Abstract
The presence of Campylobacter spp. was investigated in 41 Antarctic fur seals (Arctocephalus gazella) and 9 Weddell seals (Leptonychotes weddellii) at Deception Island, Antarctica. Infections were encountered in six Antarctic fur seals. The isolates, the first reported from marine mammals in the Antarctic region, were identified as Campylobacter insulaenigrae and Campylobacter lari.
The Antarctic and sub-Antarctic regions are often regarded as pristine landscapes, unaffected by human activity. A limited number of surveys have been carried out to investigate the possible occurrence of zoonotic enteropathogens and if certain bacteria could be used as tools for evaluating biological pollution in this area (4, 11). In the case of Campylobacter species, there have been only three reports in the literature, but in all of them Campylobacter was isolated from marine seabirds but not from marine mammals. Campylobacter jejuni was isolated in Antarctic and sub-Antarctic areas from Macaroni penguins (Eudyptes chrysolophus) (4), and Campylobacter lari was isolated from Brown skuas, South Polar skuas, and Adelie penguins (2, 11).
Reports of Campylobacter species isolated from marine mammals are rare. Campylobacter insulaenigrae was isolated from three harbor seals (Phoca vitulina) and a harbor porpoise (Phocoena phocoena) in Scotland (7). The isolation of C. jejuni, C. lari, and an unknown Campylobacter species from juvenile northern elephant seals (Mirounga angustirostris) in California was also reported (22). Finally, 71 isolates of C. insulaenigrae and 1 isolate similar to but distinct from both Campylobacter upsaliensis and Campylobacter helveticus were isolated from northern elephant seals in California (23). In the South Georgia Archipelago, fecal swabs were taken from 206 Antarctic fur seal pups, but no isolates could be obtained (4). In this study, we successfully isolated C. lari from 7.3% of Antarctic fur seals (Arctocephalus gazella) sampled and C. insulaenigrae from a further 7.3%. On the other hand, Campylobacter was not detected in the nine Weddell seals (Leptonychotes weddellii) sampled. To our knowledge, this is the first report on the isolation of C. lari and C. insulaenigrae from marine mammals in the Antarctic region.
Fieldwork was conducted at Deception Island (latitude of 62°58′S and longitude of 60°40′W), in the South Shetland Islands. During January to February 2007, Antarctic fur seals (Arctocephalus gazella) and Weddell seals (Leptonychotes weddellii) were captured and fecal samples were collected by insertion of sterile cotton wool swabs into the rectum of the marine mammals. A total of 41 Antarctic fur seals and 9 Weddell seals were sampled. The distribution by ages was of 7 adults (over 4 years of age with breeding activity), 19 subadults (2 to 4 years of age), and 15 juvenile Antarctic fur seals (less than 2 years of age), and 8 adult Weddell seals and 1 juvenile. All animals presented a good body condition and showed no symptoms at the time of sampling.
Three swabs were taken from each animal and were placed in FBP medium (8) with 0.5% active charcoal (Sigma Ltd.), Amies transport medium with charcoal, and Cary Blair transport medium, respectively. All samples were kept at +4 to 8°C until culture in the lab. The number of days between sampling and cultivation varied from 96 to 124 days, with a median value of 105 days.
Each swab was placed in 10 ml of Campylobacter enrichment broth (Lab M) with 5% laked horse blood and CAT supplement (cefoperazone [8 μg/ml], teicoplanin [4 μg/ml], and amphotericin B [10 μg/ml]) at 37°C. The broth was incubated at 37°C for 48 h and 5 days in 3.5-liter anaerobic containers using CampyGen sachets (Oxoid), before an aliquot of 100 μl was plated onto CAT agar and the plates were incubated at 37°C for 72 h in a microaerobic atmosphere. In addition, a 47-mm-diameter cellulose membrane with 0.60-μm pores was placed on the surface of an anaerobe agar base (Oxoid) with 5% laked horse blood. Eight to 10 drops of enrichment broth (200 μl) were placed onto the surface of the membrane. The membrane was left for 20 to 30 min on the agar surface at room temperature until all of the fluid had passed through (20). The plates were incubated as described above, but for 5 days to isolate the less common, slower growing species.
