Abstract
Despite the recognized potential of long-term survival or even growth of fecal indicators bacteria (FIB) in marine sediments, this compartment is largely ignored by health protection authorities. We conducted a large-scale study over approximately 50 km of the Marche coasts (Adriatic Sea) at depths ranging from 2 to 5 m. Total and fecal coliforms (FC) were counted by culture-based methods. Escherichia coli was also quantified using fluorescence in situ hybridization targeting specific 16S rRNA sequences, which yielded significantly higher abundances than culture-based methods, suggesting the potential importance of viable but nonculturable E. coli cells. Fecal coliforms displayed high abundances at most sites and showed a prevalence of E. coli. FC isolates (n = 113) were identified by API 20E, additional biochemical tests, and internal transcribed spacer-PCR. E. coli strains, representing 96% of isolates, were then characterized for genomic relatedness and phylogenetic group (A, B1, B2, and D) of origin by randomly amplified polymorphic DNA and multiplex-PCR. The results indicated that E. coli displayed a wide genotypic diversity, also among isolates from the same station, and that 44 of the 109 E. coli isolates belonged to groups B2 and D. Further characterization of B2 and D isolates for the presence of 11 virulence factor genes (pap, sfa/foc, afa, eaeA, ibeA, traT, hlyA, stx1, stx2, aer, and fyuA) showed that 90% of B2 and 65% of D isolates were positive for at least one of these. Most of the variance of both E. coli abundance and assemblage composition (>62%) was explained by a combination of physical-chemical and trophic variables. These findings indicate that coastal sediments could represent a potential reservoir for commensal and pathogenic E. coli and that E. coli distribution in marine coastal sediments largely depends upon the physical and trophic status of the sediment. We conclude that future sampling designs aimed at monitoring the microbiological quality of marine coastal areas should not further neglect the analysis of the sediment and that monitoring of these environments can be improved by including molecular methods as a complement of culture-based techniques.
Marine environments contaminated by fecal material, derived from human or animal waste, may contain a large variety of pathogenic microorganisms. Health protection and monitoring programs analyze the contamination of aquatic ecosystems (20) but, due to technical and practical difficulties, the search of fecal indicator bacteria (FIB) is routinely preferred to the systematic search of all potential pathogens to assess the sanitary risk of a water body (17). Recreational seawaters are, for instance, classified on the basis of the concentration of Escherichia coli and Enterococcus spp. (21, 33, 40), assumed to be indicators of fecal contamination and of the presence of other pathogenic enteric bacteria. Exposure to waters contaminated with E. coli and Enterococcus spp. have been associated with an increased risk of contracting gastrointestinal and respiratory illnesses (10, 24, 31, 62, 64). Although most E. coli strains are harmless, some strains can cause a variety of intestinal and extraintestinal diseases (11, 57, 58, 62) such as diarrhea, urinary tract infections, bacteremia, sepsis, and meningitis (57). Phylogenetic analyses have shown that E. coli includes four main phylogroups (A, B1, B2, and D) and that most virulent extraintestinal strains belong to the groups B2 and D (11, 23, 46).
The microbiological quality of marine waters is typically based exclusively on the water column, whereas sediments have received attention only recently (7, 14, 27, 45). Fecal coliforms (FC) and enterococci have been reported from marine sediments (5, 19, 41), and it has been also proposed that FIB accumulated in the sediments have the potential to contaminate the overlying waters by resuspension of sediment particles (35). There is evidence that FIB and pathogenic bacteria can survive longer in aquatic sediments than in the overlying water column (12, 34). However, the available knowledge on the environmental factors influencing the ecology of pathogenic bacteria in marine sediments is still extremely scant, and there are only few detailed studies on the pathogenic potential, genetic diversity, or population structure of FIB in sediments (1, 63).
The development of molecular methods has permitted a range of new approaches to monitor the safety of recreational waters (2). Among the available molecular methods, the fluorescence in situ hybridization (FISH) based on probes specific to 16S or 23S rRNA can be utilized to detect and enumerate specific prokaryotic taxa (16, 59). Since the number of ribosomes varies, generally between 103 and 105 per cell, depending on the species and physiological state, FISH has also been used to provide evidence of an active metabolic state of the detected cells (2, 8). FISH can thus represent a good complement to culture-based methods, and provides reliable quantitative data in a short time (within 4 h). With regard to FIB, the use of FISH to detect total coliforms (TC) has proven to be difficult, due to their high phylogenetic heterogeneity (55). Conversely, the use of species-specific probes for the detection of single species, such as E. coli, is routinely used (22, 47, 53); however, it has been never tested on marine sediments.
The objective of the present study was to investigate the microbiological quality of coastal marine sediments along a large area of the Adriatic Sea (Central Mediterranean Sea) and to evaluate the presence and distribution of specific bacterial genotypes associated with different marine areas. More specifically, it was our aim to evaluate whether marine sediments may be a potential reservoir of active pathogenic E. coli and thus represent a risk for human health. To do this, we analyzed (i) the abundance and distribution of TC and FC; (ii) the abundance and distribution of E. coli strains, along with their genetic relatedness; and (iii) the presence of extraintestinal pathogenic E. coli carrying virulence gene factors. To determine bacterial abundance, culture-dependent (the membrane filtration [MF] technique) and culture-independent (the FISH technique) approaches were used. Finally, to identify the factors potentially responsible for the accumulation and survival of E. coli in the benthic environment, we investigated the environmental variables possibly related to the distribution of FIB.
MATERIALS AND METHODS
Study area and sampling activities.
Sediments were collected from seven sites along ca. 50 km of the coasts of the Central Adriatic Sea (Fig. 1) in the period from May to June 2008. Each site comprised from two to four transects, each consisting of two stations, located at 2-m (ca. 10- to 30-m distance from the coastline) and 5-m (ca. 200- to 300-m distance from the coastline) depths, for a total of 46 sampled stations. Sites CM, FE, CU, and GR comprised four sampling transects each; site PE comprised three sampling transects, and sites CF and MS comprised two sampling transects (Fig. 1). Stations at a depth of 2 m were close or inside artificial reefs, deployed in the 1980s to protect the coastline and prevent sediment erosion. Sediment samples were collected by using a Van Veen grab sampler (capacity, 30 liters). Between each sampling station and prior to each deployment, the Van Veen grab was first rinsed with seawater (to remove sediment residuals) and then washed with a 10% HCl solution in deionized water and rinsed again using sterile deionized water to remove any acid residual. At each station, a Sea-Bird Electronics SBE 9/11 Plus instrument equipped with a sensor SBE43 was deployed to measure the main environmental characteristics of the water column, such as temperature, salinity, oxygen content, pH, and turbidity. All measurements were carried out at the water-sediment interface (a few centimeters above the sediment surface) and are thus likely to represent those influencing the bacterial assemblages in the superficial sediment layers investigated in the present study.
FIG. 1.
Study area, with zooming on the location of sampling sites.
For culture-based analyses of FIB (i.e., total coliforms and fecal coliforms), aliquots of ca. 20 g of sediments (n = 3 for each station, collected from different grab deployments and homogenized by mixing) were withdrawn by using a sterile spatula and stored in a sterile 50-ml Falcon test tube. Sediments were then held at 5 to 10°C and analyzed within 6 h from sampling. For FISH analyses, aliquots consisting of ca. 50 g of sediment, collected and homogenized as reported above, were immediately fixed using a prefiltered (0.2-μm pore size) and buffered 2% formalin solution. Within 24 h from fixation, sediments were washed (three times) with phosphate-buffered saline (PBS) and then stored in a PBS-ethanol mix (1:1) at −20°C until further processing (29). For total prokaryotic abundance, sediment samples (ca. 1 g), collected and homogenized as reported above, were transferred into sterile test tubes, fixed with 4 ml of prefiltered (0.2-μm pore size) and buffered 2% formalin, and stored at 4°C (36). Additional sediment samples were collected for the analysis of grain size and phytopigments (as chlorophyll a and phaeopigments) and of protein, carbohydrates, and lipid content, as a measure of organic resources for bacterial metabolism (50). For organic matter determinations, sediments were homogenized by mixing, put into a petri dish, and stored at −20°C until analysis. Analyses of phytopigments and organic matter content (such as protein, carbohydrates, and lipid content) were carried out as previously described (50, 51). Sediment grain size, expressed as a percentage of coarse sand, sand, and mud (sum of silts and clays), was measured by using standard methods.
