263E and L263. The bent arrows indicate the direction of transcription. (B) Integration of the transfected plasmids at the pfatp6 locus was confirmed by PCR analyses of the wild-type (WT) parental (7G8 and D10) lines and the recombinant clones (C17G8, C1D10, M17G8, M27G8, M1D10, and M2D10) with primers specific for the wild-type locus (p1 plus p5), the upstream truncated fragment (p1 plus p2), and the downstream recombinant, full-length allele (p3 plus p5). (C) Southern blot hybridization of XbaI/NcoI-digested genomic probe revealed a 4.4-kb band in the wild-type strains. The recombinant clones lacked the wild-type band and showed 5.4-kb and 3.2-kb bands consistent with recombination at the pfatp6 locus. The 4.2-kb band indicated integration of tandem plasmid copies into this locus.
We constructed the 8.0-kb pLN_E263 transfection plasmid to contain the bsd-selectable marker cassette and a 3′-truncated 2.5-kb pfatp6 fragment encoding the L263E mutation. This fragment (denoted pfatp6Δ3) was placed downstream of a functional 1.0-kb calmodulin 5′ UTR (Fig. 1A). Single-site crossover between this insert and the homologous pfatp6 sequence was predicted to replace the endogenous pfatp6 gene with a recombinant, full-length allele under the control of the calmodulin 5′ UTR. Crossover events downstream of position 263 resulted in the introduction of the L263E mutation into this recombinant, functional allele. The calmodulin promoter was selected, as calmodulin and pfatp6 have nearly identical transcriptional profiles, with maximal transcription at 32 to 34 h, as demonstrated in two independent, genomewide microarray studies with highly synchronized parasites (5, 30; http://www.plasmodb.org). As a control experiment, a pLN_L263 plasmid containing the same pfatp6 fragment but encoding the wild-type L263 residue was also transfected.
Recombination and single-site crossover events also created an upstream truncated fragment expressing the first 2.5 kb of the pfatp6 coding sequence, under the control of the endogenous pfatp6 promoter but lacking a 3′ UTR. This fragment, contained within exon 1 of this 3-exon gene, was 1.2 kb shorter than the full-length pfatp6 coding sequence (corresponding to amino acids 1 to 833 of the 1,228-amino-acid full-length protein and predicted to encode the first 5 of the protein's 10 transmembrane helices). The absence of a 3′ UTR has been previously shown in recombinant Plasmodium parasites to be incompatible with protein expression (9, 39, 58). Furthermore, analysis of the structural model and predicted topology of PfATP6 and comparison with the known structure of the orthologous rabbit SERCA1 molecule indicated that the removal of the five C-terminal transmembrane helices and of key functional regions (including the predicted phosphorylation site Asp359, analogous to Asp351 in rabbit SERCA1, and the putative Ca2+-binding sites in helices M4 to M6 and M8 [51-53]) would render any translated product nonfunctional.
7G8 and D10 parasites transfected with pLN_L263 and pLN_E263 were screened for plasmid integration into the pfatp6 locus by PCR. This included the use of primers specific for the recombinant upstream truncated and downstream full-length loci (p1 plus p2 and p3 plus p5, respectively). We also tested for the presence of the original wild-type locus using the primer pair p1 plus p5 (primer locations are shown in Fig. 1A). By day 86 postelectroporation, lines that were positive with the p1 plus p2 and p3 plus p5 primer pairs diagnostic for recombination had been identified. From these lines, recombinant clones were obtained by limiting dilution. The clones generated from transfection of the control pLN_L263 plasmid were named C17G8, C27G8, C1D10, and C2D10, and those generated from transfection of the mutant pLN_E263 plasmid were named M17G8, M27G8, M37G8, M47G8, M1D10, M2D10, and M3D10.
PCR analyses of these clones showed loss of the 2.7-kb wild-type-specific band (p1 plus p5) in the recombinant controls and mutants and acquisition of PCR bands specific for the upstream truncated fragment (2.7 kb; p1 plus p2) and the downstream recombinant full-length allele (2.7 kb; p3 plus p5) (shown for a subset of clones in Fig. 1B; other clones produced equivalent results [data not shown]). These PCR products were excised and sequenced, revealing that the integration event in the mutant clones occurred in such a way that the L263E mutation was downstream in the full-length allele (Fig. 1A). Southern blot hybridization of XbaI/NcoI-digested genomic-DNA samples with a pfatp6 probe confirmed the expected gene organization in all clones, revealing the loss of a 4.4-kb band present in the wild-type strains and showing 5.4-kb and 3.2-kb bands consistent with single-site recombination at the pfatp6 locus (Fig. 1C). The 4.2-kb band indicated integration of two or more tandem copies of the transfection plasmid into the pfatp6 locus in all recombinant clones, with the exception of C17G8, which harbored a single plasmid copy in the integrated locus.