Isolates were examined by dark-field microscopy to determine morphology and motility and tested to determine whether oxidase was produced. For each sample, five isolates from each of the solid media that had typical morphology and motility and for which the oxidase test was positive were frozen at −80°C in FBP medium (8) until they were tested by phenotypic and genotypic methods.
Original Campylobacter identification was done by Gram staining, catalase activity, hippurate hydrolysis, ability to hydrolyze indoxyl acetate, urease activity, H2S production on triple-sugar iron slants, growth at 25°C and 42°C in a microaerophilc environment, growth at 37°C in an aerobic atmosphere, and agglutination with Microscreen latex (Microgen, Camberley, United Kingdom).
No differences between the strains were observed in any of the phenotypic tests used. All isolates showed a Gram-negative, slender, curved, seagull wing-like morphology under light microscopy and positive reactions in the catalase test. They were negative for hippurate and indoxyl acetate hydrolysis and urease and did not show H2S production. In addition, they grew at 42°C but did not grow at 25°C or 37°C in an aerobic atmosphere. Finally, all of them were positive in the agglutination test.
Because phenotypic results commonly lead to misidentification of Campylobacter species, it is recommended that a molecular method be included in the identification scheme for Campylobacter (5, 15). Identification of the isolates was performed using 16S rRNA gene PCR and sequence analysis (15, 21). Forward and reverse conserved 16S rRNA eubacterial primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were used to amplify the 16S rRNA according to the protocol described by Jang et al. (9). Forward and reverse sequencing reactions were performed by the Laboratorio Central de Veterinaria's DNA sequencing facility (LCV Algete, Madrid, Spain). Three strains were identified as C. lari and the other three as C. insulaenigrae based on both forward and reverse sequence analysis.
Molecular characterization of strains was carried out using a combination of pulsed-field gel electrophoresis (PFGE) using KpnI enzyme and multilocus sequence typing (MLST). Preparation of intact Campylobacter DNA for PFGE was performed following the Pulsenet protocol (17, 24). PFGE for the restriction enzyme KpnI (Takara, Conda, Spain) was performed following the protocol described by Ribot et al. (17). DNA fragments were resolved on 0.9% Seakem Gold agarose gels (Iberlabo, Spain) with a Bio-Rad CHEF DRIII system (Bio-Rad, Spain) at 14°C and 6 V/cm. Electrophoresis was carried out for 22 h with pulse times ramping from 4 s to 20 s. The fingerprinting experiments were analyzed using the InfoQuest FP software (Bio-Rad, Spain), and the dendrogram was constructed using the unweighted-pair group method using average linkages (UPGMA).
MLST of C. lari strains was performed as described by Miller et al. (13). In the case of C. insulaenigrae strains, MLST was performed following the protocol described by Stoddard et al. (23). All amplicons were sequenced by the Sequencing Service of the Instituto de Salud Carlos III (Madrid, Spain). Sequence data were collated, and alleles were assigned using the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/). Novel alleles and sequence types were submitted for allele and sequence type (ST) designations when appropriate.
Regarding the age distribution of animals, C. lari was isolated from 1 of 7 adult (14.3%), 1 of 19 subadult (5.3%), and 1 of 15 juvenile (6.6%) Antarctic fur seals. C. insulaenigrae was isolated from 1 of 7 adults (14.3%) and 2 of 19 of subadults (10.5%) but not from juvenile animals (Table 1). All strains were obtained from the swabs kept in FBP transport medium.
TABLE 1.
Source of Campylobacter isolates
| Animal | Age category and sex | Date (mo/day/yr) of: |
Campylobacter sp. and isolate no. | |
|---|---|---|---|---|
| Sampling | Culture | |||
| L 06/56 | Adult male | 2/15/07 | 5/30/07 | C. insulaenigrae FR-07 |
| L 06/78 | Subadult male | 2/15/07 | 5/30/07 | C. insulaenigrae FR-15 |
| L 06/102 | Subadult male | 2/22/07 | 5/30/07 | C. lari FR-28 |
| L 06/134 | Juvenile male | 2/21/07 | 5/30/07 | C. lari FR-36 |
| L 06/146 | Subadult male | 2/22/07 | 5/30/07 | C. insulaenigrae FR-38 |
| L 06/48 | Adult male | 2/22/07 | 5/30/07 | C. lari FR-48 |
Campylobacter is very sensitive to excessive amounts of oxygen and has little capacity to survive in the environment. It is therefore possible that the prevalence of Campylobacter species in Antarctic fur seals is greater than that obtained in our survey and that we have isolated more-resistant strains with a larger ability to survive a prolonged transport. Nevertheless, we think that the freezing medium described by Gorman and Adley (8) modified by the addition of 0.5% of activated charcoal is a very good transport medium since the bacteria remained viable for 3 months at refrigeration temperature, whereas they did not survive in the transport media routinely used for the preservation of fecal samples such as Amies and Cary Blair media.