Total prokaryotic abundance.
Total prokaryotic (TP) counts were performed by using the Sybr green direct count (SGDC) procedure described by Luna et al. (36). Samples were sonicated three times (Branson Sonifier 2200, 60 W) for 1 min, diluted 250 to 500 times with 0.2-μm-pore-size prefiltered formalin (2% final concentration), and then concentrated on 0.2-μm-pore-size aluminum oxide filters (Anodisc). Filters were then stained with Sybr green (Molecular Probes) by supplementing on each filter 20 μl of the stock solution (previously diluted 1:20 with filtered [0.2-μm pore size] Milli-Q water), washed twice with 3 ml of sterilized Milli-Q water, and mounted onto microscope slides. Filters were analyzed by using epifluorescence microscopy (Zeiss Axioskop 2; magnification, ×1,000). For each filter, at least 20 microscope fields were observed, and at least 400 cells were counted. The data were normalized to sediment dry weight after desiccation (48 h at 60°C).
Cultivation and enumeration of TC and FC.
To extract bacteria from sediments, triplicate 5-g aliquots of wet sediment were suspended in 20 ml of sterile seawater, vigorously shaken and sonicated in ice bath (n = 3, 1 min each with 30-s intervals within each cycle; Branson Sonifier 2200, 60 W). Preliminary laboratory tests indicated that this procedure was necessary to dislodge bacteria from the sediments and did not affect the subsequent growth of target bacteria, as previously demonstrated (13, 19). Aliquots (1 ml), along with 10-fold serial dilutions, of the resulting seawater were then analyzed by using the MF technique according to standard methods (6). TC were enumerated using mENDO agar plates (BD), incubated 24 h at 35°C. Fecal coliforms were then enumerated using mFC agar plates (BD) incubated 24 h at 44.5°C. The abundance of total coliforms and fecal coliforms was reported as CFU per g (dry weight) of sediment.
Identification of FC isolates.
Colonies grown on mFC agar plates were at first streaked on EMB agar and E. coli/coliform chromogenic agar (Oxoid, Basingstoke, United Kingdom) for the isolation and presumptive identification of coliforms. The definitive species identification was performed by PCR of 16S-23S ribosomal DNA intergenic regions (ITS-PCR) (30), and isolates with doubtful profiles (n = 22) were further identified by using the indole production test and API 20E system (API; bioMérieux, Marcy l'Etoile, France).
Determination of E. coli phylogenetic group.
Phylogenetic grouping of E. coli isolates was determined by a multiplex PCR using a combination of three DNA markers—chuA, yjaA, and an anonymous DNA fragment (TspE4.C2)—as described by Clermont et al. (11). On the basis of the presence or absence of the three DNA fragments, strains were assigned to phylogenetic groups (A, B1, B2, and D).
Detection of virulence factor genes (VFGs).
The genes encoding fimbrial adhesins (pap, sfa/foc, and afa), an intimin (eaeA), a transmembrane protein involved in neonatal meningitis (ibeA), an outer membrane protein for host serum survival (traT), toxins (hlyA, stx1, and stx2) and iron acquisition systems (aer and fyuA) were detected by PCR as previously described (32, 46, 52).
RAPD typing of E. coli strains.
All strains identified as E. coli (n = 109 colonies) were also analyzed by using RAPD [random(ly) amplified polymorphic DNA] typing. Chromosomal DNA, used as a template in PCRs, was obtained from crude lysate of bacterial overnight culture in brain hearth broth (Oxoid). Two random primers, primer 1 (5′-CCGCAGCCAA) and primer 2 (5′-AAGAGCCCGT), described by Regua-Mangia et al. (54) were used separately in each PCR assay performed in a 50-μl final reaction volume containing 1× buffer (10 mM Tris-HCl [pH 8.8], 3 mM MgCl2, 50 mM KCl, 0.1% Triton X-100), 200 μM concentrations of each deoxynucleoside triphosphate, 25 pmol of primer, 2 U of DyNAzyme DNA polymerase (Finnzymes, Espoo, Finland), and 3 μl of bacterial lysate. The amplification program was as follows: 4 cycles of 5 min at 94°C, 5 min at 37°C, and 5 min at 72°C, followed by 30 cycles of 1 min at 94°C, 1 min at 37°C, and 1 min at 72°C, with a final extension step of 10 min at 72°C). Rarefaction curves and the expected genotype richness (normalized for a sample including 100 isolates [EG100]) for each investigated sampling site were calculated from RAPD data using the Primer software (Primer-E, Ltd., United Kingdom). The similarity between the RAPD patterns was determined on the basis of the Dice similarity coefficient and the generated matrix was subjected to clustering by the unweighted pair group method of averages (UPGMA) using the TreeCon software (Bioinformatics and Evolutionary Genomics, Ghent, Belgium).
FISH analysis.
The FISH procedure included the typical four steps: (i) cell fixation, (ii) hybridization, (iii) posthybridization washing (to remove unbound or nonspecifically bound material), and (iv) the visualization using epifluorescence microscopy (44). This analysis was performed on a subset of triplicate sediment samples from 11 stations, selected on the basis of culture-based results with the aim of analyzing samples characterized by a wide range of E. coli abundance. The selected sediment samples were from stations CM3 (2 m) within the CM site, FE8 (2 m) and FE8 (5 m) within the FE site, PE9 (2 m) and PE11 (5 m) within the PE site, CF12 (2 m) at the CF site, MS14 (2 m) at the MS site, CU16 (5 m) and CU18 (5 m) at the CU site, and GR20 (5 m) and GR22 (2 m) at the GR site. In these samples, E. coli abundance ranged from no growth (such as the CM site at both depths and the CU site at the 5-m depth) to the highest abundance observed in the entire study (the PE site). Briefly, aliquots of each selected sample (fixed and sonicated as described above) were properly diluted and filtered through 0.2-μm-pore-size black polycarbonate filters (Nuclepore). The filters were then hybridized using a Cy3-labeled probe under appropriate hybridization conditions and washed using the appropriate washing buffer. For the specific detection of E. coli, we used the probe Colinsitu described by Roslev et al. (56), targeting a 16S rRNA sequence. Probes EUB338 and NON338 were used as positive and negative controls, respectively (44). Each filter was counterstained with DAPI (4′,6′-diamidino-2-phenylindole; 0.5 μg ml−1), mounted onto microscopic slides, and observed under epifluorescence microscopy (Zeiss Axioskop 2; magnification, ×1,000). The data were expressed as E. coli cells g−1 (dry weight) of sediment.
Statistical analyses.
Differences in the microbiological variables (total prokaryotes, total and fecal coliforms) between different areas and sampling depths were tested by using two-way analysis of variance (ANOVA). The analysis included seven sampling areas (treated as a random factor) and two depths (treated as a fixed factor), with n = 3 for the combination of factors. Prior to the ANOVA, the data were checked for variance homogeneity using the Cochran's C test and log transformed whenever needed. For those variables for which the variance remained heterogeneous even after transformation, to reduce type I error, the level of significance was cautionary set to P < 0.001 (60).