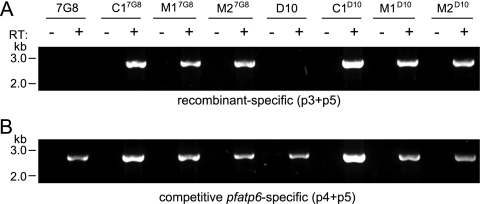
RT-PCR analysis of pfatp6 recombinant clones.
To confirm the expression of pfatp6 from the 1.0-kb calmodulin 5′ UTR, RT-PCR was performed on RNA samples prepared from trophozoite stage parasite samples. These reactions used a calmodulin 5′ UTR-specific primer (p3) and a pfatp6-specific primer (p5), which yielded a 2.7-kb band (Fig. 2 A). Mock reactions prepared without RT showed no product, demonstrating the lack of detectable genomic-DNA contamination in the RNA samples. Sequencing of the PCR products from a competitive RT-PCR utilizing primers specific for full-length pfatp6 (p4 plus p5), which can amplify from a recombinant full-length allele or a wild-type allele [Fig. 1A]), demonstrated that the clones expressed only a single functional allele (from the downstream locus in the case of the recombinant lines) (Fig. 1A and 2B).
FIG. 2.
RT-PCR analysis of pfatp6 recombinant clones. (A) Expression of the recombinant pfatp6 locus from the 1.0-kb calmodulin 5′ UTR was confirmed by RT-PCR analysis of RNA prepared from trophozoite stage parasites using a calmodulin 5′ UTR-specific primer (p3) and a pfatp6-specific primer (p5), which revealed a 2.7-kb band. Reaction mixtures were prepared with and without RT to confirm the absence of detectable genomic-DNA contamination in the samples. (B) Sequencing of competitive RT-PCR products obtained with primers specific for full-length pfatp6 (p4 plus p5) demonstrated full-length expression of only the downstream recombinant locus in the E263 mutant clones, as demonstrated by sequence identification of only the mutant codon in the RT-PCR product. The primers are indicated in Fig. 1.
Drug responses of pfatp6 recombinant clones.
We tested the pfatp6 recombinant clones against ART, DHA (the active in vivo metabolite of many clinically useful ART compounds [57]), and AS. IC50s were also obtained for the 7G8 and D10 wild-type strains, as well as the recombinant control and mutant lines generated in each strain. The ART IC50s (means ± standard errors of the mean [SEM]) for 7G8 and D10 were 16.5 ± 2.9 and 56.4 ± 5.2 nM, respectively, determined from 5 independent assays performed in duplicate. These baseline values were comparable to those from an earlier study of these strains that had reported IC50s of 11 nM and 42 nM, respectively (41). Other studies have also obtained similar IC50s with ART (36, 44, 49).
In total, each line was run on average 7 times (range, 4 to 9) against each derivative. Table 1 shows the mean (±SEM) IC50s calculated for the set of L263 and 263E lines grouped per strain background and artemisinin compound (IC50s for individual assays are provided in Table S1 in the supplemental material). Statistical analysis revealed no significant differences between the two sets of lines, as calculated by one-way ANOVA tests comparing mutants to controls for a given strain and a given compound.
The trend toward slightly elevated IC50s in the D10 263E mutant lines (Table 1), and the evidence from the literature that IC50s do not readily detect altered parasite susceptibility to ART derivatives (7, 13, 56) led us to examine our assays in greater detail. Figure 3 shows the mean (±SEM) values of percent inhibition for each line assayed against each compound at each concentration tested in these dose-response assays. Broadly, the results showed similar dose-response patterns between lines in a given genetic background. In the 7G8 background, the least susceptible across all artemisinins was the M47G8 line, while the C17G8 line appeared typically to be the most sensitive. In D10, there was a trend toward the 263E lines being less susceptible, and the C1D10 line was typically the most sensitive of all. These assays revealed reduced susceptibility of D10 parasites in general compared to the 7G8 lines, with this difference being the most evident for ART (Table 1).
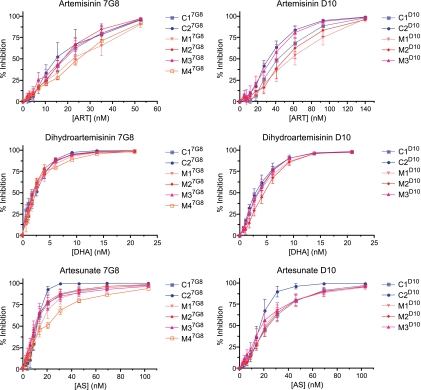
FIG. 3.
Dose-response assays for L263 and 263E congenic lines tested against ART, DHA, and AS. The results were obtained from 4 to 9 independent assays with duplicate wells per line and per concentration. Percent inhibition values are shown as means ± SEM.
