Abstract
Campylobacter jejuni is a major food-borne pathogen. Despite causing enteritis in humans, it is a well-adapted intestinal microorganism in animals, hardly ever generating disease symptoms. Nevertheless, as a true microaerophilic microorganism it is still puzzling how Campylobacter cells can survive on chicken meat, the main source of human infection. In this study, we demonstrate that C. jejuni is able to withstand conditions of atmospheric oxygen tension when cocultured with Pseudomonas species, major food-spoiling bacteria that are frequently found on chicken meat in rather high numbers. Using an in vitro survival assay, interactions of 145 C. jejuni wild-type strains and field isolates from chicken meat, broiler feces, and human clinical samples with type strains and food isolates of Pseudomonas spp., Proteus mirabilis, Citrobacter freundii, Micrococcus luteus, and Enterococcus faecalis were studied. When inoculated alone or in coculture with Proteus mirabilis, Citrobacter freundii, Micrococcus luteus, or Enterococcus faecalis type strains, Campylobacter cells were able to survive ambient oxygen levels for no more than 18 h. In contrast, Campylobacter bacteria inoculated with type strains or wild-type isolates of Pseudomonas showed a prolonged aerobic survival of up to >48 h. This microbial commensalism was diverse in C. jejuni isolates from different sources; isolates from chicken meat and humans in coculture with Pseudomonas putida were able to use this survival support better than fecal isolates from broilers. Scanning electron microscopy revealed the development of fiberlike structures braiding P. putida and C. jejuni cells. Hence, it seems that microaerophilic C. jejuni is able to survive ambient atmospheric oxygen tension by metabolic commensalism with Pseudomonas spp. This bacterium-bacterium interaction might set the basis for survival of C. jejuni on chicken meat and thus be the prerequisite step in the pathway toward human infection.
Campylobacter food-borne infections are the most prevalent bacterial enteric infections in humans in industrialized and developing countries (1). It has been shown that most human infections are related to poultry meat and food produced from cattle or sheep (34, 41). Campylobacter jejuni, the species most frequently causing human disease, can be isolated from the animal intestinal tract at levels of up to 109 CFU per gram of feces and can thus be called a well-adapted intestinal microorganism (30, 37). Nevertheless, because it causes human disease as a food-borne pathogen, it has to survive outside the gut. By cross-contamination at the level of the abattoir, Campylobacter bacteria hit the meat surface and have to adapt to different environmental challenges. C. jejuni is a true microaerophilic bacterium; thus, on the one hand it requires oxygen, but on the other hand it cannot grow under normal atmospheric oxygen tension conditions (15). Despite its sensitivity to high oxygen tension in vitro, viable and culturable Campylobacter bacteria can be isolated from nonskinned chicken meat at frequencies of 104 CFU/g (9, 19). Assumptions on the mechanisms by which Campylobacter cells survive on meat surfaces are diverse, for example, by growing in biofilms, entering a “viable but nonculturable state,” or interacting with other microorganisms.
For instance, C. jejuni is able to resist protozoa digestion and can parasitize inside protozoa, e.g., Tetrahymena pyriformis (35). This mechanism provides survival in harsh environments and resistance to antimicrobial substances and thus enhances the potential for transmission. But bacterium-bacterium interaction has also been demonstrated to be of a high level of importance for intestinal survival and uptake (20). Accordingly, members of Campylobacter have been identified to initiate cellular uptake of commensal bacteria into enterocytes (14). However, a bacterial community can also mean competition, e.g., bacteriocin production by Lactobacillus salivarius that is effective against Campylobacter colonization (36).
Meat surfaces harbor numerous bacterial species (24). Some of these bacteria have adapted to this specific environmental niche and are well-known spoilage bacteria. Most relevant species belong to the family Pseudomonadaceae. But also different members of the Enterobacteriaceae can be found on meat. To date, information regarding the interaction between spoilage bacteria and pathogens is of increasing importance for public health safety measures.