Distance-based permutational multivariate analyses of variance (PERMANOVA [3, 38]) were used to test for differences in the main physical-chemical variables, the biochemical composition of organic matter, and the composition of the E. coli assemblage in the sediments of the investigated area. The PERMANOVA test is an analogous to the multivariate analysis of variance (MANOVA), which is too stringent in its assumptions for most ecological multivariate data sets (3). Nonparametric methods based on permutation tests such as the one performed by the PERMANOVA tool are preferable since they allow to partition the variability in the data according to a complex design or model and to base the analysis on a multivariate distance measure that is reasonable for ecological data sets (38). The design included two factors: the site (seven levels, random) and the water depth (two levels, fixed and orthogonal to site), with n = 3 for the combination of factors. The analysis was based on Euclidean distances of non-normalized data, using 4,999 random permutations of the appropriate units (4). The analysis was run using the FORTRAN-written PERMANOVA.exe program. The pseudo-multivariate variance components for each term in the model were calculated using direct multivariate analogues to the univariate ANOVA estimators.
A correlation analysis was carried out to ascertain significant relationships between all of the investigated variables (data not shown). Those variables not significantly correlated each other were grouped into a physical-chemical (including temperature, salinity, pH, dissolved oxygen, and turbidity in the overlying waters and the percentage of mud in the sediment) and a trophic (including concentrations of chlorophyll a, protein, carbohydrate, and lipid in the sediment) set of variables. Then, a multivariate, multiple regression analysis was used to ascertain the relative proportions of variation of E. coli abundance and assemblage composition explained cumulatively by the sets of physical-chemical and trophic variables. All analyses were carried out with the routine distance-based multivariate analysis for a linear model (DISTLM forward). P values were obtained with 4,999 permutations of residuals under the reduced model (3).
RESULTS
Physical-chemical and trophic variables in the investigated areas.
Results for the main physical-chemical (temperature, salinity, pH, oxygen content, turbidity, and grain size, expressed as coarse sand, sand, and mud) and trophic (chlorophyll a, phaeopigment, protein, carbohydrate, and lipid sediment content) variables, averaged (± the standard error) for each investigated site and sampling depth (Fig. 1), are reported in Table 1. The PERMANOVA tests revealed a significant effect of interaction site × depth either on the physical-chemical or trophic variables. This result suggests that differences in physical-chemical and trophic conditions between the two water depths varied across the seven sites (Table 2), for instance with physical-chemical conditions being significantly different between water depths in FE and CU and not significantly different in the remaining sites. Most of the variance in the trophic and physical-chemical characteristics of the study area was explained by differences between sites (52 to 84%, respectively).
TABLE 1.
Main environmental variables in the investigated sitesa
| Site | Depth (m) | Temp |
Salinity |
pH |
Oxygen |
Turbidity |
Chl-a |
Phaeo |
Prt |
Cho |
Lip |
Coarse sand |
Sand |
Mud |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| °C | SE | PSU | SE | Avg | SE | ml/liter | SE | FTU | SE | μg/g | SE | μg/g | SE | mg/g | SE | mg/g | SE | mg/g | SE | % | SE | % | SE | % | SE | ||
| CM | 2 | 18.45 | 0.06 | 36.53 | 0.01 | 8.141 | 0.006 | 5.36 | 0.05 | 4.34 | 1.45 | 0.72 | 0.23 | 0.95 | 0.25 | 0.12 | 0.03 | 0.10 | 0.02 | 0.03 | 0.01 | 0.03 | 0.03 | 99.58 | 0.18 | 0.40 | 0.19 |
| 5 | 18.07 | 0.06 | 36.80 | 0.08 | 8.143 | 0.014 | 5.53 | 0.04 | 0.88 | 0.16 | 1.64 | 0.37 | 3.32 | 0.63 | 0.31 | 0.02 | 0.20 | 0.03 | 0.03 | 0.01 | 0.00 | 0.00 | 97.83 | 0.08 | 2.18 | 0.08 | |
| FE | 2 | 18.27 | 0.10 | 36.61 | 0.02 | 8.189 | 0.012 | 5.26 | 0.01 | 7.72 | 2.17 | 0.63 | 0.08 | 0.86 | 0.07 | 0.25 | 0.04 | 0.07 | 0.02 | 0.03 | 0.00 | 0.00 | 0.00 | 99.08 | 0.26 | 0.93 | 0.26 |
| 5 | 18.39 | 0.06 | 36.68 | 0.02 | 8.222 | 0.001 | 5.29 | 0.03 | 1.47 | 0.07 | 1.49 | 0.24 | 2.69 | 0.29 | 0.38 | 0.05 | 0.14 | 0.03 | 0.04 | 0.01 | 0.00 | 0.00 | 97.10 | 0.27 | 2.90 | 0.27 | |
| PE | 2 | 22.73 | 0.12 | 34.16 | 0.06 | 8.204 | 0.014 | 5.25 | 0.06 | 3.99 | 1.36 | 1.53 | 0.34 | 4.04 | 1.11 | 0.87 | 0.41 | 0.27 | 0.17 | 0.05 | 0.01 | 0.00 | 0.00 | 78.80 | 13.70 | 21.20 | 13.71 |
| 5 | 22.51 | 0.10 | 34.23 | 0.06 | 8.200 | 0.011 | 5.28 | 0.06 | 1.50 | 0.30 | 1.10 | 0.16 | 2.01 | 0.31 | 0.43 | 0.05 | 0.21 | 0.06 | 0.06 | 0.01 | 0.00 | 0.00 | 90.73 | 0.66 | 9.27 | 0.73 | |
| CF | 2 | 22.50 | 0.03 | 34.23 | 0.06 | 8.173 | 0.011 | 5.11 | 0.11 | 4.03 | 2.30 | 0.64 | 0.13 | 1.14 | 0.22 | 0.22 | 0.04 | 0.11 | 0.01 | 0.04 | 0.01 | 0.00 | 0.00 | 82.90 | 14.20 | 17.10 | 14.20 |
| 5 | 22.22 | 0.03 | 34.29 | 0.02 | 8.192 | 0.003 | 5.31 | 0.01 | 0.61 | 0.03 | 1.22 | 0.02 | 2.31 | 0.26 | 0.33 | 0.00 | 0.15 | 0.02 | 0.04 | 0.01 | 0.00 | 0.00 | 96.85 | 0.15 | 3.15 | 0.15 | |
| MS | 2 | 23.17 | 0.23 | 34.03 | 0.07 | 8.216 | 0.012 | 5.20 | 0.17 | 4.88 | 2.50 | 1.24 | 0.12 | 2.06 | 0.29 | 0.26 | 0.03 | 0.12 | 0.01 | 0.03 | 0.01 | 0.00 | 0.00 | 98.25 | 0.15 | 1.75 | 0.15 |
| 5 | 22.89 | 0.04 | 33.96 | 0.01 | 8.226 | 0.001 | 5.22 | 0.05 | 1.69 | 0.81 | 0.70 | 0.23 | 1.92 | 0.49 | 0.41 | 0.08 | 0.15 | 0.01 | 0.04 | 0.01 | 0.00 | 0.00 | 96.05 | 0.25 | 3.95 | 0.25 | |
| CU | 2 | 22.77 | 0.15 | 34.25 | 0.03 | 8.196 | 0.006 | 4.94 | 0.03 | 5.34 | 0.79 | 5.04 | 2.83 | 6.49 | 3.37 | 0.37 | 0.03 | 0.24 | 0.07 | 0.03 | 0.00 | 0.00 | 0.00 | 83.78 | 7.23 | 16.25 | 7.29 |
| 5 | 22.56 | 0.08 | 34.24 | 0.01 | 8.230 | 0.004 | 5.12 | 0.02 | 2.64 | 0.35 | 2.09 | 0.43 | 3.31 | 0.38 | 0.44 | 0.11 | 0.36 | 0.10 | 0.06 | 0.01 | 0.00 | 0.00 | 95.15 | 0.36 | 4.85 | 0.81 | |
| GR | 2 | 22.92 | 0.18 | 30.48 | 0.03 | 8.291 | 0.013 | 4.87 | 0.11 | 6.37 | 1.28 | 2.91 | 1.92 | 7.74 | 6.22 | 0.72 | 0.55 | 0.47 | 0.31 | 0.12 | 0.07 | 0.00 | 0.00 | 72.75 | 18.40 | 27.25 | 18.65 |
| 5 | 22.77 | 0.04 | 30.56 | 0.06 | 8.304 | 0.005 | 4.90 | 0.04 | 9.37 | 2.80 | 1.38 | 0.29 | 2.01 | 0.46 | 0.25 | 0.03 | 0.25 | 0.06 | 0.03 | 0.01 | 0.00 | 0.00 | 94.43 | 0.36 | 5.58 | 0.63 | |
Measurements for temperature, salinity, pH, oxygen, and turbidity were obtained in situ at the water-sediment interface using a CTD probe. The results are averaged for each site. Abbreviations: FTU, formazin turbidity unit; Chl-a, chlorophyll a; Phaeo, phaeopigments; Prt, proteins; Cho, carbohydrates; Lip, lipids. Coarse sand and sand = fraction < 2,000 μm and > 63 μm; Mud = fraction < 63 μm.