To analyze these data further, we organized the dose-response data into six groups, corresponding to the data for ART, AS, and DHA for the 7G8 and D10 backgrounds, and performed hierarchical clustering analysis. For this, we converted the dose-response data for a particular line and compound in a given assay into a heat map that visually represented the dose-response profile (ranging from 0% inhibition, depicted in blue, to 50%, in green, and 100%, in red). We then plotted each line and assay vertically down a set of rows corresponding to the drug concentration (set on a scale of 1 to 10 for ART and DHA and 1 to 11 for AS; with 1 being zero drug and the highest number representing the highest drug concentration). The data were then clustered from left to right so that the leftmost data set represented the most sensitive dose-response profile and the rightmost data set the least sensitive profile (Fig. 4). This allowed a visual representation of how lines compared in their dose responses. As an example, with DHA and the 7G8 lines, one could observe that the most sensitive response was the C17G8 line tested in assay 4, whereas the four least sensitive responses were observed with mutant lines M17G8 (assay 5), M27G8 (assay 6), and M37G8 (assays 9 and 10). This analysis allowed us to peruse all the data without regard to the initial IC50 and to compare across genetic backgrounds and compounds.
Visual inspection of these data revealed some trends toward decreased susceptibility in 263E lines, manifested as a preponderance of these lines (shown in green in the top row above the heat maps) toward the right of the clustered distribution. We tested this statistically using a hypergeometric test that calculated the probability that a given type of line (L263 or 263E) would be represented at a particular place on the spectrum of IC50s clustered from least to most sensitive (i.e., from right to left in the top panels in Fig. 4; note that more assays were performed with 263E than with L263 lines because more mutant lines were included in our assays). The null hypothesis in this case was that the distribution was random. Statistical tests for individual experimental groups approached but did not attain significance (Table 2). Nonetheless, we noted that combining responses for two or more drugs, or combining drugs for 7G8 and D10, resulted in significant differences between mutant and control lines for most combinations (Table 2).
Two further points emerged from this analysis. First, the genetic background (7G8 versus D10) influenced the drug sensitivity phenotype of the L263E mutation, with the D10 line most likely to show differences between 263E and L263 lines. Second, susceptibility to the clinically employed derivatives AS and DHA produced a highly significant difference between mutant and control lines when data for both genetic backgrounds were combined.
These results led us to examine our data across lines and compounds by normalizing our IC50s so that data could be analyzed across groups irrespective of host strain-specific differences in the degree of susceptibility to individual compounds. First, we calculated the mean IC50 for all isogenic control lines (harboring the L263 allele) for a given set of concurrently run independent assays and then used these mean values to calculate normalized IC50s for control (L263) and mutant (263E) lines by dividing their raw IC50s in a given assay by the mean IC50 obtained from the control lines. This method of normalization enabled comparisons to be made both within and between experiments. Normalized mean (±SEM) IC50s for individual assays are listed in Table S2 in the supplemental material.
Results from the set of normalized IC50 determinations, presented in Fig. 5, revealed significant differences in the distributions of wild-type versus mutant lines. As shown in Fig. 5A, normalized IC50s for congenic control lines showed normal distributions (Shapiro-Wilks and Shapiro-Francia P values, >0.05 for both groups), whereas the distributions were not normal for the E263 lines (Shapiro-Wilks and Shapiro-Francia P values, <0.01 for both groups and tests; see Table S3 in the supplemental material for the data distribution represented in Fig. 5A). This pronounced distributional difference between mutant and control cell lines resulted predominantly from some relatively high values for normalized IC50s observed in the mutant lines (see Table S2 in the supplemental material). When the data for all three compounds were aggregated, the difference between the highest and lowest normalized IC50s was 5.7-fold in congenic control strains for the larger aggregated data set (range, 0.34 to 1.93 times the mean value for control strains) and 13.3-fold for transgenic lines carrying the L263E mutation (range, 0.26 to 3.47 times the mean value for control strains). The mean increase in IC50s was 22.9% (95% confidence interval [CI], 1.11 to 1.36) in the aggregated E263 parasites compared with controls (95% CI, 0.89 to 1.1) with corresponding median (interquartile range) values for L263 of 1.0 (0.67 to 1.3) and for E263 of 1.1 (0.7 to 1.6), representing an approximate 11% increase in median normalized IC50s for E263 parasites (P = 0.038 [one-tailed Mann-Whitney U test] and P = 0.003 [unpaired t test with Welch's correction]). Table S4 in the supplemental material provides a listing of the normalized data from each drug assay that comprise the individual data points illustrated in Fig. 5B. Normalized mean (±SEM) IC50s for the two genetic strains and for each ART derivative are illustrated in Table 1, which also shows the nonnormalized data. We note that it would have been preferable to compare these results with a comparator drug, such as chloroquine, for which no difference would have been expected. However, no other drug was included, as the emphasis was placed on generating a large number of replicate assays in order to increase the sensitivity of our analysis of ART responses in our pfatp6-modified lines.
