Abstract
Vinylogous ureas 2-amino-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophene-3-carboxamide and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (compounds 1 and 2, respectively) were recently identified to be modestly potent inhibitors of the RNase H activity of HIV-1 and HIV-2 reverse transcriptase (RT). Both compounds shared a 3-CONH2-substituted thiophene ring but were otherwise structurally unrelated, which prevented a precise definition of the pharmacophore. We have therefore examined a larger series of vinylogous ureas carrying amide, amine, and cycloalkane modifications of the thiophene ring of compound 1. While cycloheptane- and cyclohexane-substituted derivatives retained potency, cyclopentane and cyclooctane substitutions eliminated activity. In the presence of a cycloheptane ring, modifying the 2-NH2 or 3-CONH2 functions decreased the potency. With respect to compound 2, vinylogous ureas whose dimethylthiophene ring contained modifications of the 2-NH2 and 3-CONH2 functions were investigated. 2-NH2-modified analogs displayed potency equivalent to or enhanced over that of compound 2, the most active of which, compound 16, reflected intramolecular cyclization of the 2-NH2 and 3-CONH2 groups. Molecular modeling was used to define an inhibitor binding site in the p51 thumb subdomain, suggesting that an interaction with the catalytically conserved His539 of the p66 RNase H domain could underlie inhibition of RNase H activity. Collectively, our data indicate that multiple functional groups of vinylogous ureas contribute to their potencies as RNase H inhibitors. Finally, single-molecule spectroscopy indicates that vinylogous ureas have the property of altering the reverse transcriptase orientation on a model RNA-DNA hybrid mimicking initiation plus-strand DNA synthesis.
Current success in treating HIV infection and AIDS can be attributed to effective combination antiretroviral therapy involving a cocktail of inhibitors directed primarily against the retroviral protease and reverse transcriptase (RT) (22). Inhibition of RT function is achieved either directly, by incorporating chain-terminating nucleoside derivatives (nucleoside RT inhibitors [NRTIs]), or indirectly, by nonnucleoside RT inhibitors (NNRTIs) that occupy a hydrophobic pocket at the base of the p66 thumb to interrupt the chemical step of DNA synthesis (25) and to influence the binding orientation and binding position of RT on substrates (1, 20). Although they are not presently in clinical use, the 1-[2′,5′-bis-O-(t-butyldimethylsilyl)-β-d-pentofuranosyl]-3′-spiro-5"-(4"-amino-1",2"-oxathiole-2",2"-dioxide)pyrimidine (TSAO) derivatives targeting the interface of the p66-p51 RT heterodimer (5, 29) and nucleoside-competing RT inhibitors (15) are also showing promise as anti-HIV drugs. However, the absolute dependence on RT-encoded RNase H activity for virus replication (32) suggests that inhibitors targeting this critical function could be added to the armament of antiviral drugs, especially if synergy with the NRTI-NNRTI combinations currently in clinical use could be achieved.
A limited number of candidate RNase H inhibitors have been reported, including N-hydroxyimides (28), N-acyl hydrazones (6), diketo acids (28), and pyrimidinol carboxylic acids (16). High-throughput screening of National Cancer Institute libraries of natural products also identified the hydroxytropolone β-thujaplicinol (7), phenolic glycosides (4), 1,3,4,5-tetragalloylaopiitol (31), and dimeric lactones (10) to be moderately potent RNase H inhibitors, with 50% inhibitory concentrations (IC50s) ranging from 0.25 to 1.5 μM for both the HIV-1 and HIV-2 enzymes. Screening of a library of 100,000 synthetic compounds identified the vinylogous ureas 2-amino-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophene-3-carboxamide and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (Fig. 1, compounds 1 and 2, respectively) to be members of a novel class of RNase H inhibitors (34) (Fig. 1). Rather than chelating divalent metal in the RNase H active site, which is the proposed mechanism for many RNase H inhibitors (12, 16), compound 1 was postulated to interact with the p51 thumb subdomain, possibly affecting the conformation of the immediately adjacent p66 RNase H domain (2, 9, 11, 14). Unlike β-thujaplicinol, which can be displaced from the RNase H active site by nucleic acid, the data presented in this communication illustrate that vinylogous ureas have the advantage that they are still effective in the presence of substrate (3, 35). However, the structural similarities between compounds 1 and 2 were insufficient to identify the pharmacophore, which has hampered the design of more potent and selective inhibitors.
FIG. 1.
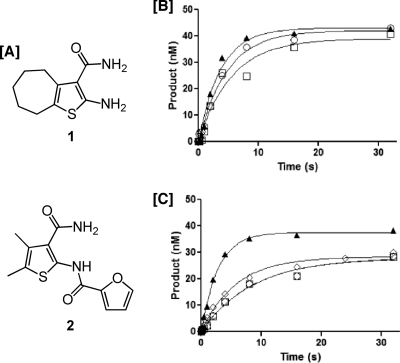
(A) Structures of the HIV-1 RNase H inhibitors 2-amino-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophene-3-carboxamide (compound 1) and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (compound 2). (B and C) Effects of order-of-addition on RNase H activity of compound 1. (B) No inhibitor. ○, preincubation of RT with Mg2+, hydrolysis initiated with RNA-DNA hybrid; □, RT alone, hydrolysis initiated with DNA-RNA hybrid and Mg2+; ▴, RT preincubated with DNA-RNA hybrid, hydrolysis initiated by adding Mg2+. (C) With inhibitor. ○, preincubation of RT with Mg2+, hydrolysis initiated by adding RNA-DNA hybrid; □, preincubation of RT with inhibitor, hydrolysis initiated by adding RNA-DNA hybrid and Mg2+; ▴, preincubation of RT with DNA-RNA hybrid, hydrolysis initiated by adding Mg2+ and inhibitor; ⋄, preincubation of RT with DNA-RNA hybrid and inhibitor, hydrolysis initiated by adding Mg2+.