TABLE 2.
Results of the PERMANOVA test on the physical-chemical and trophic variables of the study areaa
| Variable type and source | df | SS | MS | Pseudo-F | Significance (P) | % of explained variance |
|---|---|---|---|---|---|---|
| Physical-chemical variables | ||||||
| Site | 6 | 206.2 | 34.360 | 16.329 | <0.001 | 84.2 |
| Depth | 1 | 25.7 | 25.657 | 4.131 | <0.01 | 8.8 |
| Site × depth | 6 | 37.3 | 6.211 | 2.952 | <0.001 | 6.3 |
| Residuals | 28 | 58.9 | 2.104 | 0.7 | ||
| Total | 41 | 328.0 | ||||
| Trophic variables | ||||||
| Site | 6 | 69.6 | 11.604 | 4.245 | <0.001 | 51.9 |
| Depth | 1 | 2.6 | 2.632 | 0.963 | NS | 3.8 |
| Site × depth | 6 | 56.2 | 9.368 | 3.427 | <0.01 | 42.3 |
| Residual | 28 | 76.5 | 2.733 | 2.0 | ||
| Total | 41 | 205.0 |
The results of the PERMANOVA test on the physical-chemical (temperature, salinity, pH, oxygen, turbidity, and sediment grain size) and trophic (chlorophyll a, phaeopigment, protein, carbohydrate, and lipid sediment contents) variables of the study area are presented. df, degrees of freedom; SS, sum of squares; MS, mean square; Pseudo-F, statistic F; NS, not significant.
Prokaryote abundance and distribution.
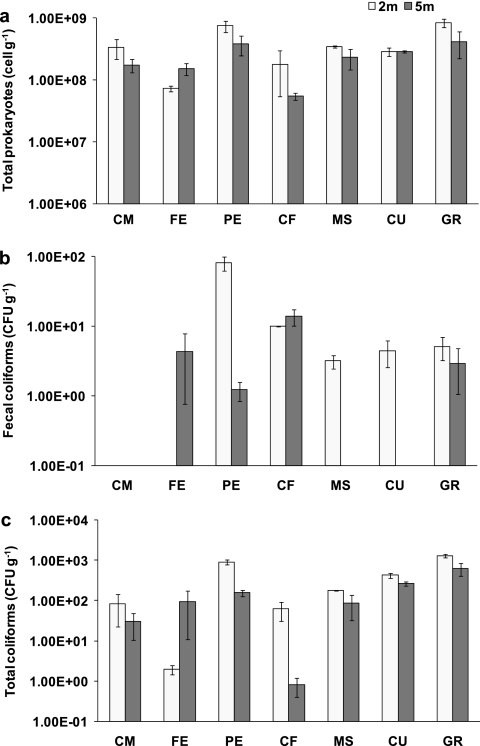
The average abundance (± the standard error) of TP, FC, and TC at each investigated site is shown in Fig. 2. The ANOVA revealed a significant effect of interaction site × depth on the abundance of TP and FC, suggesting that, for these two variables, differences between water depths varied across the investigated sites (Table 3). The factor “site” explained most of the variation in the total prokaryotic and FC abundances (78 and 49%, respectively) (Table 3). The abundance of TP in PE and GR at a depth of 2 m was higher than in all other sites and depths (Fig. 2a). At the FE site, FC were found only at a depth of 5 m, while at the MS and CU sites they were detected only at a depth of 2 m (Fig. 2b). TC were detected at all sites, and the ANOVA revealed that the factor site explained significantly the largest proportion (78%) of the variance (Table 3), whereas no effects of the factor depth were observed (Table 3). TC abundance in PE and GR sediments at a depth of 2 m displayed the highest values in the whole sampling area (Fig. 2c).
FIG. 2.
Abundance of total prokaryotes (a), fecal coliforms (b), and total coliforms (c) in the investigated sites. Reported are the average (calculated as arithmetic mean) ± the standard error for each investigated site and sampling depth.
TABLE 3.
Results of the ANOVA test on the total prokaryote, total coliform, and fecal coliform abundance (log transformed) in the study areaa
| Variable and sourceb | SS | DF | MS | F | Significance (P) | % of explained variance |
|---|---|---|---|---|---|---|
| Total prokaryotes | ||||||
| Site | 18.373 | 6 | 3.062 | 25.85 | <0.0001 | 78.4 |
| Depth | 1.677 | 1 | 1.677 | 14.16 | <0.0001 | 7.6 |
| Site × depth | 3.059 | 6 | 0.509 | 4.3 | <0.0001 | 13.5 |
| Residual | 3.317 | 28 | 0.119 | 0.5 | ||
| Total | 26.426 | 41 | ||||
| Total coliforms | ||||||
| Site | 127.470 | 6 | 21.245 | 24.9 | <0.0001 | 78.4 |
| Depth | 3.620 | 1 | 3.620 | 4.24 | NS | 2.7 |
| Site × depth | 29.198 | 6 | 4.866 | 5.7 | <0.001 | 18.4 |
| Residual | 23.890 | 28 | 0.853 | 0.5 | ||
| Total | 184.179 | 41 | ||||
| Fecal coliforms | ||||||
| Site | 30.934 | 6 | 5.156 | 18.38 | <0.0001 | 49.2 |
| Depth | 5.003 | 1 | 5.003 | 17.83 | <0.0001 | 8.3 |
| Site × depth | 26.327 | 6 | 4.388 | 15.64 | <0.0001 | 42.0 |
| Residual | 7.855 | 28 | 0.281 | 0.4 | ||
| Total | 70.119 | 41 |
df, degrees of freedom; SS, sum of squares; MS, mean square; Pseudo-F, statistic F; NS, not significant.
Cochran's test results were as follows: total prokaryotes, C = 0.4569 (P < 0.05); total coliforms, C = 0.5744 (P < 0.01); and fecal coliforms, C = 0.6551 (P < 0.01).
Identification of the isolates.
Isolates grown on mFC medium were identified by biochemical and molecular methods. The results indicated that 109 of 113 (96%) fecal coliform isolates were E. coli (Table 4). These isolates were recovered from all of the investigated sites, with the exception of CM (negative to FC counts) and MS, at both 2- and 5-m depths. Of the four remaining isolates, two (one from the MS site at 2 m and one from the FE site at 5 m) were identified as Klebsiella spp., and two (one from the CU site at 2 m and the other from the FE site at 5 m) as Citrobacter spp.
TABLE 4.