As shown in Fig. 5B, there was no significant increase in the median value for IC50 results between mutant and control lines for AS (P = 0.4), whereas differences between these parasite lines were significant for DHA (P = 0.02; one-tailed Mann-Whitney U test). These values reflect a skewed distribution of results for mutant lines compared with controls. This aspect of the results can also be illustrated by considering the proportion of results for normalized IC50s that fall >60% above the mean of control values. These differences were maintained for ART (0% [0/23 in L263 parasites] compared with 27% [11/41 in E263 parasites]; P = 0.005) and for DHA (4.2% [1/24 in L263 parasites] compared with 27% [14/55 in E263 parasites]; P = 0.029; all two-sided Fisher's exact tests). When the DHA and ART data were combined, a significantly higher proportion (26%; 25/96) of values lying >60% above the mean control IC50 was also noted in the 263E lines compared with the L263 parasites (3.8%; 1/26; P = 0.0002; Fisher's exact test).
DISCUSSION
Recent reports of delayed parasite clearance times following artemisinin treatment of P. falciparum infections in western Cambodia provide evidence of emerging resistance (13, 37) and add urgency to studies that aim to examine the mechanism of action of this critical class of first-line antimalarial drugs. How ARTs might act against parasites, against other infections, and as agents to treat cancer progression remains a subject of considerable controversy (14, 29).
Allelic-exchange experiments with P. falciparum are powerful tools to examine mechanisms of antimalarial action and parasite resistance (16, 41, 44, 54). Their value is complemented by combining data from heterologous expression studies, for example, in the X. laevis oocyte or Saccharomyces cerevisiae expression models, when testing the hypothesis that PfATP6, or its orthologs in other organisms, might constitute an important target for the activity of artemisinins. Several points emerge from our allelic-exchange study that illuminate possible mechanisms of action of this class of antimalarial and that may carry significance for clinical studies on emerging ART resistance. First, in our parasite assays, the magnitude of any change in artemisinin sensitivity associated with the L263E mutation was relatively small (between 10 and 20% change in the mean absolute IC50) (Table 1). One interpretation of these data is that PfATP6 is not a primary target of artemisinin action. Clearly, further studies will be needed to confirm the precise contribution of PfATP6 to artemisinin drug resistance or drug action.
Nonetheless, a similar magnitude of change in IC50 for DHA has been observed in field isolates from patients in Pailin, western Cambodia, who carry parasites that manifest reduced parasite clearance times compared to those of patients from Wanh Pha, Thailand, where clearance times are shorter (from these two regions, mean IC50s for DHA were 2.3 nM and 1.5 nM, respectively [13]). When analyzed using aggregated data that increased the number of assays used for comparison, our studies with isogenic lines provide evidence that the introduction of a single amino acid mutation (L263E) into PfATP6 affected parasite sensitivity to ARTs in several measurable ways.
Second, our drug assays revealed substantial variability in normalized IC50s for ART and DHA (up to approximately 13-fold between the highest and lowest values), with this variability being higher in parasites bearing the E263 mutant than in congenic control lines. This variability suggests a degree of phenotypic plasticity that distinguishes the two sets of parasite lines. A significantly higher proportion of parasites (∼2-fold) expressing the E263 mutant PfATP6 had relatively elevated IC50s (by up to 300% in some assays) (Fig. 5B) compared to the mean values for control lines assayed simultaneously. Field isolates show similar dispersion patterns in IC50s (13, 37). For example, for DHA assays from Pailin (Cambodia), the difference observed between the highest and lowest values was ∼14-fold, and for parasites from Wang Pha (Thailand), it was ∼10-fold (see Fig. 3 in reference 13). In those assays, the contribution of variability in genetic backgrounds was not controlled as it is in transgenic laboratory parasites. This high dispersion in IC50s for ART has important implications for assessing resistance, as based on our calculations, a minimum sample size of 84 (partially resistant) Cambodian and 42 (sensitive) Thai isolates would have been needed to detect a significant difference between the Pailin and Wang Pha groups (2-sided alpha; power, 0.8). This calculation assumes a similar fold difference is observed with the reported data with results from approximately 20 isolates from each site. Correlating sensitivities with single nucleotide polymorphisms (SNPs) in putative resistance genes under these circumstances, where no difference was seen in the study by Dondorp et al. (13), may well also require larger numbers of assays.
Third, not all ARTs affected parasites bearing the L263E mutation to similar extents, as there was no discernible effect on the distributions of the normalized IC50s measured for AS compared with those for ART and DHA (Fig. 5B). This lack of modulation of AS sensitivity by the L263E mutation may reflect functional differences between drugs, as the results of sufficient assays have probably been determined, according to our power calculations. Of note, the field isolates from western Cambodia that showed significantly increased IC50 estimates for DHA, when taken from the cohort of patients with prolonged parasite clearance times, had IC50s for AS that were also not significantly increased (see Fig. 3 in reference 13).