Hence, experimental data on the survival of C. jejuni isolates in the presence of selected meat-spoiling bacteria were analyzed and clearly demonstrated a specific interaction with type strains and isolates of Pseudomonas putida, Pseudomonas fragi, and Pseudomonas fluorescens from chicken meat surfaces.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The type strains and isolates of Campylobacter, Pseudomonas, and meat-spoiling bacteria used in this study are listed in Table 1. All strains were kept as glycerol stock culture broths at −80°C. Strains of Pseudomonas spp. were plated on glutamate-starch-phenol red (GSP) agar (Merck, Darmstadt, Germany) according to Kielwein (16) and incubated aerobically at 25°C for 48 h for further use. Strains of C. jejuni and Campylobacter coli were grown on Campylosel agar (bioMerieux, Marcy l'Etoile, France) or on modified charcoal-cefoperazone-deoxycholate (mCCD; CM739) agar with SR155E supplement (both from Oxoid, Basingstoke, England) and grown at 42°C for 48 h under microaerobic conditions (10% CO2, 5% O2, and 85% N2) by using a microbiological culture jar equipped with a Campy gas generating kit (BR0060A; Oxoid) unless stated otherwise. All other strains used were grown on Luria-Bertani (LB) agar (Merck, Darmstadt, Germany) and incubated at the appropriate temperature and time for further use.
TABLE 1.
Bacterial strains used in this study
| Species | Strain namea | Originc |
|---|---|---|
| Campylobacter jejuni | DSM 4688T | DSMZ |
| NCTC 11168 | NCTC | |
| NCTC 12662 | NCTC | |
| Campylobacter coli | DSM 4689 | DSMZ |
| FC56 | This study | |
| Citrobacter freundii | ATCC 8090T | ATCC |
| Enterococcus faecalis | ATCC 19433T | ATCC |
| Micrococcus luteus | ATCC 4698T | ATCC |
| Proteus mirabilis | ATCC 29906T | ATCC |
| Pseudomonas chlororaphis | FP64d | This study |
| Pseudomonas fluorescens | 151/3 | This study |
| 223/2Ab | This study | |
| 224/PS3 | This study | |
| 409/1 | This study | |
| 409/2 | This study | |
| 427/PS2 | This study | |
| FP22a | This study | |
| FP56aII | This study | |
| Pseudomonas fragi | DSM 3456T | DSMZ |
| Pseudomonas putida | DSM 50198 | DSMZ |
| FP56bII | This study | |
| FP64b | This study | |
| FP64c | This study |
T, type strain.
API 20 NE identification only 63%.
ATCC, American Type Culture Collection, Rockville, MD; DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; NCTC, National Collection of Type Cultures, Colindale, London, United Kingdom.
Field isolates of C. jejuni (n = 142) have been isolated in different previous studies in Austria from 2001 to 2008.
Identification of Campylobacter jejuni.
Identification of Campylobacter jejuni isolated from chicken meat and broiler feces was done by hippurate hydrolysis according to the methods of Hwang and Ederer (12). Additionally, all isolates were further characterized by PCR analysis of the hippuricase gene hip (18) and the flagellin gene fla (40). The Campylobacter strains C. jejuni DSM 4688 and C. coli DSM 4689 from the German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ], Braunschweig, Germany) were used as positive and negative controls, respectively. For DNA extraction and detection, standard molecular biological techniques (33) were used.
flaA SVR sequence typing.
The method described by Meinersmann et al. (22) and primers and conditions reported by Dingle et al. (7) were used to amplify a 621-bp fragment of the gene flaA by PCR analysis, and the 321-bp short variable region (SVR) sequence as well as the peptide sequence encoded by the SVR nucleotide sequence were used to type six C. jejuni isolates (see Fig. 2). The sequence was determined by using the BigDye Terminator v3.1 cycle sequencing kit and an Applied Biosystems 310 ABI Prism genetic analyzer.
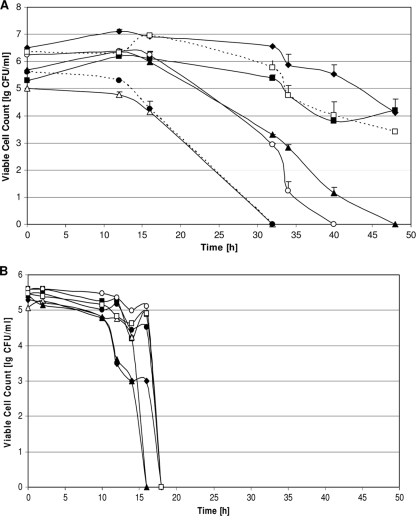
FIG. 2.