We therefore extended our screening efforts to investigate an additional 22 vinylogous ureas carrying 2-NH2, 3-CONH2, and cycloalkane modifications of the thiophene ring common to the parent inhibitors. Although an optimal size for the cycloalkyl substituent could be demonstrated, the improved potencies of compounds 14 and 16 indicated that this functionality was less important than modifications of the 2-amino group. Compound 16 has a dihydrothieno[2,3-d]pyrimidine moiety derived from intramolecular cyclization of the 2-(4′-nitrobenzoyl)-amino and 3-carboxamide groups on thiophene. Eliminating the —NO2 group of compound 16 (compound 23) or replacing it with an electron-donating group, —OCH3 (compound 24), abolished the activity, suggesting that the NO2 group of compound 16 contributes to binding to the p51 thumb by either directly engaging in a hydrogen-bonding interaction or indirectly facilitating a π-π stacking interaction between the benzene ring and an aromatic side chain of the enzyme.
In conjunction with our recent mass spectrometric footprinting studies (34), data from this communication have been used to define a putative inhibitor binding site in the p51 thumb. This exercise indicated that docking the cycloheptane ring of compound 1 within a hydrophobic pocket permitted hydrogen bonding between its 2-NH2 group and the catalytically conserved His539 of the RNase H domain (32). Modeling studies suggested that compound 16 adopted a similar orientation, with its benzene ring occupying the hydrophobic pocket and with its 4-NO2 substituent being within hydrogen-bonding distance to Arg284 of the p51 thumb. Compound 16 thus represents a member of a new class of vinylogous ureas for further optimization as RNase H inhibitors. A comparative thermodynamic analysis of binding of compound 1 and the hydroxylated tropolone β-thujaplicinol (7) was performed in order to gain insight into their mechanisms of action. In contrast to β-thujaplicinol, compound 1 may induce a conformational change in HIV-1 RT by reordering a number of bonds. Finally, single-molecule spectroscopy studies suggest that compound 1 influences the RT orientation on a polypurine tract (PPT)-containing RNA-DNA hybrid, while compounds 14 and 16 possibly induce either a conformational change in RT or slippage on the duplex.
MATERIALS AND METHODS
Chemicals and enzymes.
All chemicals were purchased from Sigma (St. Louis, MO) and used without further purification, unless otherwise stated. Vinylogous ureas were purchased from ChemBridge Corporation (San Diego, CA). The bacterial expression vector pRSET was from Invitrogen (Carlsbad, CA). Purified oligonucleotides for the fluorescence-based RNase H and DNA polymerase assays were purchased from TriLink Biotechnologies (San Diego, CA). His-p66/His-p51 HIV-1 RT, derived from the HIV-1HXB2 isolate (23), was expressed and purified as reported previously (7). This sequence of this enzyme is identical to that of the residues discussed in modeling studies.
IC50 determination.
IC50 determination was carried out as reported previously (7), with minor modifications, using an 18-nucleotide 3′-fluorescein-labeled RNA sequence (5′-GAU CUG AGC CUG GGA GCU-fluorescein-3′) annealed to a complementary 18-nucleotide 5′-dabsyl-labeled DNA sequence (5′-dabsyl-AGC TCC CAG GCT CAG ATC-3′). Briefly, to a 96-well plate was added 1 μl of each compound (in dimethyl sulfoxide [DMSO]), followed by addition of 5 μl of RT (50 ng/μl) in reaction buffer. Hydrolysis was initiated by adding 10 μl of the RNA-DNA hybrid (2.5 μM). The final concentrations of the reagents in the assay were 50 mM Tris·HCl, pH 8.0, 60 mM KCl, 10 mM MgCl2, 1% DMSO, 250 ng RT, and 250 nM substrate with various concentrations of each inhibitor. Wells containing DMSO alone or no RT were used as negative controls and background, respectively. Plates were incubated at 37°C in a Spectramax Gemini EM fluorescence spectrometer for 10 min, and the fluorescence (λ excitation = 475 nm; λ emission = 520 nm) was measured every minute, such that linear initial rates could be measured in the presence (vi) and absence (vo) of inhibitor. Percent inhibition was calculated as 100 × [(vo − vi)/vo] and then plotted against log inhibitor concentration, and IC50s were determined using SigmaPlot software. All assays were performed in triplicate.
Antiviral activity.
The antiviral activities of 17 of the vinylogous ureas reported here were determined via the 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-5-[(phenylamino)carbonyl]-2H tetrazolium hydroxide (XTT)-based cell viability assay of Weislow et al. (33), using the HIV-1RF isolate and human T-cell line CEM-SS.
DNA-dependent DNA polymerase assay.
DNA-dependent DNA synthesis was measured on a fluorescently labeled duplex DNA generated by annealing a 42-nucleoptide (nt) template (5′-TAC ATA CCC ATA CAT AAA TCC TAA CCT TGA AGA ACT CGT CAC-3′) to the 5′ Cy5-labeled primer 5′-ATG TAT GGG TAT GTA TTT AGG-3′. Polymerization was initiated by adding 1 μl of 2 mM deoxynucleoside triphosphates (dNTPs) to 9 μl of a mixture containing 30 ng enzyme, 200 nM substrate, 5 or 50 μM inhibitor in 10 mM Tris-HCl (pH 7.8), 80 mM KCl, 1 mM dithiothreitol (DTT), 10 mM MgCl2, and 10% DMSO at 37°C and was terminated after 10 min by adding an equal volume of a formamide-based gel-loading buffer. The reaction products were analyzed by denaturing polyacrylamide gel electrophoresis and fluorescent imaging (Typhoon Trio+; GE Healthcare).
Thermodynamic evaluation of RNase H inhibitors.
Dose-response curves were generated for compound 1 and the metal-chelating hydroxytropolone RNase H inhibitor β-thujaplicinol (7) as a function of temperature. Experiments were conducted at 20, 25, 30, and 35°C in a total volume of 50 μl, with enzyme and RNA-DNA hybrid being present at 4 nM and 250 nM, respectively. Hydrolysis was initiated by adding substrate and, following 30 min of incubation at the indicated temperature, was quenched with 25 μl of 500 mM EDTA, pH 8.0. Product fluorescence was determined with a Safire fluorimeter (Tecan US, Durham NC), as described previously (7). Quadruplicate dose-response curves were determined for each assay temperature. In order to determine the equilibrium inhibition constant (Ki) for each compound, data were plotted and analyzed using the following equation for simple nontight binding inhibition: (vi/vo) = 1/[1 + ([I]/Ki)], where vi/vo is the initial velocity of the enzyme reaction, [I] is the inhibitor concentration, and Ki is the equilibrium dissociation constant for noncompetitive inhibitor binding to the RNase H domain. For Van't Hoff analysis, the values of Ki at each temperature were plotted as ln(Ki) versus 1/temperature (T; in Kelvin). The affinities of both compounds showed a linear relationship with temperature change, allowing determination of the enthalpy (ΔH) and entropy (ΔS) values from the simplified Van't Hoff equation: ln(Ki) = [(ΔH/R) − (1/T)] − ΔS/R, where ln(Ki) is the natural log of the equilibrium dissociation constant; ΔH and ΔS are the enthalpy and entropy, respectively, of inhibitors interacting with the RNase H domain; and R is the molar gas constant (8.314 kJ−1 mol−1). This equation allows estimates of ΔH and ΔS to be made from the slope and y-axis intercept of the line, respectively, by assuming that ΔH and ΔS are constants within the temperature range investigated.