Number of FC and E. coli isolates in the investigated sitesa
| Site | Depth (m) | No. of isolates |
|
|---|---|---|---|
| Fecal | E. coli | ||
| CM | 2 | NI | NI |
| 5 | NI | NI | |
| FE | 2 | NI | NI |
| 5 | 16 | 14 | |
| PE | 2 | 59 | 59 |
| 5 | 5 | 5 | |
| CF | 2 | 15 | 15 |
| 5 | 2 | 2 | |
| MS | 2 | 1 | NI |
| 5 | NI | NI | |
| CU | 2 | 7 | 6 |
| 5 | NI | NI | |
| GR | 2 | 6 | 6 |
| 5 | 2 | 2 | |
| Total | 113 | 109 | |
Isolates were obtained from three replicated sediments collected from each sampling station and depth. All 109 E. coli isolates were then subjected to RAPD analysis and determination of phylogroup (A, B1, B2, and D). NI, not isolated.
Determination of E. coli phylogenetic group.
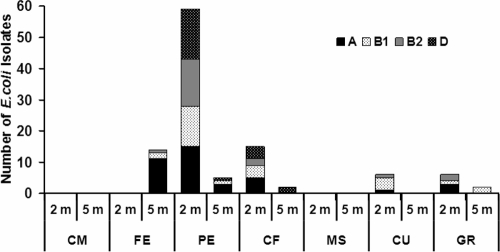
The assignment of all 109 E. coli isolates to a specific phylogroup was performed by the triplex PCR technique described by Clermont et al. (11). This technique allows distinguishing four major phylogroups of E. coli strains: A and B1, including most of the commensal strains, and B2 and D, including most of the virulent extraintestinal strains. Overall, ca. 60% of the strains belonged to groups A (35%) and B1 (25%), while 21% belonged to group D and 19% belonged to group B2. The relative abundance of the four phylogenetic groups changed among sites and depths (Fig. 3), with the only exception being site PE (2-m depth), where the four groups were equally represented (from 22 to 27%). At the same site at a depth of 5 m, extraintestinal group B2 and D isolates were negligible (no isolates and only one isolate, respectively) and commensal groups A and B1 were present in low concentrations (only three isolates and one isolate, respectively). At the FE, CU, and GR sites, at both 2-m and 5-m depths, isolates belonging to group D were never detected. These results were confirmed by the PERMANOVA test, which revealed that the composition of E. coli assemblage (A, B1, B2, and D phylogroups) varied significantly between sites but not between sampling depths (Table 5).
FIG. 3.
Spatial distribution of E. coli phylogroups (A, B1, B2, and D) at the different sites and sampling depths.
TABLE 5.
Results of the PERMANOVA test on E. coli assemblage composition (phylogroups A, B1, B2, and D) in the study areaa
| Source | df | SS | MS | Pseudo-F | Significance (P) |
|---|---|---|---|---|---|
| Site | 6 | 19.571 | 3.2619 | 68.5 | <0.001 |
| Depth | 1 | 2.619 | 2.619 | 1.068 | NS |
| Site × depth | 6 | 14.714 | 2.4524 | 51.5 | <0.001 |
| Residual | 28 | 1.3333 | 0.047619 | ||
| Total | 41 | 38.238 |
df, degrees of freedom; SS, sum of squares; MS, mean square; Pseudo-F, statistic F; Pseudo-F, probability level; NS, not significant.
Detection of VFGs.
All E. coli strains belonging to B2 (n = 21) and D (n = 23) phylogroups were tested for the presence of 11 VFGs (pap, sfa/foc, afa, eaeA, ibeA, traT, hlyA, stx1, stx2, aer, and fyuA) frequently associated with extraintestinal diseases. The results are shown in Table 6. Overall, a large proportion of the tested strains was shown to contain VFGs, which were more frequent in group B2 than in group D strains. Indeed, 90% of group B2 and 65% of group D strains contained at least one VFG. The most frequently detected was traT (54.5%), followed by fyuA (50%) and ibeA (41%). stx1 and stx2 were never found. Notably, 71% of B2 and 35% of D strains carried two or more (up to six and four, respectively) VFGs (data not shown).
TABLE 6.
Frequency of virulence factor genes in E. coli isolates belonging to the B2 and D phylogroupsa
| VFG | Gene product (category) | Activity/effect | Pathotype(s) | No. of isolates (% prevalence) |
|
|---|---|---|---|---|---|
| Group B2 (n = 21) | Group D (n = 23) | ||||
| sfa/foc | Adhesin | Adhesion and colonization of uroepithelium and endothelium | UPEC, MNEC | 3 (14) | 0 |
| afa | Adhesin | Adhesion and colonization of uroepithelium, cytopathic effects | DAEC, UPEC | 0 | 0 |
| pap | Adhesin | Induction of cytokine expression, adhesion and colonization of uroepithelium | UPEC | 2 (9.5) | 0 |
| eaeA | Intimin | Adhesion and colonization of epithelial cells, induction of TH1 immune responses | EPEC, STEC | 2 (9.5) | 2 (8.7) |
| fyuA | Siderophore | Iron acquisition | Various | 15 (71) | 7 (30) |
| aer | Siderophore | Iron acquisition | EIEC, MNEC | 2 (9.5) | 3 (13) |
| stx1 | Toxin | Inhibition of protein synthesis, apoptosis induction | STEC | 0 | 0 |
| stx2 | Toxin | Inhibition of protein synthesis, apoptosis induction | STEC | 0 | 0 |
| hlyA | Alpha-hemolysin | Cell lysis | UPEC | 2 (9.5) | 0 |
| ibeA | Membrane protein | Promotes invasion | MNEC, UPEC | 13 (62) | 5 (22) |
| traT | Membrane protein | Serum survival | MNEC | 11 (52) | 13 (56) |
VFG, virulence factor gene; UPEC, uropathogenic E. coli, MNEC, sepsis-and meningitis-associated E. coli; DAEC, diffusely adherent E. coli; EPEC, enteropathogenic E. coli; STEC, Shiga toxin-producing E. coli; EIEC, enteroinvasive E. coli.
RAPD typing and genotype diversity estimates of E. coli isolates.
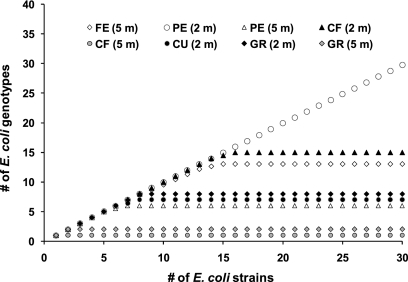
All 109 E. coli isolates were typed by RAPD to establish their genetic relationships, as well as to assess the presence of specific E. coli clones, potentially fitted to survive in the investigated sediments. Strains relatedness was represented by a dendrogram which refers to all E. coli isolates (see Fig. S1 in the supplemental material). Overall, a high genetic diversity among E. coli isolates, within the same site and within replicate samples in the same sampling station, was observed, and no specific phylogroups were clearly associated with specific sites. Saturated rarefaction curves for all but one of the investigated sites (the PE site at 2-m depth) indicated that the genotype diversity in the study area was almost entirely described (Fig. 4). The diversity of E. coli assemblages, calculated as the expected genotype richness (EG100, Fig. 5), ranged from 1 (CF, 5-m depth) to 57 (PE, 2-m depth) genotypes.
FIG. 4.
Rarefaction curves based on genetic relatedness of all (n = 109) E. coli isolates, as determined by RAPD analysis.
FIG. 5.
Diversity index (calculated as the expected genotype richness [EG100]) of E. coli assemblages, based on the genetic relatedness of all (n = 109) E. coli isolates.
FISH enumeration of E. coli.