Fourth, our findings remain consistent with PfATP6 being a potential target for ARTs, because the change in normalized IC50s in parasites is associated with a change in an amino acid that, based on structural modeling, occupies a key position (53). Nevertheless, there is evident overlap in the results for IC50s of control and mutant lines (see Tables S1 and S2 in the supplemental material), and the mean IC50s did not differ significantly. This may be attributable to several factors. Inherent variability in assays is one possibility, although this would be expected to affect results from both control and mutant lines similarly and would thus seem highly unlikely to account for the distributional differences between the two experimental groups (Fig. 5). The introduction of mutations in PfATP6 may alter expression phenotypes, particularly if the mutations can subtly shift the processing efficiencies of nascent proteins. An alternative explanation is that a secondary, non-PfATP6 target becomes important in ART activity when the sensitivity of PfATP6 as a primary target diminishes, or PfATP6 may be one target but not the primary target. Our data nonetheless indicate that in most assays, IC50s of L263 and 263E parasites do not differ significantly, and such a difference is observed only in a subset of assays that reveal relatively higher IC50s in 263E parasites (Fig. 5; see Table S1 in the supplemental material). This could potentially reflect an altered susceptibility to ARTs that is not adequately captured using these [3H]hypoxanthine-based measurements.
We note that the L263E mutation, selected based on modeling and homology data to study the role of PfATP6 in ART susceptibility, has not been identified in field isolates. Nevertheless, further support for an important role for PfATP6 in ART action comes from identification of different mutations in this gene in field isolates of P. falciparum. This includes several examples of field isolates that displayed increased IC50s for particular ART derivatives (reviewed in reference 29). In most cases where PfATP6 polymorphisms have been described (3, 11, 25, 33, 48), corresponding phenotypic results are lacking, and in any case, interpretations may be confounded by some of the factors discussed above, as well as difficulties in adapting field isolates to laboratory culture for phenotype-genotype comparisons.
When expressed in yeast and either highly purified or reconstituted, PfATP6 is inhibited only by very high concentrations of thapsigargin (∼30 μmol) and not by ARTs (6a). These findings suggest that the methodological aspects of the experimental system being used can impact significantly on the physiological properties of malarial transporters and studies of their inhibition. In support of this, when the Toxoplasma gondii ortholog of PfATP6 was expressed in yeast without purification, it complemented a yeast SERCA-deficient mutant and rendered it sensitive to ART (34). Methodologies used to study mammalian SERCA also influence the assessment of sensitivity to ARTs, because rabbit muscle SERCA was inhibited by DHA and not by ART in our earlier assays (50), whereas others have shown ART-mediated inhibition of SERCAs purified by immunoprecipitation from cancer cell lines (42). A third group has reported sensitivity of mammalian SERCAs to ART dimers, but not monomers (47). This evaluation of the SERCA hypothesis across multiple experimental systems is commensurate with the growing importance of ARTs across diverse areas of medicine. For malarial parasites, the results from this study are consistent with the proposition that ARTs might act, at least in part, via inhibition of PfATP6. Undoubtedly, many variables that influence the assessment of ART sensitivity remain to be identified. Furthermore, ART resistance may arise in ways other than alterations in susceptibility of a target. Further investigations are clearly required to delineate how resistance mechanisms will occur in the field and to apply that knowledge to restricting their spread and clinical impact.
Supplementary Material
Acknowledgments
This work was supported in part by institutional funds from Columbia University and the Albert Einstein College of Medicine provided to D.A.F. S.K. is funded by the European Commission projects ANTIMAL (grant no. 018834) and MALSIG (grant no. 223044), as well as the Wellcome Trust (grant no. 074395). S.G.V. gratefully acknowledges funding support from the NIH Training Program in Molecular Pathogenesis of Infectious Diseases (5T32AI007506; director, Arturo Casadevall).
A.-C.U. and S.K. originally conceived the allelic-exchange idea and provided initial versions of the transfection constructs; S.G.V. and D.A.F. designed the experimental investigations; S.G.V. performed the transfections, molecular analyses, and drug assays; S.G.V., D.A.F., D.S., and S.K. analyzed the data; D.S. performed the heat map analysis; S.K. normalized the IC50 data and implemented the statistical tests; and S.G.V., D.A.F., and S.K. wrote the manuscript.
Footnotes
Published ahead of print on 21 June 2010.