Single-molecule FRET measurements.
A 21-nt PPT:D2 RNA-DNA primer (5′-uuuuaaaagaaaaggggggAC-3′, DNA nucleotides are in uppercase) was annealed to the biotinylated 50-nt template (5′-ATTAGATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCGTG GC-3′) at 1.2:1 ratio. The fluorescent resonance energy transfer (FRET) acceptor fluorophore Cy5 was attached near the 3′ end of the template, and the FRET donor fluorophore Cy3 was attached to the RNase H C terminus of the p66 RT subunit. The interactions between RT and the primer/template substrates were monitored by single-molecule FRET, as described previously (1, 20). Nevirapine or RNase H inhibitor 1, 14, or 16 was added at a final concentration of 10 μM.
Inhibitor docking.
Molecular docking was performed with AutoDockTools, version 4.2 (ADT 4.2), software (30). Receptor coordinates were obtained from Protein Data Bank (PDB) entry 1HMV (24), and inhibitor coordinates were generated de novo using the Build and Clean Geometry functions in Discovery Studio, version 2.0, software (Accelyrs, San Diego, CA). Flexible inhibitors were docked onto rigid, unliganded HIV-1 RT within a cube 50 by 50 by 50 Å centered near the junction between the p66 RNase H domain and the p51 thumb subdomain (i.e., on the α carbon of p51 residue Val276) using the AutoDock, version 4.2, Lamarckian genetic algorithm. Of the 250,000 complexes evaluated for each inhibitor, the 20 lowest-energy conformers were retained, clustered, and evaluated.
RESULTS
Nucleic acid fails to displace compound 1 from the RNase H active site.
We recently demonstrated that although the hydroxylated tropolone β-thujaplicinol, a metal-chelating RNase H inhibitor, was almost 10-fold more potent than compound 1, it could be displaced from its binding site by the RNA-DNA hybrid (3). In order to determine whether vinylogous ureas displayed this property, similar order-of-addition experiments were performed, the results of which are presented in Fig. 1B and C. In the absence of inhibitor, Fig. 1B indicates that the order in which the assay components are added does not affect RNase H activity. The data in Fig. 1C show that, in contrast to β-thujaplicinol, preincubation of enzyme with the RNA-DNA hybrid and inhibitor compound 1, followed by addition of divalent metal, compound 1 is still inhibitory. The exception to this was preincubation of enzyme with the RNA-DNA hybrid, after which hydrolysis was initiated by adding inhibitor and Mg2+, where compound 1 failed to inhibit. One possibility for this observation is that under the short time period allowed under pre-steady-state conditions, the vinylogous urea had insufficient time to bind.
Optimal size for cycloalkyl-substituted 2-amino-thiophene-3-carboxamides.
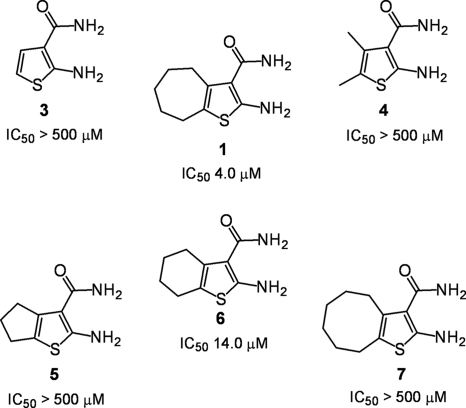
The flexible cycloheptane ring of compound 1 suggested that HIV-1 RNase H activity might be inhibited through docking of the cycloalkyl moiety into a hydrophobic pocket in the p51 thumb, thereby indirectly altering the coordination geometry of a divalent metal(s) at the p66 RNase H active site. This notion was in part supported by the data presented in Fig. 2, where 2-amino-thiophene-3-carboxamides that either lacked a cycloalkane ring (compound 3) or replaced it with a 4,5-dimethyl function (compound 4) were completely inactive. Compounds 5 and 7, which contain cyclopentane and cyclooctane substitutions, respectively, were also inactive, while an IC50 of 14 μM was determined for the cyclohexane-substituted compound, compound 6. The data presented in Fig. 2 thus suggest that when the 2-amino- and 3-carboxamide functions of compound 1 are not derivatized, there is an optimal size for cycloalkane substitution of the thiophene ring. In the current study, no attempts were made to examine whether more extensive modification of the cycloheptane ring of compound 1 enhanced or interfered with inhibitor potency.
FIG. 2.
Inhibition of HIV-1 RNase H activity by vinylogous ureas containing cycloalkyl-substituted 2-amino-thiophene-3-carboxamides. IC50 values are reported only for HIV-1 RT and are the averages of triplicate assays.
Substitutions on the 2-NH2 and/or 3-CONH2 groups of compound 1 decrease the efficacy of RNase H inhibition.