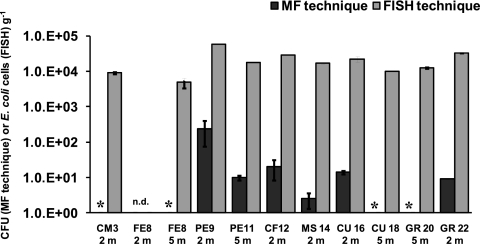
The FISH technique was used to enumerate E. coli in the investigated sediments. The analysis was performed on a subset of 11 sediment samples, which were selected on the basis of FIB abundance (as detected by culture-based methods) with the aim of analyzing samples with a wide range of E. coli concentrations. FISH counts ranged from 0 (at the FE8 station, 2-m depth) to 5.86 ± 0.12 × 104 cell g−1 (PE9 station, 2-m depth) (Fig. 6). The comparison between FISH and counts obtained by MF (assumed to represent E. coli counts, since 96% of the FC isolates were accounted by this species) revealed the presence of a significant and positive correlation between the two measures (r2 = 0.63, P < 0.05). However, the FISH technique resulted always in significantly higher E. coli abundances (ANOVA, P < 0.01, data not shown), with values up to 4 logs higher than those obtained by culture-based techniques.
FIG. 6.
Abundance of E. coli in selected sediment samples, as resulting from the FISH and the culture-based (growth on mFC agar) methods. Reported are the averages for each sediment sample ± the standard deviation. *, no colonies recovered on agar; n.d., not detected.
Relationships between E. coli assemblage composition and environmental and trophic conditions.
The multivariate multiple regression analyses revealed that the largest proportion of variance of the total abundance and assemblage composition of E. coli was significantly explained by either physical-chemical (temperature, salinity, pH, dissolved oxygen, turbidity, and the percentage of mud) and trophic (concentrations of chlorophyll a, phaeopigment, protein, carbohydrate, and lipid in the sediment) sets of variables. The physical-chemical set of variables alone accounted for >40% of the observed variations of both E. coli abundance and assemblage composition (Table 7).
TABLE 7.
Effects of environmental and trophic sets of variables on E. coli abundance and assemblage compositiona
| Conditional (sequential) test | Variable | SS | Pseudo-F | P | Explained variance |
|
|---|---|---|---|---|---|---|
| % | Cumulative | |||||
| E. coli abundance | Physical-chemical | 5.359 | 3.975 | 0.002 | 40.5 | 40.5 |
| Trophic | 2.964 | 3.627 | 0.016 | 22.4 | 62.9 | |
| E. coli assemblage composition | Physical-chemical | 54.680 | 4.809 | 0.002 | 45.2 | 45.2 |
| Trophic | 21.081 | 2.796 | 0.008 | 17.4 | 62.6 | |
Variables not significantly correlated with each other were grouped into two multivariate sets of explanatory variables: physical-chemical (including temperature, salinity, pH, dissolved oxygen, turbidity, and percentage of mud) and trophic (including sediment contents of chlorophyll a, phaeopigment, protein, carbohydrate, and lipid).
DISCUSSION
The presence of FIB in marine sediments has been documented in coastal areas worldwide (12, 19, 37). Recent studies have also suggested that the environment outside the gastrointestinal tracts of warm-blooded animals (such as water, sediment, and soil) can represent a “secondary habitat” for FIB (63). This has been also confirmed by studies conducted in subtropical areas, where E. coli and enterococci display higher growth and survival rates in sediments than in the overlying seawater (26). It can be therefore hypothesized that marine coastal sediments can be “reservoirs” of metabolically active FIB (45).
The sediments investigated in the present study were characterized by high concentrations of phytopigments and organic matter, which reflect the relatively high trophic status of the coastal sediments of this portion of the Adriatic Sea (15). Similarly high values were observed in terms of total prokaryotic abundance.
The abundance of total and fecal coliforms in the Adriatic sediments measured by cultivation methods are comparable with those previously reported in coastal sediments from other coastal areas of the Adriatic Sea (45), the North Western England (41), and Australia (13). However, when based on the counts obtained using FISH, the abundances of E. coli were significantly higher than those reported in any previous study. Comparative analyses between culture-based and molecular methods for E. coli counting have been already conducted in freshwaters, drinking waters (53), and wastewaters (22), but never thus far in marine sediments. The observed differences between counts carried out using the two techniques are somehow expected and likely due to the ability of FISH to detect also viable but not culturable bacteria (2, 44). The high concentration of solids and organic matter in the investigated sediments, which may have led to underestimations of CFU due to clogging of the filters or to the presence of substances inhibiting bacterial growth, could be involved as well. However, such a negative effect should be mitigated by the serial dilutions we performed on the sediment suspensions before filtering. Moreover, Yamahara et al. (65) and Vezzulli et al. (61) have recently demonstrated in marine sediments that the use of molecular methods, such as quantitative PCR, yielded Enterococcus spp. and Vibrio spp. counts up to 4 orders of magnitude higher than those obtained by cultivation on selective agar. On the other hand, it is known that FIB released in “secondary habitats,” such as marine sediments, while preserving their viability, can lose their ability to grow on culture media (22). E. coli pathogenic cells encountered in marine sediments can be nonculturable while maintaining their virulence, thus representing a higher risk for human health (39, 42). The FISH technique we applied in the present study could thus be incorporated in monitoring programs for a more accurate assessment of the microbiological quality of coastal marine systems.
Almost all of the FC isolated from the investigated area were E. coli. RAPD analyses indicated, for the first time, that the E. coli from coastal marine sediments shows a large genotypic diversity not only among isolates from different sites but also among those from the same site. Such a large variability is as wide as the one reported in freshwater and lake sediments (1, 9, 48, 63). These results suggest that the investigated marine sediments, because of their large heterogeneity in both chemical and environmental characteristics, do not select specific genotypes. The resulting high genetic heterogeneity of E. coli could be caused by the multiple sources of fecal contamination (since the coastal area investigated included river inputs, waste waters, sediment, and urban discharges) and by horizontal gene transfer (2). This hypothesis is supported by the presence of the highest abundance and diversity of E. coli in the PE site (station 9), which received either river waters (from the Aso river) or uncontrolled urban discharges within a relatively small area. To support this hypothesis, we also investigated the role of the major environmental variables in the distribution and assemblage composition of E. coli. The results of the multivariate multiple regression analyses revealed that E. coli abundance and assemblage composition were significantly related to a combination of environmental and trophic variables (overall explaining more than 60% of variance), thus suggesting that variations in E. coli abundance and assemblage composition are to a large extent controlled by either the local environmental characteristics (including sediment grain size and oxygen availability) and the availability of labile (51) organic substrates. These results strengthen previous speculations, based on correlative analyses, about the potential importance of the water physical-chemical parameters (e.g., salinity [43]) and the availability of organic substrates in marine and lacustrine sediments (14, 25). In addition, we stress here for the first time, using multivariate statistical analyses, that these factors could control also the composition of E. coli assemblages, thus potentially modulating the exposure of humans to different levels of pathogenic risk.
Our results highlighted the presence of a large fraction (ca. 40%) of extraintestinal E. coli belonging to phylogroups B2 and D, most of which harbored one or more virulence factors. In particular, the B2 phylogroup alone accounted for ca. 20% of all E. coli isolates. It is worth noting that while the importance of phylogroup D has been previously underlined in aquatic ecosystems (62), group B2, the most involved in extraintestinal infections, is thus far reported to be rare or negligible (typically less than 3%) (28). The high frequency of B2 and D strains carrying VFGs, including those involved in meningitis (ibeA) and sepsis (traT), poses serious questions about the potential risk for humans once in contact with contaminated marine sediments. This question is even more crucial if we consider that some of the investigated sites (i.e., the CF, CU, and GR), according to the EU monitoring program carried out by the Local Authorities, met the bathing waters regulatory criteria. Moreover, the high genetic diversity of E. coli isolates belonging to the phylogroup B2 suggests that coastal marine sediments can represent a reservoir of genetically diversified extraintestinal pathogenic E. coli (ExPEC). These findings let us hypothesize that, as recently suggested for other environments (18), this highly diversified genetic pool can contribute to the development and spread of pathogenic E. coli strains.