Viable cell counts of C. jejuni strains coincubated with P. putida DSM 50198. (A) C. jejuni strains DSM 4688 (filled diamonds), EuC300 (open squares), GC3 (filled squares), EuC196 (filled triangles), GC162 (open circles), EuC245 (filled circles), and GC8 (open triangles) were coincubated with P. putida DSM 50198, and some survived the aerobic growth conditions for more than 48 h. (B) When incubated without the support of P. putida, none of the C. jejuni strains could survive 18 h of aerobic incubation.
For phylogenetic analysis, sequences were aligned using ClustalW, and a dendrogram was generated using neighbor-joining analysis of MEGA v4.0 (38).
Isolation and identification of Pseudomonas spp.
Pseudomonas spp. were isolated from chicken meat surfaces by serial dilution of the meat rinsates (25 g chicken meat in 250 ml peptone water) and by plating 10−2/10−4 dilutions onto GSP agar plates. After an aerobic incubation at 25°C for 48 h, single colonies were picked and identified by the API 20 NE system (bioMerieux, Marcy l'Etoile, France).
Survival assays.
For the survival assay, suspensions of C. jejuni and the respective supporter strains were made: from a C. jejuni culture freshly grown on Campylosel agar plate, bacterial cells were harvested and suspended in 500 μl Mueller-Hinton (MH) broth (Merck, Darmstadt, Germany) to give a final optical density at 600 nm (OD600) of 0.3 as determined by the use of a Biomek plate reader (Beckman Coulter, Krefeld, Germany). A culture of the supporter strain adjusted to a final OD600 of 0.4 was achieved the same way. A starting OD600 of 0.3 and 0.4 for the Campylobacter and supporter strains, respectively, was chosen as the best from a series of different combinations of various bacterial concentrations.
The bacterial suspensions were further diluted 1:10 (vol/vol), and 10 μl was used for each assay. The survival assays were performed in 96-well microtiter plates, and each assay was performed in duplicate. For the assay, 10-μl samples each of C. jejuni and of the supporter strains to be tested, alone or in combination, were transferred to the wells and filled up to 100 μl with MH broth. The microtiter plates were incubated aerobically at 35°C for 48 h. Viable cell counts at selected time points were determined by removing 10-μl aliquots, and serial dilutions were plated on GSP agar and mCCD agar. Controls of Campylobacter isolates alone and Pseudomonas isolates alone were included in duplicate for each assay and for every isolate tested. Mean CFU values were calculated from four independent assays.
For mass survival assays of large series of Campylobacter and supporter strains, the same microtiter plate format was chosen, but instead of following the CFU over time only two time points were chosen. Ten-microliter samples collected after 32 and 48 h of aerobic incubation were plated on GSP agar and on mCCD agar and incubated (either aerobically at 25°C [GSP] or microaerobically at 42°C [mCCD], respectively). In each experiment, control samples of each Pseudomonas and Campylobacter strain were plated on GSP and mCCD agar. Thus, possible growth of Pseudomonas cells on mCCD or Campylobacter cells on GSP agar would have been detected as well as survival of culturable Campylobacter spp. at the different time points. All Campylobacter spp. were also grown without the support of Pseudomonas, and they were tested for viability at the 18-h time point.
Swarming ability.
A statistically significant set of C. jejuni isolates was tested for swarming ability performed on soft agar plates composed of Bolton broth (Oxoid, Basingstoke, England) containing 0.22% agar (LP0011; Oxoid). Colonies were harvested, suspensions were made (1:1,000 [wt/vol]) in sterile saline, and the OD600 was determined. For each suspension, aliquots of 5 μl were spotted onto the center of the soft agar plates. Swarming ability was measured in millimeter diameter growth from the center after 24 and 48 h of microaerobic incubation at 42°C and 35°C.
Scanning electron microscopy (SEM).