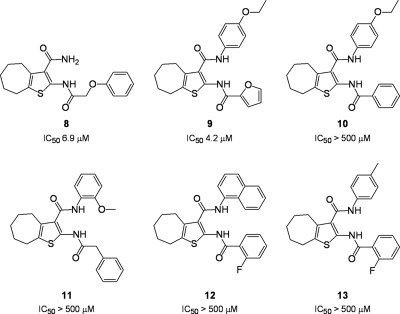
Our previous demonstration that compound 2 inhibited RNase H activity even though it lacked a cycloalkane-substituted thiophene ring (34) suggested that the 2-NH2 and 3-CONH2 groups also contributed to RNase H inhibition. We therefore evaluated several analogs of compound 1 whose thiophene ring contained substitutions on the 2-NH2 and 3-CONH2 groups, and the results are presented in Fig. 3. An IC50 of 6.9 μM was determined for compound 8, which contains a 2-phenoxyacetylamino substitution, suggesting that introducing the aromatic function was reasonably well tolerated. A similar IC50 (4.2 μM) was determined for compound 9, which contains furanoyl and 4′-ethoxyphenyl substitutions of the 2-NH2 and 3-CONH2 groups, respectively. Surprisingly, replacing the furan ring with benzene completely abolished the activity (compound 10). Such a loss of activity does not appear to be attributed solely to introducing the relatively rigid benzene ring, since compound 8 carries a phenoxyacetyl substitution of the 2-NH2 moiety yet retained activity. Moreover, the data in the following section indicate that nitrobenzene and methylbenzene modifications of the 2-NH2 group were well tolerated, provided the thiophene ring lacks the cycloheptane ring. Thus, when the cycloheptane ring of compound 1 is retained and the 3-carboximide group is functionalized, introducing a rigid benzene ring onto the 2-NH2 group prevents inhibitor binding. The potency of compound 9, which differs minimally from that of compound 10, may also reflect involvement of the furanyl oxygen in hydrogen bonding. In support of our rationale for the lack of potency of compound 10, vinylogous ureas whose 2-NH2 group carried a phenylacetyl substitution (compound 11) or a fluorobenzoyl substitution (compounds 12 and 13) were also inactive.
FIG. 3.
Inhibition of HIV-1 RNase H activity by 2-amino- and 3-carboxamide-substituted cyclohepta[b]thiophenes. IC50s are reported only for HIV-1 RT and are the averages of triplicate assays.
2-Acylamino 4,5-dimethylthiophene-3-carboxamides.
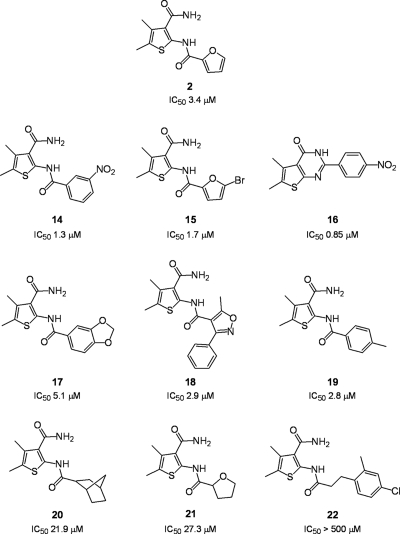
Since compound 2 lacked a cycloalkane ring yet inhibited RNase H activity (IC50 = 3.4 μM) (Fig. 1), we next evaluated 4,5-dimethylthiophene-3-carboxamides containing a variety of 2-NH2 modifications. As shown in Fig. 4, these derivatives could be classified as having a relatively rigid aryl substitution (compounds 14, 15, 16, 17, 18, and 19) or a more flexible bulky alkyl substitution (compounds 20 and 21). The exception to this was compound 22, which contained a 4-chloro-2-methylphenoxyacetyl substitution. The data in Fig. 4 indicate that aryl substitutions in most cases improved the potency. The most promising of these was compound 16, whose IC50 (0.85 μM) represented a 4-fold improvement over that of compound 2 (3.4 μM). The structures of compounds 14 and 16 suggest that the potency of the latter is related to intramolecular cyclization of the 2-NH2 and 3-CONH2 groups to generate a pyrimidine ring with even greater rigidity. In contrast, the more flexible acyl substitutions of the 2-NH2 group were less acceptable, decreasing the IC50s to 21.0 μM (compound 20) and 27.3 μM (compound 21). Finally, a 4-chloro-2-methylphenoxyacetyl substitution rendered compound 22 inactive.
FIG. 4.
Inhibition of HIV-1 RNase H activity by 2-amino-substituted 4,5-dimethylthiophene-3-carboxamides. IC50s are reported only for HIV-1 RT and are the averages of triplicate assays.
To better understand what might contribute to the improved activity of compound 16, derivatives whose benzene ring lacked the 4′-NO2 group (compound 23) or replaced it with —OCH3 (compound 24) were examined. Figure 5 A indicates that both modifications severely reduced the inhibitory potency, suggesting an important contribution from hydrogen bonding between the —NO2 group of compound 16 and critical catalytic residues of the RNase H domain. At the same time, the DNA polymerase activity of HIV-1 RT was unaffected in the presence of 50 μM compound 16 (Fig. 5B), indicating specificity for the RNase H domain.
FIG. 5.
(A) Eliminating the 4-NO2 of compound 16 (compound 23) or replacing it with an —OCH3 function (compound 24) eliminates its inhibitory potency. (B) Compound 16 specifically inhibits the RNase H activity of HIV-1 RT. DNA-dependent DNA polymerase activity was determined at inhibitor concentrations of 5 μM (lane 3) and 50 μM (lane 4). Lane 1, no enzyme; lane 2, no inhibitor. The migration positions of the unextended and fully extended primer are indicated.
Comparative thermodynamic evaluation of RNase H inhibitors.
Attempts to derive thermodynamic parameters for vinylogous urea binding via isothermal titration calorimetry were unsuccessful (data not shown). As an alternative, the effect of temperature on the binding affinity of compound 1 was determined by Van't Hoff analysis and was compared with the effect on the binding affinity of the metal-chelating RNase H inhibitor β-thujaplicinol (7). Both inhibitors displayed decreased binding affinities as the assay temperature was increased, which was linear when they were displayed in the final Van't Hoff plots in Fig. 6 A. A simplified Van't Hoff equation was fit to the data, yielding estimates of the free energy (ΔG), ΔH, and ΔS of inhibitor binding (Fig. 6B). A steeper, more pronounced decrease in binding affinity was obtained with compound 1, suggesting a large favorable enthalpy of binding, despite having the lower affinity of the two compounds. In contrast, β-thujaplicinol showed a very shallow, almost horizontal, decreasing slope, indicative of a relatively small favorable binding enthalpy. The data from Fig. 6B were used to construct thermodynamic binding profiles for the two classes of RNase H inhibitors (Fig. 6C), from which differences in the binding thermodynamics were clearly evident. A large unfavorable (positive) entropic contribution was observed for compound 1, balancing out the large favorable (negative) value for enthalpy. This pattern might indicate that binding of compound 1 induces a conformational change in HIV-1 RT, reordering bonds within the protein and shedding a number of shell water molecules into the bulk solvent. In contrast, β-thujaplicinol produced a small favorable value for ΔH, as expected from the almost flat line in the Van't Hoff plot, with the remaining binding energy being derived from a large favorable contribution from ΔS.