REFERENCES
- 1.Akompong, T., J. VanWye, N. Ghori, and K. Haldar. 1999. Artemisinin and its derivatives are transported by a vacuolar-network of Plasmodium falciparum and their anti-malarial activities are additive with toxic sphingolipid analogues that block the network. Mol. Biochem. Parasitol. 101:71-79. [DOI] [PubMed] [Google Scholar]
- 2.Asawamahasakda, W., I. Ittarat, Y. M. Pu, H. Ziffer, and S. R. Meshnick. 1994. Reaction of antimalarial endoperoxides with specific parasite proteins. Antimicrob. Agents Chemother. 38:1854-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertaux, L., H. Quang Le, V. Sinou, N. X. Thanh, and D. Parzy. 2009. New PfATP6 mutations found in Plasmodium falciparum isolates from Vietnam. Antimicrob. Agents Chemother. 53:4570-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhisutthibhan, J., and S. R. Meshnick. 2001. Immunoprecipitation of [(3)H]dihydroartemisinin translationally controlled tumor protein (TCTP) adducts from Plasmodium falciparum-infected erythrocytes by using anti-TCTP antibodies. Antimicrob. Agents Chemother. 45:2397-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray, P. G., S. A. Ward, and P. M. O'Neill. 2005. Quinolines and artemisinin: chemistry, biology and history, p. 444. In D. Sullivan and S. Krishna (ed.), Malaria: drugs, disease and post-genomic biology, 1st. ed., vol. 295. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 6a.Cardi, D. 2009. Ph.D. thesis. Université Paris XI, Paris, France.
- 7.Chavchich, M., L. Gerena, J. Peters, N. Chen, Q. Cheng, and D. E. Kyle. 2010. Role of pfmdr1 amplification and expression in induction of resistance to artemisinin derivatives in Plasmodium falciparum. Antimicrob. Agents Chemother. 54:2455-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coghi, P., N. Basilico, D. Taramelli, W. C. Chan, R. K. Haynes, and D. Monti. 2009. Interaction of artemisinins with oxyhemoglobin Hb-Fe(II), Hb-Fe(II), carboxyHb-Fe(II), heme-Fe(II), and carboxyheme Fe(II): significance for mode of action and implications for therapy of cerebral malaria. ChemMedChem. 4:2045-2053. [DOI] [PubMed] [Google Scholar]
- 9.Combe, A., D. Giovannini, T. G. Carvalho, S. Spath, B. Boisson, C. Loussert, S. Thiberge, C. Lacroix, P. Gueirard, and R. Menard. 2009. Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe 5:386-396. [DOI] [PubMed] [Google Scholar]
- 10.Crabb, B. S., and A. F. Cowman. 1996. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 93:7289-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlström, S., M. I. Veiga, P. Ferreira, A. Martensson, A. Kaneko, B. Andersson, A. Bjorkman, and J. P. Gil. 2008. Diversity of the sarco/endoplasmic reticulum Ca2+-ATPase orthologue of Plasmodium falciparum (PfATP6). Infect. Genet. Evol. 8:340-345. [DOI] [PubMed] [Google Scholar]
- 12.del Pilar Crespo, M., T. D. Avery, E. Hanssen, E. Fox, T. V. Robinson, P. Valente, D. K. Taylor, and L. Tilley. 2008. Artemisinin and a series of novel endoperoxide antimalarials exert early effects on digestive vacuole morphology. Antimicrob. Agents Chemother. 52:98-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dondorp, A. M., F. Nosten, P. Yi, D. Das, A. P. Phyo, J. Tarning, K. M. Lwin, F. Ariey, W. Hanpithakpong, S. J. Lee, P. Ringwald, K. Silamut, M. Imwong, K. Chotivanich, P. Lim, T. Herdman, S. S. An, S. Yeung, P. Singhasivanon, N. P. Day, N. Lindegardh, D. Socheat, and N. J. White. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eastman, R. T., and D. A. Fidock. 2009. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat. Rev. Microbiol. 7:864-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckstein-Ludwig, U., R. J. Webb, I. D. Van Goethem, J. M. East, A. G. Lee, M. Kimura, P. M. O'Neill, P. G. Bray, S. A. Ward, and S. Krishna. 2003. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424:957-961. [DOI] [PubMed] [Google Scholar]
- 16.Fidock, D. A., R. T. Eastman, S. A. Ward, and S. R. Meshnick. 2008. Recent highlights in antimalarial drug resistance and chemotherapy research. Trends Parasitol. 24:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidock, D. A., T. Nomura, and T. E. Wellems. 1998. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol. Pharmacol. 54:1140-1147. [DOI] [PubMed] [Google Scholar]
- 18.Gelb, M. H. 2007. Drug discovery for malaria: a very challenging and timely endeavor. Curr. Opin. Chem. Biol. 11:440-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golenser, J., J. H. Waknine, M. Krugliak, N. H. Hunt, and G. E. Grau. 2006. Current perspectives on the mechanism of action of artemisinins. Int. J. Parasitol. 36:1427-1441. [DOI] [PubMed] [Google Scholar]