Campylobacter jejuni alone or in conjunction with Pseudomonas putida (DSM 50198) from microtiter plates at time point 32 h were spotted onto poly-l-lysine-coated cover slides. The cover slides were then washed three times with phosphate-buffered saline (PBS) followed by 2 washes with cacodylate buffer (0.1 M sodium cacodylate, pH 7.4) for 10 min. Samples were fixed in 2.5% glutaraldehyde in cacodylate buffer for 2 h at 4°C and washed three times in cacodylate buffer. The samples were then dehydrated with graded series of ethanol concentrations. Next, the specimens were critical point dried in a Bal-Tec CPD030 (Leica Microsystems GmbH, Wetzlar, Germany), and after being mounted, specimens were sputter coated with gold-palladium in a Polaron SC7640 (Quorum Technologies Ltd., Newhaven, United Kingdom). The samples were finally viewed on a Jeol JSM 5410LV scanning electron microscope (Jeol Ltd., Tokyo, Japan), operated at 10 to 15 kV. Samples of four independent experiments were analyzed.
Statistical analysis.
The abilities of two different Pseudomonas species isolates to improve aerobic survival of Campylobacter were compared by the chi-square test (32). Also the influence of the source of the Campylobacter isolates on their improved aerobic tolerance was statistically analyzed by the chi-square test. The significance of differences in swarming ability of Campylobacter isolates showing different aerotolerance when coincubated with Pseudomonas was assessed by comparing box plots and the 95% confidence intervals of the medians (StatGraphics; Statistical Graphics Corp., Princeton, NJ).
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under accession numbers HM581947 to HM581952.
RESULTS
Interaction between spoilage bacteria and Campylobacter jejuni.
To study the relevance of bacterial interactions for the survival of C. jejuni under aerobic growth conditions, field isolates and type strains of C. jejuni and spoilage bacteria commonly found on meat (Table 1) were coincubated and the viability was determined at different time points.
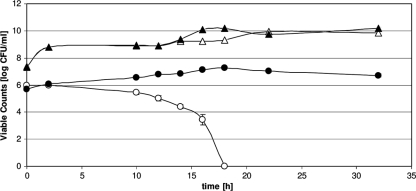
Incubation of C. jejuni type strain DSM 4688 with type strains of Proteus mirabilis, Citrobacter freundii, Micrococcus luteus, and Enterococcus faecalis did not result in a prolonged survival of the aerobic growth conditions for C. jejuni DSM 4688. However, when Pseudomonas putida DSM 50198 was present in the mixed growth culture, high numbers of viable C. jejuni DSM 4688 cells were detected even after 32 h, whereas 18 h of aerobic growth conditions sufficed to eliminate all Campylobacter cells (all type strains and all isolates used) grown alone (Fig. 1). Viability of P. putida DSM 50198 seemed unaffected by coincubation with C. jejuni. We also included two Campylobacter coli strains, DSM 4689 and one field isolate, FC56, in the study. Both strains were able to survive under the tested conditions with P. putida DSM 50198 for more than 42 h but did not survive without P. putida DSM 50198 more than 18 h.
FIG. 1.
Viable cell counts of C. jejuni and P. putida under aerobic growth conditions. C. jejuni DSM 4688 (circles) and P. putida DSM 50198 (triangles) were grown at 35°C under aerobic conditions alone (open symbols) or in combination (filled symbols). Coincubation with P. putida prolonged the survival of C. jejuni for at least 14 h. Viability of P. putida seemed unaffected by coincubation with C. jejuni. Data correspond to mean values of four independent experiments.
Survival ability is dependent on Campylobacter jejuni and Pseudomonas strains.
To analyze if the enhanced aerotolerance is a general phenomenon in C. jejuni, selected strains isolated from chicken meat were examined. When cocultivated with P. putida DSM 50198, all tested C. jejuni strains exhibited an increased aerobic tolerance but differed in the overall extent (Fig. 2A). Field isolates GC8 and EuC245 survived the aerobic incubation for only a short period of time, having lost viability after 32 h, while GC162 and EuC196 survived for at least 40 h. Field isolates GC3 and EuC300 as well as type strain DSM 4688 revealed a growth phase for the first 18 h and only a modest decrease in viability for less than 2 log10 units at time point 48 h (Fig. 2A). However, despite the fact that aerobic survival in the presence of P. putida was not of the same quality in all C. jejuni strains, all strains benefitted from the coincubation, as no C. jejuni strain was able to stay alive for more than 18 h when grown alone (Fig. 2B).
flaA SVR sequencing of these field isolates revealed that both GC8 and EuC245, the two isolates having lost viability already after 32 h, belong to the same flaA SVR group although they were isolated from very different sources, turkey meat and chicken feces. The other field isolates all belonged to different flaA SVR groups (GenBank accession no. HM581947 to HM581952).