FIG. 6.
(A) Effect of temperature change upon affinity of compound 1 (•) and β-thujaplicinol (○) for p66/p51 HIV-1 RT. (B) ΔG, ΔH, and ΔS estimates for inhibitor binding and thermodynamic profiles for inhibition of HIV-1 RNase H by compound 1 and β-thujaplicinol. ΔG, ΔH, and ΔS are represented by open, gray, and black rectangles, respectively.
Vinylogous ureas influence the HIV-1 RT orientation on the polypurine tract.
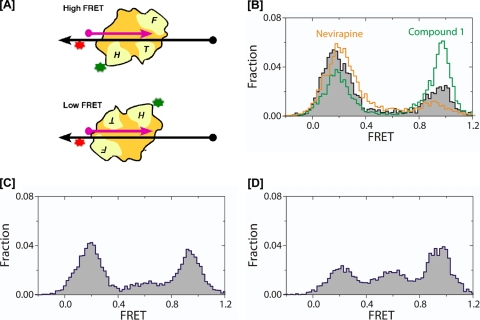
We recently exploited single-molecule spectroscopy to demonstrate that the NNRTI nevirapine induced a conformational rearrangement of HIV-1 RT on a model PPT substrate that promoted positioning of the RNase H domain at the PPT-U3 junction (1). Since our original analysis of vinylogous ureas implied inhibition via an interaction with the p51 thumb subdomain rather than a direct interaction with RNase H active-site residues (34), we next examined whether vinylogous urea-induced inhibition reflected an altered enzyme orientation and/or altered conformational dynamics. The experimental setup is depicted in Fig. 7 A, and the results of this analysis are presented in Fig. 7B to D.
FIG. 7.
DNA polymerase and RNase H inhibitors can influence HIV-1 RT orientation on the PPT-containing RNA-DNA hybrid. (A) Cartoon depicting the high and low FRET states assumed by RT on the nucleic acid duplex. RNA and DNA strands are depicted in pink and black, respectively. 5′ termini are indicated by filled circles, and 3′ termini are indicated by arrows. RNase H-deficient HIV-1 RT was derivatized with Cy3 (green) on the p66 RNase H C terminus and bound to a surface-immobilized hybrid labeled with Cy5 near the DNA template 3′ terminus (red). The fingers, thumb, and RNase H domains of RT are indicated F, T, and H, respectively. (Upper cartoon) Enzyme adopting a polymerizing orientation with the fingers domain of the polymerase subunit over the PPT 3′ terminus brings Cy3 and Cy5 into proximity, yielding a high FRET signal. (Lower cartoon) separation of the fluorophores when the RNase H domain is in the vicinity of the primer 3′ terminus produces a low FRET signal (1, 20). (B) FRET histograms depicting enzyme orientation in the absence of inhibitor (black trace), in the presence of nevirapine (gold trace), or in the presence of RNase H inhibitor compound 1 (green trace). (C and D) FRET histograms depicting RT orientation in the presence of compounds 14 and 16, respectively.
In keeping with previous observations (1), RT alone bound the model PPT:D2 substrate predominantly with its C-terminal RNase H domain over the PPT-U3 junction, giving a major low FRET peak at a FRET value of 0.17 (Fig. 7B, black trace). In contrast to observations with nevirapine, which enhanced this binding orientation (Fig. 7B, gold trace), the substantial shift of the distribution to the high FRET peak near a FRET value of 0.95 observed in the presence of compound 1 suggests augmented enzyme binding in a polymerization-competent mode, which places the polymerase active site over the PPT 3′ terminus (Fig. 7B, green trace). Such a small-molecule-induced reorientation of the enzyme is not unprecedented, as we showed previously that mimicking ternary complex formation with a chain-terminated PPT primer and the incoming dNTP favored binding in the polymerization-competent mode (1).
A more surprising result was obtained with compounds 14 and 16. Figure 7C and D indicates that both inhibitors give rise to an additional, intermediate FRET peak of about 0.6. This effect was more pronounced with compound 16, which Fig. 4 shows is the more potent of the two derivatives. Although further analysis is necessary, the intermediate FRET state may be caused by (i) an inhibitor-mediated conformational change in RT or (ii) an inhibitor-induced slippage of the enzyme over the single-stranded DNA template. This notion will be discussed in more detail later, but it could indicate that vinylogous urea-induced inhibition of PPT cleavage may in part reflect displacement of RT from the PPT-U3 junction.
Molecular modeling studies.
Several vinylogous ureas of this study were subjected to molecular docking onto unliganded HIV-1 RT (24) using AutoDockTools 4.2 (30). Although these compounds bind liganded RT as well, nucleic acid does not fully extend through the RNase H domain in published RT cocrystal structures, making them inappropriate for docking studies targeting regions encompassing the extreme C terminus of p66. Initially, a docking cube 50 by 50 by 50 Å centered on the α carbon of Val276 of the p51 thumb was instantiated using AutoGrid (a component of AutoDockTools), such that most of the p51 thumb subdomain, the p66 RNase H domain, and their interface were encompassed. A total of 250,000 potential binding sites for compound 1 within this cube were sampled using the AutoDock Lamarckian genetic algorithm, and the 20 lowest-energy conformers were identified and evaluated.
Nineteen of the 20 lowest-energy compound 1 docking instances placed the inhibitor at the p66 RNase H/p51 thumb interface, as depicted in Fig. 8 A. In each instance, the compound 1 conformer is oriented such that the tip of the cycloheptane ring occupied a small binding pocket formed by Gly541 of the RNase H domain at the base of the pocket and p51 residues Cys280 and Arg284, with the Arg284 side chain protruding into the solvent (Fig. 8C and D). This binding mode would be consistent with protection of Cys280 from biotinylation observed earlier by mass spectrometric protein footprinting (34). In the most energetically favorable docking instance, the substituted thiophene ring of compound 1 forms three hydrogen bonds (Fig. 8D). The 3-CONH2 group forms hydrogen bonds with the carbonyl oxygen of Val276 (p51) and side chain amine of Lys275 (p51). More intriguingly, the 2-NH2 group forms a hydrogen bond with the backbone carbonyl oxygen of p66 His539, a conserved, highly flexible amino acid important for RNase H activity (27). While the precise mechanism remains unclear and hydrogen bonding with His539 does not occur in all docking instances, altering the His539 geometry may constitute an important feature of compound 1 activity.