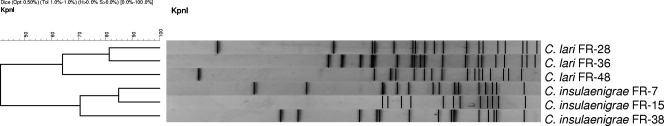
PFGE is a useful tool for conducting epidemiological studies of Campylobacter species. We used digestion with KpnI because it has been reported to have greater power of discrimination than digestion with SmaI (16). All isolates showed very different patterns (Fig. 1), indicating different sources of infection and circulation of different clones on Deception Island. These data were confirmed by the results of MLST, in which each strain belonged to a different ST, none of which had been previously reported. We submitted to the MLST database 12 new sequences of alleles tested for C. insulaenigrae and 10 new sequences of C. lari obtained (Table 2).
FIG. 1.
UPGMA dendrogram of PFGE profiles.
TABLE 2.
Alelle numbers and sequence types of Campylobacter isolates
| Species and isolate no. | ST | Allele no.a |
||||||
|---|---|---|---|---|---|---|---|---|
| aspA or adk | atpA | glnA | glyA | pgi | pgm | tkt | ||
| C. insulaenigrae | ||||||||
| FR-7 | 41 | 2 (aspA) | 16* | 12* | 2 | 15* | 15* | 11* |
| FR-15 | 42 | 4 (aspA) | 10 | 11* | 12* | 14* | 15* | 12* |
| FR-38 | 43 | 7 (aspA) | 17* | 11* | 13* | 14* | 3 | 13* |
| C. lari | ||||||||
| FR-28 | 17 | 52* (adk) | 57* | 2 | 50* | 56* | 51* | 31* |
| FR-36 | 16 | 52* (adk) | 57* | 2 | 2 | 56* | 52* | 31* |
| FR-48 | 18 | 53* (adk) | 58* | 1 | 2 | 57* | 52* | 32* |
Asterisks indicate new alleles.
The introduction of C. lari in the Antarctic fur seal colonies may have occurred through seabirds. C. lari has been isolated from Adelie penguins (Pygoscelis adeliae), kelp gull (Larus dominicanus), Brown skuas (Stercorarius antarctica lonnbergi), and South Polar skuas (Stercorarius maccormicki) in Hope Bay (11) and in the Antarctic Peninsula (2). Gulls can travel between South America and Antarctica and are potential carriers of enteric pathogens (1). Thus, C. lari has been isolated from kelp gulls in southern Chile (6). Also, South Polar skuas have been reported in Greenland and the Aleutian Islands and Brown skuas move around the Antarctic coast. Therefore, it is possible that these birds acquire infectious organisms when they move to areas with high levels of human activity. These birds have been reported on Deception Island (10), and it is common to find skuas and giant petrels on beaches where Antarctic fur seal colonies rest. The carrier birds could eliminate Campylobacter and pollute these areas. Alternatively, these birds could be occasional prey for Antarctic fur seals.
C. insulaenigrae is a new Campylobacter species whose host range might be restricted to marine mammals (23). It could be hypothesized that C. insulaenigrae evolved from C. lari based on the presence of both species in sea lions and their sharing other characteristics such as the absence of the citrate synthase gene (23). In addition, considering that C. insulaenigrae has not been isolated from seabirds or shellfish and the migration ranges of sea lions are generally not very large, Antarctic fur seals could have been initially infected with C. lari, and subsequently this species has evolved, adapting to mammals. Alternatively both species could share an ancestor and have adapted to different hosts.