The results from the present study provide evidence that coastal marine sediments represent a potential reservoir of pathogenic E. coli. Moreover, since the surface layer of coastal sediments can be easily resuspended (e.g., by rough sea conditions and trawling fishing activities) (49), these pathogenic strains could potentially be released into the water column. This suggests that the assessment of microbiological quality of coastal areas should take into consideration the analysis of the sediment. Moreover, our results stress that the use of culture-based methods can severely underestimate the risks for human health, which can be more reliably identified by integrating culture-based and molecular methods. If our results would be confirmed on larger spatial scales and on different marine coastal areas worldwide, future monitoring programs should not longer neglect the synoptic analysis of microbiological (using culture-based and molecular tools) and environmental properties of the sediment.
Supplementary Material
Acknowledgments
This study was carried out in the framework of a research project funded by the Istituto Superiore per la Ricerca e Protezione Ambientale (ISPRA).
We acknowledge S. Bianchelli (Polytechnic University of Marche) for help with the sampling activities and laboratory analyses.
Footnotes
Published ahead of print on 2 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alm, E. W., J. Burke, and E. Hagan. 2006. Persistence and potential growth of the fecal indicator bacteria, Escherichia coli, in shoreline sand at Lake Huron. J. Great Lakes Res. 32:401-405. [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, M. J. 2001. Permutation tests for univariate or multivariate analysis of variance and regression. Can. J. Fish. Aquat. Sci. 58:626-639. [Google Scholar]
- 4.Anderson, M. J., and C. J. F. ter Braak. 2003. Permutation tests for multi-factorial analysis of variance. J. Stat. Comput. Simul. 73:85-113. [Google Scholar]
- 5.Anderson, S. A., S. J. Turner, and G. D. Lewis. 1997. Enterococci in the New Zealand environment: implications for water quality monitoring. Water Sci. Technol. 35:325-331. [Google Scholar]
- 6.APHA/AWWA/WEF. 1998. Standard methods for examination of water and wastewater, 20th ed. APHA, Washington, DC.
- 7.Boehm, A. B., J. Griffith, C. McGee, T. A. Edge, H. M. Solo-Gabriele, R. Whitman, Y. Cao, M. Getrich, J. A. Jay, D. Ferguson, K. D. Goodwin, C. M. Lee, M. Madison, and S. B. Weisberg. 2009. Faecal indicator bacteria enumeration in beach sand: a comparison study of extraction methods in medium to coarse sands. J. Appl. Microbiol. 107:1740-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvier, T., and P. A. del Giorgio. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Rev. 44:3-15. [DOI] [PubMed] [Google Scholar]
- 9.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lake watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 10.Cabelli, V. J., A. P. Dufour, M. A. Levin, and L. J. McCabe. 1982. Swimming-associated gastroenteritis and water quality. Am. J. Epidemiol. 115:606-616. [DOI] [PubMed] [Google Scholar]
- 11.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crabill, C., R. Donald, J. Snelling, R. Foust, and G. Southam. 1999. Impact of sediment fecal coliform reservoirs on seasonal water quality in Oak Creek, Arizona. Water Res. 33:2163-2171. [Google Scholar]
- 13.Craig, D. L., J. Fallowfield, and N. J. Cromar. 2002. Enumeration of fecal coliforms from recreational coastal sites: evaluation of techniques for the separation of bacteria from sediments. J. Appl. Microbiol. 93:557-565. [DOI] [PubMed] [Google Scholar]
- 14.Davies, C. M., J. A. H. Long, M. Donald, and N. J. Ashbolt. 1995. Survival of fecal microorganisms in marine and freshwater sediments. Appl. Environ. Microbiol. 61:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dell'Anno, A., A. Pusceddu, L. Langone, and R. Danovaro. 2008. Early diagenesis of organic matter in coastal sediments influenced by riverine inputs. Chem. Ecol. 24:75-85. [Google Scholar]
- 16.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: rRNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 17.Donovan, E. P., D. F. Staskal, K. M. Unice, J. D. Roberts, L. C. Haws, B. L. Finley, and M. A. Harris. 2008. Risk of gastrointestinal disease associated with exposure to pathogens in the sediments of the Lower Passaic River. Appl. Environ. Microbiol. 74:1004-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewers, C., E.-M. Antao, I. Diehl, H.-C. Phillipo, and L. H. Wieler. 2009. Intestine and environment of the chicken as reservoirs of extra-intestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 75:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson, D. M., D. F. Moore, M. A. Getrich, and M. H. Zhowandai. 2005. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. J. Appl. Microbiol. 99:598-608. [DOI] [PubMed] [Google Scholar]
- 20.Fleming, L. E., K. Broad, A. Clement, E. Dewailly, S. Elmir, A. Knap, S. A. Pomponi, S. Smith, H. Solo Gabriele, and P. Walsh. 2006. Oceans and human health: emerging public health risks in the marine environment. Mar. Pollut. Bull. 53:545-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furet, J. P., O. Firmesse, M. Gourmelon, C. Bridonneau, J. Tap, S. Mondot, J. Doré, and G. Corthier. 2009. Comparative assessment of human and farm animal fecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68:351-362. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Armisen, T., and P. Servais. 2004. Enumeration of viable Escherichia coli in rivers and wastewaters using fluorescent in situ hybridization. J. Microbiol. Methods 58:269-279. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, D. M., O. Clermont, H. Tolley, and E. Denamur. 2008. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 10:2484-2496. [DOI] [PubMed] [Google Scholar]
- 24.Haile, R. W., J. S. Witte, M. Gold, R. Cressey, C. McGee, R. C. Millikan, A. Glasser, N. Harawa, C. Ervin, P. Harmon, J. Harper, J. Dermand, J. Alamillo, K. Barrett, M. Nides, and G. Y. Wang. 1999. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology 10:355-363. [PubMed] [Google Scholar]
- 25.Haller, L., J. Potéa, J.-L. Loizeau, and W. Wildi. 2009. Distribution and survival of faecal indicator bacteria in the sediments of the Bay of Vidy, Lake Geneva, Switzerland. Ecol. Indicat. 9:540-547. [Google Scholar]
- 26.Hartz, A., M. Cuvelier, K. Nowosielski, T. D. Bonilla, M. Green, N. Esiobu, D. S. McCorquodale, and A. Rogersond. 2008. Survival potential of Escherichia coli and enterococci in subtropical beach sand: implications for water quality managers. J. Environ. Qual. 37:898-905. [DOI] [PubMed] [Google Scholar]
- 27.Heaney, C. D., E. Sams, S. Wing, S. Marshall, K. Brenner, A. P. Dufour, and T. J. Wade. 2009. Contact with beach sand among beachgoers and risk of illness. Am. J. Epidemiol. 170:164-172. [DOI] [PubMed] [Google Scholar]
- 28.Ishii, S., and M. J. Sadowsky. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ. 23:101-108. [DOI] [PubMed] [Google Scholar]