FIG. 8.
Lowest-energy docking conformers of select vinylogous ureas on the surface of HIV-1 RT. AutoDock 4.2 positions compound 1 (A and B) and compound 16 (C and D) in the conformations shown within a small binding pocket located at the junction between the p51 thumb subdomain (green) and the RNase H domain (magenta). A surface representation of this pocket is depicted in panels A and C, while panels B and D highlight residues in close proximity to or forming hydrogen bonds with the docked conformers of compounds 1 and 16, respectively.
Compounds 5, 6, and 7, which differ from compound 1 only in the sizes of their cycloalkane ring (possessing 5-, 6-, and 8-membered rings, respectively), were similarly docked onto RT. Compound 6 (IC50 = 14 μM) displayed binding properties with respect to the site of docking, orientation, and binding energy equivalent to those of compound 1. Conversely, the binding energies of the 20 most energetically favorable docking instances of compound 5, a poor RNase H inhibitor (IC50 > 500 μM), were higher than those of compound 1, none of which positioned the inhibitor at the proposed binding site. This may indicate that the cyclopentane ring of compound 5 has neither the size nor the flexibility required for favorable docking/binding. Finally, the structural dissimilarity between compounds 1 and 16 suggested that the latter, while interacting with the same site on the p51 thumb, might adopt an alternate orientation. In support of this possibility, molecular modeling suggested that the most energetically favorable conformation positioned the 4-nitrophenyl moiety of compound 16 within the binding pocket and within hydrogen-bonding distance of Arg284 (Fig. 8C and D).
Antiviral activity of vinylogous ureas.
Finally, 17 of the compounds reported on here were examined in vivo for their ability to inhibit HIV replication (33). The most promising candidate was the parent molecule, compound 1, which displayed an IC50 of 4.3 μM and a 50% cytotoxic concentration (CC50) of 19.9 μM. In addition, compounds 17 (IC50 = 19.0 μM; CC50 = 38.0 μM) and 11 (IC50 = 54.0 μM; CC50 = 141 μM) were moderately active. All remaining compounds displayed only marginal antiviral activity.
DISCUSSION
N-Hydroxyimides (17), hydroxylated tropolones (7), pyrimidinol carboxylic acids (16), and diketo acids (28) are presently among the more promising inhibitors of HIV-1 RNase H activity. Despite differing in their complexities, these lead compounds share a metal-chelating scaffold that presumably targets the two catalytically critical divalent metals within the RNase H active site. In support of this notion, crystallographic analysis of the isolated RNase H domain in the presence of Mg2+ or Mn2+ indicates metal chelation by an N-hydroxyimide, pyrimidinol carboxylic acid, and β-thujaplicinol (16, 18). However, recent characterization of the hydroxytropolone β-thujaplicinol (3) indicated that it has an inability to inhibit RNase H activity in a preformed enzyme-RNA-DNA complex, suggesting that the duplex nucleic acid substrate prevented inhibitor from accessing the catalytic center, and vice versa. Such observations suggest that additional strategies other than directly targeting the RNase H active site should be considered, analogous to the case for the NRTI-NNRTI combinations that effectively inhibit DNA polymerase activity. In this respect, vinylogous ureas could represent a novel class of off-site allosteric inhibitors that bind to the p51 thumb, a key structural platform for the p66 RNase H domain (26, 34). However, the structures of lead compounds 1 and 2 provided insufficient information to identify the pharmacophore for further optimization. Consequently, we examined 22 derivatives with modifications of the cycloalkane, 2-NH2, and 3-CONH2 components of the thiophene ring common to compounds 1 and 2. These studies have shown that vinylogous ureas retaining a cycloheptane substitution exhibited little to no improvement in activity over that of compound 1, while modifying the dimethylthiophene core increased the potency 4- to 5-fold.
In the absence of a high-resolution crystal structure, molecular modeling was used to identify a binding site common to compounds 1 and 16 and at the same time compatible with earlier protein footprinting studies that implicated Cys280 and Lys281 in inhibitor binding (34). The models in Fig. 8B and D suggest that the cycloheptane ring of compound 1 occupies a hydrophobic pocket close to these two residues of the p51 thumb, while the 3-CONH2 group is within hydrogen-bonding distance of Lys275 and Arg277. This orientation also favors hydrogen bonding between the 2-NH2 group of compound 1 and the peptide backbone of His539, an important residue of the RNase H active site (27). While mutating His539 inhibits RNase H activity (27), subunit-selective mutagenesis (19) will allow the contributions of the p51 residues Lys275, Arg277, Cys280, and Lys281 to be investigated.
The same region of p51 appears to be the most energetically favorable compound 16 binding site, with the planar benzene ring occupying the hydrophobic pocket and its 4-NO2 substituent being within hydrogen-bonding distance of Arg284. Compound 16 is derived from intramolecular cyclization of the 2-(4′-nitrobenzoyl)-amino and 3-carboxamide groups and thus has a nitrobenzoyl moiety in common with compound 14. The similar IC50s for these two inhibitors (compound 16 IC50 = 0.85 μM; compound 14 IC50 = 1.3 μM) supports the importance of this nitrobenzoyl moiety. Furthermore, removing (compound 23) or replacing (compound 24) the 4′-NO2 group abolished the activity. One approach to testing the importance of the —NO2 group in compound 16 would be to examine a derivative of the compound in which a —CO2H is substituted for —NO2. This would yield a more negative charge density at that position while retaining the structural integrity of the functional group.
Thermodynamic profiling studies were carried out in an attempt to elucidate potential differences between the binding configurations of different compound classes. The lack of a measurable signal prevented the use of isothermal titration calorimetry (ITC), the method of choice for measuring the enthalpic signal (ΔH) associated with compound binding. While doubts regarding the accuracy of ΔH estimates derived from Van't Hoff plots have been raised (21), more recent work suggests that the careful use of Van't Hoff analysis may in fact be acceptable as a method for the measurement of the thermodynamic forces involved in the association of proteins and small molecules (13). It was believed that the dose-response data utilized in our analysis were of sufficient quality (with small associated errors) to justify the use of Van't Hoff analysis.