- 20.Goodyer, I. D., and T. F. Taraschi. 1997. Plasmodium falciparum: a simple, rapid method for detecting parasite clones in microtiter plates. Exp. Parasitol. 86:158-160. [DOI] [PubMed] [Google Scholar]
- 21.Haynes, R. K., W. C. Chan, C. M. Lung, A. C. Uhlemann, U. Eckstein, D. Taramelli, S. Parapini, D. Monti, and S. Krishna. 2007. The Fe(2+)-mediated decomposition, PfATP6 binding, and antimalarial activities of artemisone and other artemisinins: the unlikelihood of C-centered radicals as bioactive intermediates. ChemMedChem. 2:1480-1497. [DOI] [PubMed] [Google Scholar]
- 22.Haynes, R. K., B. Fugmann, J. Stetter, K. H. Rieckmann, H.-D. Heilmann, H.-W. Chan, M.-K. Cheung, W.-L. Lam, H.-N. Wong, S. L. Croft, L. Vivas, L. Rattray, L. Stewart, W. Peters, B. L. Robinson, M. D. Edstein, B. M. Kotecka, D. E. Kyle, B. Beckermann, M. Gerisch, M. Radtke, G. Schmuck, W. Steinke, U. Wollborn, K. Schmeer, and A. Romer. 2006. Artemisone—a new, highly active antimalarial drug of the artemisinin class. Angew. Chem. Int. Ed. Engl. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Haynes, R. K., and S. Krishna. 2004. Artemisinins: activities and actions. Microbes Infect. 6:1339-1346. [DOI] [PubMed] [Google Scholar]
- 24.Hoppe, H. C., D. A. van Schalkwyk, U. I. Wiehart, S. A. Meredith, J. Egan, and B. W. Weber. 2004. Antimalarial quinolines and artemisinin inhibit endocytosis in Plasmodium falciparum. Antimicrob. Agents Chemother. 48:2370-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibrahim, M. L., N. Khim, H. H. Adam, F. Ariey, and J. B. Duchemin. 2009. Polymorphism of PfATPase in Niger: detection of three new point mutations. Malar; J. 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jambou, R., E. Legrand, M. Niang, N. Khim, P. Lim, B. Volney, M. T. Ekala, C. Bouchier, P. Esterre, T. Fandeur, and O. Mercereau-Puijalon. 2005. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet 366:1960-1963. [DOI] [PubMed] [Google Scholar]
- 27.Jung, M., H. Kim, K. Y. Nam, and K. T. No. 2005. Three-dimensional structure of Plasmodium falciparum Ca2+-ATPase(PfATP6) and docking of artemisinin derivatives to PfATP6. Bioorg. Med. Chem. Lett. 15:2994-2997. [DOI] [PubMed] [Google Scholar]
- 28.Kremsner, P. G., and S. Krishna. 2004. Antimalarial combinations. Lancet 364:285-294. [DOI] [PubMed] [Google Scholar]
- 29.Krishna, S., L. Bustamante, R. K. Haynes, and H. M. Staines. 2008. Artemisinins: their growing importance in medicine. Trends Pharmacol. Sci. 29:520-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503-1508. [DOI] [PubMed] [Google Scholar]
- 31.Li, W., W. Mo, D. Shen, L. Sun, J. Wang, S. Lu, J. M. Gitschier, and B. Zhou. 2005. Yeast model uncovers dual roles of mitochondria in the action of artemisinin. PLoS Genet. 1:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mamoun, C. B., I. Y. Gluzman, S. Goyard, S. M. Beverley, and D. E. Goldberg. 1999. A set of independent selectable markers for transfection of the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 96:8716-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menegon, M., A. R. Sannella, G. Majori, and C. Severini. 2008. Detection of novel point mutations in the Plasmodium falciparum ATPase6 candidate gene for resistance to artemisinins. Parasitol. Int. 57:233-235. [DOI] [PubMed] [Google Scholar]
- 34.Nagamune, K., W. L. Beatty, and L. D. Sibley. 2007. Artemisinin induces calcium-dependent protein secretion in the protozoan parasite Toxoplasma gondii. Eukaryot. Cell 6:2147-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naik, P. K., M. Srivastava, P. Bajaj, S. Jain, A. Dubey, P. Ranjan, R. Kumar, and H. Singh. 2010. The binding modes and binding affinities of artemisinin derivatives with Plasmodium falciparum Ca2+-ATPase (PfATP6). J. Mol. Model. [Epub ahead of print.] doi: 10.1007/s00894-010-0726-4. [DOI] [PubMed]
- 36.Noedl, H., S. Krudsood, W. Leowattana, N. Tangpukdee, W. Thanachartwet, S. Looareesuwan, R. S. Miller, M. Fukuda, K. Jongsakul, K. Yingyuen, S. Sriwichai, C. Ohrt, and C. Knirsch. 2007. In vitro antimalarial activity of azithromycin, artesunate, and quinine in combination and correlation with clinical outcome. Antimicrob. Agents Chemother. 51:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noedl, H., Y. Se, K. Schaecher, B. L. Smith, D. Socheat, and M. M. Fukuda. 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359:2619-2620. [DOI] [PubMed] [Google Scholar]
- 38.O'Neill, P. M., and G. H. Posner. 2004. A medicinal chemistry perspective on artemisinin and related endoperoxides. J. Med. Chem. 47:2945-2964. [DOI] [PubMed] [Google Scholar]