The Antarctic fur seals captured showed no weight loss, diarrhea, or other symptoms at the time of sampling. However, due to the nature of our study, it is not possible to know whether the animal had been ill before the time of collection and was subsequently a carrier. Taking into account previous reports (7, 23) and our results, pinnipeds could possibly act as reservoir of C. insulaenigrae.
The presence of Campylobacter in Antarctic fur seals could also be important due to the zoonotic potential of both species (5, 12, 18, 19). Therefore, researchers should continue to exercise caution when working with these animals. In addition, C. lari has been involved in waterborne outbreaks (3) and some reports have identified this species as the most frequently isolated from surface water (25). Most of the Antarctic stations' catchwater from lakes generated by meltwater and the water treatment cannot be accomplished by chemical products to prevent marine pollution. In general, water is not treated or is treated only by filtration and UV light. Antarctic fur seals can nevertheless pollute the water of these lakes and/or infect other species such as penguins and other birds, which in turn could also act as a source of infection for humans. Furthermore, Obiri-Danso et al. (14) have reported that C. lari survives for longer in surface waters than C. jejuni and Campylobacter coli, so it would have a greater chance of surviving the water treatment. Finally, in case of infection, the therapy may be complicated because in many of the stations there are only basic medical services.
In summary, we describe here the first isolation and characterization of two species of Campylobacter, C. lari and C. insulaenigrae, from Antarctic fur seals. Further studies are needed to determine the prevalence of Campylobacter spp. in Antarctic pinnipeds, the possible sources of infection and if the presence of Campylobacter in marine mammals could be a risk for human illness or could be a result of microbial pollution associated with human activity.
Acknowledgments
This work was funded by the Spanish Ministry of Education and Science (CGL-2004-22025-E/ANT and CGL-2005-25073-E/ANT).
We thank the military personnel at the Spanish Antarctic Base “Gabriel de Castilla” for their help and assistance and the Marine Technology Unit (CSIC) and the Spanish Navy's Oceanographic Research Ship “Las Palmas” for their help, logistics, and transport.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Abulreesh, H. H., R. Goulder, and G. W. Scott. 2007. Wild birds and human pathogens in the context of ringing and migration. Ringing Migration 23:193-200. [Google Scholar]
- 2.Bonnedahl, J., T. Broman, S. Waldenström, H. Palmgren, T. Niskanen, and B. Olsen. 2005. In search of human-associated bacterial pathogens in Antarctic wildlife: report from six penguin colonies visited by tourists. Ambio 34:430-432. [PubMed] [Google Scholar]
- 3.Broczyk, A., S. Thompson, D. Smith, and H. Lior. 1987. Water-borne outbreak of Campylobacter laridis-associated gastroenteritis. Lancet i:164-165. [DOI] [PubMed] [Google Scholar]
- 4.Broman, T., S. Bergström, S. L. W. On, H. Palmgren, D. J. McCafferty, M. Sellin, and B. Olsen. 2000. Isolation and characterization of Campylobacter jejuni subsp. jejuni from Macaroni penguins (Eudyptes chrysoslophus) in the subantarctic region. Appl. Environ. Microbiol. 66:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, K., V. Gürtler, J. Montgomery, M. Fraenkel, B. C. Mayall, and M. L. Grayson. 2007. Campylobacter insulaenigrae causing septicaemia and enteritis. J. Med. Microbiol. 56:1565-1567. [DOI] [PubMed] [Google Scholar]
- 6.Fernández, H., W. Gesche, A. Montefusco, and R. Schlatter. 1996. Wild birds as reservoir of thermophilic Campylobacter species in southern Chile. 1996. Mem. Inst. Oswaldo Cruz 9:699-700. [DOI] [PubMed] [Google Scholar]