Thermodynamic profiling of β-thujaplicinol and compound 1 highlighted important differences between these two inhibitor classes. In particular, the data suggest that while perturbation of enzyme structure upon β-thujaplicinol binding is minimal, binding of compound 1 induces a conformational change within the RNase H domain. Keeping in mind the putative binding site for the vinylogous urea inhibitors, candidates for local structural rearrangements include (i) displacement of amino acid residues within or adjacent to the loop containing conserved residue His539 and/or (ii) destabilizing the interface between the p66 RNase H domain and p51 thumb. We do not believe that a more substantial rearrangement in subdomain/subunit geometry is likely, since both circular dichroism and 19F-nuclear magnetic resonance analysis of HIV-1 RT containing site-specific trifluoromethoxy-phenylalanine insertions (8) in the vicinity of the RNase H active site failed to detect conformational changes in the presence of compound 1 or 16 (S. Chung, S. Le Grice, and A. Gronenborn, unpublished data).
An unexpected observation from our study was the effect of vinylogous ureas on RT conformational dynamics, where data from Fig. 7 indicate that the orientation on the PPT RNA-DNA hybrid can be significantly influenced by inhibitor binding. These data indicate that compound 1 affects the distribution of the enzyme in the polymerization or RNase H binding orientations, while binding events that occur in the presence of compound 16 produce intermediate FRET values. While it is intriguing, the structural and/or mechanistic basis for the latter observation is not clear. It may be that compound 16 binding prevents a stable association between the RNase H domain and nucleic acid at either primer terminus and that an intermediate binding position in which neither terminus is occupied by RNase H becomes preferred. Alternatively, an inhibitor-induced enzyme conformational change that repositions the enzyme-bound FRET acceptor relative to the donor might produce the intermediate FRET state in the absence of complete enzyme repositioning. Further study is required to distinguish among these and other potential explanations for intermediate FRET.
Finally, the crystal structure of a pyrimidinol carboxylic acid with the isolated HIV-1 RNase H domain indicates an interaction between the C-2 aromatic substituent and the imidazole ring of His539 (16), in addition to chelation of divalent metals at the active site. In the current study, we propose that vinylogous ureas embedded within a hydrophobic pocket in the p51 thumb extend toward His539, making contact in the case of compound 1. The notion that His539 can be targeted by small molecules accessing it from both the RNase H active site and a neighboring hydrophobic pocket in the p51 thumb suggests that the use of bidentate inhibitors spanning both sites might be considered. This should improve the specificity relative to that of the individual molecules.
Acknowledgments
S.F.J.L.G. is supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health; M.G. is supported by a grant from the Canadian Foundation for AIDS Research; and X.Z. is supported by NIH grant GM 068518. X.Z. is a Howard Hughes Medical Institute investigator. This project was supported in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400.
The contents of this publication do not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.Abbondanzieri, E. A., G. Bokinsky, J. W. Rausch, J. X. Zhang, S. F. Le Grice, and X. Zhuang. 2008. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature 453:184-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard, W. A., S. J. Stahl, H. R. Kim, K. Bebenek, A. Kumar, M. P. Strub, S. P. Becerra, T. A. Kunkel, and S. H. Wilson. 1994. Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase. Alanine scanning mutagenesis of an alpha-helix in the thumb subdomain. J. Biol. Chem. 269:28091-28097. [PubMed] [Google Scholar]
- 3.Beilhartz, G. L., M. Wendeler, N. Baichoo, J. Rausch, S. Le Grice, and M. Gotte. 2009. HIV-1 reverse transcriptase can simultaneously engage its DNA/RNA substrate at both DNA polymerase and RNase H active sites: implications for RNase H inhibition. J. Mol. Biol. 388:462-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bokesch, H. R., A. Wamiru, S. F. Le Grice, J. A. Beutler, T. C. McKee, and J. B. McMahon. 2008. HIV-1 ribonuclease H inhibitory phenolic glycosides from Eugenia hyemalis. J. Nat. Prod. 71:1634-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonache, M. C., E. Quesada, C. W. Sheen, J. Balzarini, N. Sluis-Cremer, M. J. Perez-Perez, M. J. Camarasa, and A. San-Felix. 2008. Novel N-3 substituted TSAO-T derivatives: synthesis and anti-HIV-evaluation. Nucleosides Nucleotides Nucleic Acids 27:351-367. [DOI] [PubMed] [Google Scholar]
- 6.Borkow, G., R. S. Fletcher, J. Barnard, D. Arion, D. Motakis, G. I. Dmitrienko, and M. A. Parniak. 1997. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry 36:3179-3185. [DOI] [PubMed] [Google Scholar]
- 7.Budihas, S. R., I. Gorshkova, S. Gaidamakov, A. Wamiru, M. K. Bona, M. A. Parniak, R. J. Crouch, J. B. McMahon, J. A. Beutler, and S. F. Le Grice. 2005. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 33:1249-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cellitti, S. E., D. H. Jones, L. Lagpacan, X. Hao, Q. Zhang, H. Hu, S. M. Brittain, A. Brinker, J. Caldwell, B. Bursulaya, G. Spraggon, A. Brock, Y. Ryu, T. Uno, P. G. Schultz, and B. H. Geierstanger. 2008. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 130:9268-9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba, J., A. Yamaguchi, Y. Suzuki, M. Nakano, W. Zhu, H. Ohba, A. Saito, H. Shinagawa, Y. Yamakawa, T. Kobayashi, and T. Kurata. 1996. A novel neutralization epitope on the ‘thumb’ subdomain of human immunodeficiency virus type 1 reverse transcriptase revealed by a monoclonal antibody. J. Gen. Virol. 77(Pt 12):2921-2929. [DOI] [PubMed] [Google Scholar]