- 29.Ishii, K., M. Mussmann, B. J. MacGregor, and R. Amann. 2004. An improved fluorescence in situ hybridization protocol for the identification of Bacteria and Archaea in marine sediments. FEMS Microbiol. Ecol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 30.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay, D., J. M. Fleisher, R. L. Salmon, F. Jones, M. D. Wyer, A. F. Godfree, J. Zelenauch, and R. Shore. 1994. Predicting likelihood of gastroenteritis from sea bathing: results from randomized exposure. Lancet 344:905-909. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J. Y., S. H. Kim, N. H. Kwon, W. K. Bae, J. Y. Lim, H. C. Koo, J. M. Kim, K. M. Noh, W. K. Jung, K. T. Park, and Y. H. Park. 2005. Isolation and identification of Escherichia coli O157:H7 using different detection methods and molecular determination by multiplex PCR and RAPD. J. Vet. Sci. 6:7-19. [PubMed] [Google Scholar]
- 33.Korajkic, A., B. D. Badgley, M. J. Brownell, and V. J. Harwood. 2009. Application of microbial source tracking methods in a Gulf of Mexico field setting. J. Appl. Microbiol. 107:1518-1527. [DOI] [PubMed] [Google Scholar]
- 34.LaLiberte, P., and D. J. Grimes. 1982. Survival of Escherichia coli in lake bottom sediment. Appl. Environ. Microbiol. 43:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Fevre, N. M., and G. D. Lewis. 2003. The role of resuspension in enterococci distribution in water at an urban beach. Water Sci. Technol. 47:205-210. [PubMed] [Google Scholar]
- 36.Luna, G. M., E. Manini, and R. Danovaro. 2002. Large fraction of dead and inactive bacteria in coastal marine sediments: comparison of protocols for determination and ecological significance. Appl. Environ. Microbiol. 68:3509-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marino, R. P., and J. J. Gannon. 1991. Survival of fecal coliforms and fecal streptococci in storm drain sediment. Water Res. 25:1089-1098. [Google Scholar]
- 38.McArdle, B. H., and M. J. Anderson. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290-297. [Google Scholar]
- 39.Na, S. H., K. Miyanaga, H. Unno, and Y. Tanji. 2006. The survival response of Escherichia coli K-12 in a natural environment. Appl. Microbiol. Biotechnol. 72:386-392. [DOI] [PubMed] [Google Scholar]
- 40.Noble, R. T., D. F. Moore, M. K. Leecasterc, C. D. McGeed, and S. B. Weisberg. 2003. Comparison of total coliform, fecal coliform, and Enterococcus bacterial indicator response for ocean recreational water quality testing. Water Res. 37:1637-1643. [DOI] [PubMed] [Google Scholar]
- 41.Obiri-Danso, K., and K. Jones. 2000. Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae, and fecal indicators in three EU recognized bathing waters in north west England. Water Res. 34:519-527. [Google Scholar]
- 42.Oliver, J. D. 2000. The public health significance of viable but nonculturable bacteria, p. 277-300. In R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, DC.
- 43.Ortega, C., H. M. Solo-Gabriele, A. Abdelzaher, M. Wright, Y. Deng, and L. M. Stark. 2009. Correlations between microbial indicators, pathogens, and environmental factors in a subtropical estuary. Mar. Pollut. Bull. 58:1374-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes, p. 207-226. In J. Paul (ed.), Methods in microbiology. Academic Press, Inc., San Diego, CA.
- 45.Pianetti, A., F. Bruscolini, L. Sabatini, and P. Colantoni. 2004. Microbial characteristics of marine sediments in bathing area along Pesaro-Gabicce coast (Italy): a preliminary study. J. Appl. Microbiol. 97:682-689. [DOI] [PubMed] [Google Scholar]
- 46.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Power, M. L., J. Littlefield-Wyer, D. M. Gordon, D. A. Veal, and M. B. Slade. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631-640. [DOI] [PubMed] [Google Scholar]
- 49.Pusceddu, A., C. Fiordelmondo, P. Polymenakou, T. Polychronaki, A. Tselepides, and R. Danovaro. 2005. Effects of bottom trawling on the quantity and biochemical composition of organic matter in coastal marine sediments (Thermaikos Gulf, northwestern Aegean Sea). Cont. Shelf Res. 25:2491-2505. [Google Scholar]
- 50.Pusceddu, A., S. Fraschetti, S. Mirto, M. Holmer, and R. Danovaro. 2007. Effects of intensive mariculture on sediment biochemistry. Ecol. Appl. 17:1366-1378. [DOI] [PubMed] [Google Scholar]
- 51.Pusceddu, A., A. Dell'Anno, M. Fabiano, and R. Danovaro. 2009. Quantity and bioavailability of sediment organic matter as signatures of benthic trophic status. Mar. Ecol. Prog. Ser. 375:41-52. [Google Scholar]
- 52.Ramos, N. L., M. L. Saayman, T. A. Chapman, J. R. Tucker, H. V. Smith, J. Faoagali, J. C. Chin, A. Brauner, and M. Katouli. 2010. Genetic relatedness and virulence gene profiles of Escherichia coli strains isolated from septicemic and uroseptic patients. Eur. J. Clin. Microbiol. Infect. Dis. 29:15-23. [DOI] [PubMed] [Google Scholar]
- 53.Regnault, B., S. Martin-Delautre, M. Lejay-Collin, M. Lefevre, and P. A. D. Grimont. 2000. Oligonucleotide probe for the visualization of Escherichia coli/Escherichia fergusonii cells by in situ hybridization: specificity and potential application. Res. Microbiol. 151:521-533. [DOI] [PubMed] [Google Scholar]
- 54.Regua-Mangia, A. H., J. R. C. Andrade, A. G. M. Gonzales, V. Zahner, A. M. F. Cerqueira, and L. M. Teixeira. 2008. Genetic relatedness of non-motile variant O157 enteropathogenic Escherichia coli (EPEC) strain and E. coli strains belonging to pathogenic related groups. Microbiol. Res. 163:225-233. [DOI] [PubMed] [Google Scholar]
- 55.Romprè, A., P. Servais, J. Baudart, M. R. de-Roubin, and P. Laurent. 2002. Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J. Microbiol. Methods 49:31-54. [DOI] [PubMed] [Google Scholar]
- 56.Roslev, P., L. A. Bjergbaek, and M. Hesselsoe. 2004. Effect of oxygen on survival of fecal pollution indicators in drinking water. J. Appl. Microbiol. 96:938-945. [DOI] [PubMed] [Google Scholar]
- 57.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect. 5:449-456. [DOI] [PubMed] [Google Scholar]
- 58.Scott, T. M., S. Parveen, K. M. Portier, J. B. Rose, M. L. Tamplin, S. R. Farrah, A. Koo, and J. Lukasik. 2003. Geographical variation in ribotype profiles of Escherichia coli isolates from humans, swine, poultry, beef, and dairy cattle in Florida. Appl. Environ. Microbiol. 69:1089-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sekar, R., A. Pernthaler, J. Pernthaler, F. Warnecke, T. Posch, and R. Amann. 2003. An improved protocol for quantification of freshwater actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Underwood, A. J. 1997. Experiments in ecology: their logical design and interpretation using analysis of variance. Cambridge University Press, Cambridge, United Kingdom.
- 61.Vezzulli, L., E. Pezzati, M. Moreno, M. Fabiano, L. Pane, C. Pruzzo, and the VibrioSea Consortium. 2009. Benthic ecology of Vibrio spp. and pathogenic Vibrio species in a coastal Mediterranean environment (La Spezia Gulf, Italy). Microb. Ecol. 58:808-818. [DOI] [PubMed] [Google Scholar]
- 62.Wade, T. J., N. Pai, J. N. S. Eisenberg, and J. M. Colford, Jr. 2003. Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ. Health Perspect. 111:1102-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walk, S. T., E. W. Alm, L. M. Calhoun, M. J. Mladonicky, and T. S. Whittam. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274-2288. [DOI] [PubMed] [Google Scholar]
- 64.Whitman, R. L., and M. B. Nevers. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 69:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamahara, K. M., S. P. Walters, and A. B. Boehm. 2009. Growth of enterococci in unaltered, unseeded beach sands subjected to tidal wetting. Appl. Environ. Microbiol. 75:1517-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.