- 39.Omara-Opyene, A. L., P. A. Moura, C. R. Sulsona, J. A. Bonilla, C. A. Yowell, H. Fujioka, D. A. Fidock, and J. B. Dame. 2004. Genetic disruption of the Plasmodium falciparum digestive vacuole plasmepsins demonstrates their functional redundancy. J. Biol. Chem. 279:54088-54096. [DOI] [PubMed] [Google Scholar]
- 40.Pandey, A. V., B. L. Tekwani, R. L. Singh, and V. S. Chauhan. 1999. Artemisinin, an endoperoxide antimalarial, disrupts the hemoglobin catabolism and heme detoxification systems in malarial parasite. J. Biol. Chem. 274:19383-19388. [DOI] [PubMed] [Google Scholar]
- 41.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. PgH1 modulates sensitivity and resistance of multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 42.Riganti, C., S. Doublier, D. Viarisio, E. Miraglia, G. Pescarmona, D. Ghigo, and A. Bosia. 2009. Artemisinin induces doxorubicin resistance in human colon cancer cells via calcium-dependent activation of HIF-1alpha and P-glycoprotein overexpression. Br. J. Pharmacol. 156:1054-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robert, A., F. Benoit-Vical, C. Claparols, and B. Meunier. 2005. The antimalarial drug artemisinin alkylates heme in infected mice. Proc. Natl. Acad. Sci. U. S. A. 102:13676-13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sidhu, A. B., A. C. Uhlemann, S. G. Valderramos, J. C. Valderramos, S. Krishna, and D. A. Fidock. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidhu, A. B., S. G. Valderramos, and D. A. Fidock. 2005. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913-926. [DOI] [PubMed] [Google Scholar]
- 46.Sidhu, A. B., D. Verdier-Pinard, and D. A. Fidock. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stockwin, L. H., B. Han, S. X. Yu, M. G. Hollingshead, M. A. ElSohly, W. Gul, D. Slade, A. M. Galal, and D. L. Newton. 2009. Artemisinin dimer anticancer activity correlates with heme-catalyzed reactive oxygen species generation and endoplasmic reticulum stress induction. Int. J. Cancer 125:1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahar, R., P. Ringwald, and L. K. Basco. 2009. Molecular epidemiology of malaria in Cameroon. XXVIII. In vitro activity of dihydroartemisinin against clinical isolates of Plasmodium falciparum and sequence analysis of the P. falciparum ATPase 6 gene. Am. J. Trop. Med. Hyg. 81:13-18. [PubMed] [Google Scholar]
- 49.ter Kuile, F., N. J. White, P. Holloway, G. Pasvol, and S. Krishna. 1993. Plasmodium falciparum: in vitro studies of the pharmacodynamic properties of drugs used for the treatment of severe malaria. Exp. Parasitol. 76:85-95. [DOI] [PubMed] [Google Scholar]
- 50.Toovey, S., L. Y. Bustamante, A. C. Uhlemann, J. M. East, and S. Krishna. 2008. Effect of artemisinins and amino alcohol partner antimalarials on mammalian sarcoendoplasmic reticulum calcium adenosine triphosphatase activity. Basic Clin. Pharmacol. Toxicol. 103:209-213. [DOI] [PubMed] [Google Scholar]
- 51.Toyoshima, C., and T. Mizutani. 2004. Crystal structure of the calcium pump with a bound ATP analogue. Nature 29:529-535. [DOI] [PubMed] [Google Scholar]
- 52.Toyoshima, C., M. Nakasako, H. Nomura, and H. Ogawa. 2000. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature 405:647-655. [DOI] [PubMed] [Google Scholar]
- 53.Uhlemann, A. C., A. Cameron, U. Eckstein-Ludwig, J. Fischbarg, P. Iserovich, F. A. Zuniga, M. East, A. Lee, L. Brady, R. K. Haynes, and S. Krishna. 2005. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat. Struct. Mol. Biol. 12:628-629. [DOI] [PubMed] [Google Scholar]
- 54.Valderramos, S. G., J. C. Valderramos, L. Musset, L. A. Purcell, O. Mercereau-Puijalon, E. Legrand, and D. A. Fidock. 2010. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog. 6:e1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White, N. J. 2008. Qinghaosu (artemisinin): the price of success. Science 320:330-334. [DOI] [PubMed] [Google Scholar]
- 56.Witkowski, B., J. Lelievre, M. J. Barragan, V. Laurent, X. Z. Su, A. Berry, and F. Benoit-Vical. 2010. Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother. 54:1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodrow, C. J., R. K. Haynes, and S. Krishna. 2005. Artemisinins. Postgrad Med. J. 81:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, K. D., W. S. Lee, J. Wey, D. Bungard, and J. Lytton. 1995. Localization and quantification of endoplasmic reticulum Ca2+-ATPase isoform transcripts. Am. J. Physiol. 269:C775-784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.