- 10.Dat, N. T., K. Bae, A. Wamiru, J. B. McMahon, S. F. Le Grice, M. Bona, J. A. Beutler, and Y. H. Kim. 2007. A dimeric lactone from Ardisia japonica with inhibitory activity for HIV-1 and HIV-2 ribonuclease H. J. Nat. Prod. 70:839-841. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, L. L., M. J. McWilliams, K. Das, E. Arnold, and S. H. Hughes. 2009. Mutations in the thumb allow human immunodeficiency virus type 1 reverse transcriptase to be cleaved by protease in virions. J. Virol. 83:12336-12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himmel, D. M., S. G. Sarafianos, S. Dharmasena, M. M. Hossain, K. McCoy-Simandle, T. Ilina, A. D. Clark, Jr., J. L. Knight, J. G. Julias, P. K. Clark, K. Krogh-Jespersen, R. M. Levy, S. H. Hughes, M. A. Parniak, and E. Arnold. 2006. HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem. Biol. 1:702-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horn, J. R., D. Russell, E. A. Lewis, and K. P. Murphy. 2001. Van't Hoff and calorimetric enthalpies from isothermal titration calorimetry: are there significant discrepancies? Biochemistry 40:1774-1778. [DOI] [PubMed] [Google Scholar]
- 14.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 15.Jochmans, D., J. Deval, B. Kesteleyn, H. Van Marck, E. Bettens, I. De Baere, P. Dehertogh, T. Ivens, M. Van Ginderen, B. Van Schoubroeck, M. Ehteshami, P. Wigerinck, M. Gotte, and K. Hertogs. 2006. Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J. Virol. 80:12283-12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirschberg, T. A., M. Balakrishnan, N. H. Squires, T. Barnes, K. M. Brendza, X. Chen, E. J. Eisenberg, W. Jin, N. Kutty, S. Leavitt, A. Liclican, Q. Liu, X. Liu, J. Mak, J. K. Perry, M. Wang, W. J. Watkins, and E. B. Lansdon. 2009. RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information. J. Med. Chem. 52:5781-5784. [DOI] [PubMed] [Google Scholar]
- 17.Klumpp, K., J. Q. Hang, S. Rajendran, Y. Yang, A. Derosier, P. Wong Kai In, H. Overton, K. E. Parkes, N. Cammack, and J. A. Martin. 2003. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 31:6852-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumpp, K., and T. Mirzadegan. 2006. Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr. Pharm. Des. 12:1909-1922. [DOI] [PubMed] [Google Scholar]
- 19.Le Grice, S. F., T. Naas, B. Wohlgensinger, and O. Schatz. 1991. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 10:3905-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S., E. A. Abbondanzieri, J. W. Rausch, S. F. Le Grice, and X. Zhuang. 2008. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science 322:1092-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naghibi, H., A. Tamura, and J. M. Sturtevant. 1995. Significant discrepancies between van't Hoff and calorimetric enthalpies. Proc. Natl. Acad. Sci. U. S. A. 92:5597-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell, K., J. L. Davis, A. M. Morris, A. Chi, M. R. Bensley, and L. Huang. 2009. Survival for patients with HIV admitted to the ICU continues to improve in the current era of combination antiretroviral therapy. Chest 135:11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R. S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retroviruses 3:57-69. [DOI] [PubMed] [Google Scholar]
- 24.Rodgers, D. W., S. J. Gamblin, B. A. Harris, S. Ray, J. S. Culp, B. Hellmig, D. J. Woolf, C. Debouck, and S. C. Harrison. 1995. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 92:1222-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruane, P. J., and E. DeJesus. 2004. New nucleoside/nucleotide backbone options: a review of recent studies. J. Acquir. Immune Defic. Syndr. 37(Suppl. 1):S21-S29. [DOI] [PubMed] [Google Scholar]
- 26.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schatz, O., F. V. Cromme, F. Gruninger-Leitch, and S. F. Le Grice. 1989. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 257:311-314. [DOI] [PubMed] [Google Scholar]
- 28.Shaw-Reid, C. A., V. Munshi, P. Graham, A. Wolfe, M. Witmer, R. Danzeisen, D. B. Olsen, S. S. Carroll, M. Embrey, J. S. Wai, M. D. Miller, J. L. Cole, and D. J. Hazuda. 2003. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 278:2777-2780. [DOI] [PubMed] [Google Scholar]
- 29.Sluis-Cremer, N., N. Hamamouch, A. San Felix, S. Velazquez, J. Balzarini, and M. J. Camarasa. 2006. Structure-activity relationships of [2′,5′-bis-O-(tert-butyldimethylsilyl)-beta-d-ribofuranosyl]-3′-spiro-5″-(4″-amino-1″,2″-oxathiole-2″,2″-dioxide)thymine derivatives as inhibitors of HIV-1 reverse transcriptase dimerization. J. Med. Chem. 49:4834-4841. [DOI] [PubMed] [Google Scholar]
- 30.Spanner, M. F. 1999. Python: a programming language for software integration and development. J. Mol. Graphics Mod. 17:57-61. [PubMed] [Google Scholar]
- 31.Takada, K., A. Bermingham, B. R. O'Keefe, A. Wamiru, J. A. Beutler, S. F. Le Grice, J. Lloyd, K. R. Gustafson, and J. B. McMahon. 2007. An HIV RNase H inhibitory 1,3,4,5-tetragalloylapiitol from the African plant Hylodendron gabunensis. J. Nat. Prod. 70:1647-1649. [DOI] [PubMed] [Google Scholar]
- 32.Tisdale, M., T. Schulze, B. A. Larder, and K. Moelling. 1991. Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J. Gen. Virol. 72(Pt 1):59-66. [DOI] [PubMed] [Google Scholar]
- 33.Weislow, O. S., R. Kiser, D. L. Fine, J. Bader, R. H. Shoemaker, and M. R. Boyd. 1989. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J. Natl. Cancer Inst. 81:577-586. [DOI] [PubMed] [Google Scholar]
- 34.Wendeler, M., H. F. Lee, A. Bermingham, J. T. Miller, O. Chertov, M. K. Bona, N. S. Baichoo, M. Ehteshami, J. Beutler, B. R. O'Keefe, M. Gotte, M. Kvaratskhelia, and S. Le Grice. 2008. Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem. Biol. 3:635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendeler, M., G. L. Beilhartz, J. A. Beutler, M. Goette, and S. F. J. Le Grice. 2009. HIV ribonuclease H: continuing the search for small molecule antagonists. HIV Ther. 3:39-53. [Google Scholar]