- 7.Foster, G., B. Holmes, A. G. Steigerwalt, P. A. Lawson, P. Thorne, D. E. Byrer, H. M. Ross, J. Xerry, P. M. Thompson, and M. D. Collins. 2004. Campylobacter insulaenigrae sp. nov., isolated from marine mammals. Int. J. Syst. Evol. Microbiol. 54:2369-2373. [DOI] [PubMed] [Google Scholar]
- 8.Gorman, R., and C. C. Adley. 2004. An evaluation of five preservation techniques and conventional freezing temperatures of −20°C and −85°C for long-term preservation of Campylobacter jejuni. Lett. Appl. Microbiol. 38:306-310. [DOI] [PubMed] [Google Scholar]
- 9.Jang, S. S., J. M. Donahue, A. B. Arata, J. Goris, L. M. Hansen, D. L. Earley, P. A. Vandamme, P. J. Timoney, and D. C. Hirsh. 2001. Taylorella asinigenitalis sp. nov., a bacterium isolated from the genital tract of male donkeys (Equus asinus). Int. J. Syst. Evol. Microbiol. 51:971-976. [DOI] [PubMed] [Google Scholar]
- 10.Kendall, K. A., H. A. Ruhl, and R. C. Wilson. 2003. Distribution and abundance of marine birds and pinniped populations within Port Foster, Deception Island, Antarctica. Deep Sea Res. II 50:1883-1888. [Google Scholar]
- 11.Leotta, G., G. Vigo, and G. Giacoboni. 2006. Isolation of Campylobacter lari from seabirds in Hope Bay, Antarctica. Pol. Polar Res. 27:303-308. [Google Scholar]
- 12.Martinot, M., B. Jaulhac, R. Moog, S. De Martino, P. Kehrli, H. Monteil, and Y. Piemont. 2001. Campylobacter lari bacteremia. Clin. Microbiol. Infect. 7:96-97. [DOI] [PubMed] [Google Scholar]
- 13.Miller, W. G., S. L. W. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obiri-Danso, K., N. Paul, and K. Jones. 2001. The effects of UVB and temperature on the survival of natural populations and pure cultures of Campylobacter jejuni, Camp. coli, Camp. lari and urease-positive thermophilic campylobacters (UPTC) in surface waters. J. Appl. Microbiol. 90:256-267. [DOI] [PubMed] [Google Scholar]
- 15.On, S. L. 1996. Identification methods for campylobacters, helicobacters, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.On, S. L. W., E. M. Nielsen, J. Engberg, and M. Madsen. 1998. Validity of Sma-defined genotypes of Campylobacter jejuni examined by SalI, KpnI and BamHI polymorphisms: evidence of identical clones infecting humans, poultry and cattle. Epidemiol. Infect. 120:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosef, O., G. Johnsen, A. Stølan, and H. Klaeboe. 2008. Similarity of Campylobacter lari among human, animal, and water isolates in Norway. Foodborne Pathog. Dis. 5:33-39. [DOI] [PubMed] [Google Scholar]
- 19.Soderstrom, C., C. Schalen, and M. Walder. 1991. Septicemia caused by an unusual Campylobacter species (C. laridis and C. mucosalis). Scand. J. Infect. Dis. 23:369-371. [DOI] [PubMed] [Google Scholar]
- 20.Steele, T. W., and S. N. McDermott. 1984. The use of membrane filters applied directly to the surface of agar plates for the isolation of C. jejuni from faeces. Pathology 16:263-265. [DOI] [PubMed] [Google Scholar]
- 21.Steinhauserova, I., J. Ceskova, K. Fojtikova, and I. Obrovska. 2001. Identification of thermophilic Campylobacter spp by phenotypic and molecular methods. J. Appl. Microbiol. 90:470-475. [DOI] [PubMed] [Google Scholar]
- 22.Stoddard, R. A., F. M. D. Gulland, E. R. Atwill, J. Lawrence, S. Jang, and P. A. Conrad. 2005. Salmonella and Campylobacter spp. in northern elephant seals, California. Emerg. Infect. Dis. 11:1967-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoddard, R. A., W. G. Miller, J. E. Foley, J. Lawrence, F. M. Gulland, P. A. Conrad, and B. A. Byrne. 2007. Campylobacter insulaenigrae isolates from northern elephant seals (Mirounga angustirostris) in California. Appl. Environ. Microbiol. 73:1729-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Dyke, M. I., V. K. Morton, N. L. McLellan, and P. M. Huck. 29 March 2010, posting date. The occurrence of Campylobacter in river water and waterfowl within a watershed in Southern Ontario, Canada. J. Appl. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1365-2672.2010.04730.x. [DOI] [PubMed]