When all the different Pseudomonas strains (Table 1) were analyzed for their capacities to enhance aerobic tolerance of C. jejuni type strain DSM 4688, all Pseudomonas strains supported the aerobic survival of C. jejuni DSM 4688 for at least 48 h.
Interestingly, the interactions between C. jejuni and the supporting Pseudomonas strains seem to be very specific. When testing either P. putida DSM 50198 or the meat isolate FP56bII for enhancement of the aerobic survival of 142 C. jejuni field isolates, two isolates exhibited aerobic survival for less than 32 h with DSM 50198. When coincubated with P. putida FP56bII, the same two isolates survived for more than 48 h (Table 2). Eight additional isolates which survived for less than 32 h with DSM 50198 showed an increased aerobic survival in the presence of FP56bII, although the viable counts were highly reduced at 32 h (reduction to 10 to 100 CFU/ml).
TABLE 2.
Survival of Campylobacter jejuni strains with P. putida strain DSM 50198 or FP56bII
| Survival of C. jejuni when coincubated with DSM 50198 | No. of C. jejuni strains with indicated result when coincubated with FP56bIIa |
Total C. jejuni strains | ||
|---|---|---|---|---|
| Survival of >48 h | Highly reduced CFU at 32 hb | Survival of <32 h | ||
| Survival of >48 h | 89 | 3 | 0 | 92 |
| Highly reduced CFU at 32 h | 22 | 6 | 0 | 28 |
| Survival of <32 h | 2 | 8 | 12 | 22 |
| Total | 113 | 17 | 12 | 142 |
Chi-square = 101.45; P = 0.000.
“Highly reduced” indicates reduction to 10 to 100 CFU/ml.
Survival ability of Campylobacter jejuni isolates from different sources.
To see whether C. jejuni isolates from different sources differed in their abilities to use Pseudomonas species as viability supporters, isolates from chicken meat, broiler feces, and human clinical samples were cocultured with P. putida DSM 50198 or with the meat isolate FP56bII. C. jejuni isolates from broiler fecal samples differed significantly (P < 0.01) in aerobic survival from chicken meat isolates and from human isolates (Table 3). Whereas all 42 C. jejuni strains isolated from human clinical samples and 45 out of 47 chicken meat isolates showed prolonged aerobic survival in the presence of P. putida DSM 50198, 20 out of 53 C. jejuni strains isolated from broiler fecal samples could not even survive 32 h. Without coincubation with a Pseudomonas supporter, no strain survived 18 h of aerobic growth conditions. When coincubated with FP56bII, the results were similar.
TABLE 3.
Survival of Campylobacter jejuni isolates from different sources cocultured with either P. putida strain DSM 50198 or FP56bII
| Source of C. jejuni | No. of C. jejuni strains with indicated result when coincubated with: |
|||
|---|---|---|---|---|
| DSM 50198a |
FP56bIIb |
|||
| Survival of ≥32 hc | Survival of <32 h | Survival of ≥32 hc | Survival of <32 h | |
| Broiler feces | 33 | 20 | 42 | 11 |
| Chicken meat | 45 | 2 | 46 | 1 |
| Humans | 42 | 0 | 42 | 0 |
Chi-square = 32.26; P < 0.01. Within “Source,” broiler feces differ from chicken meat and human stool samples.
Chi-square = 16.68; P < 0.01. Within “Source,” broiler feces differ from chicken meat and human stool samples.
Combined from “Survival of >48 h” and “Highly reduced CFU at 32 h” (see Table 2).
Scanning electron microscopy of Campylobacter jejuni coincubated with Pseudomonas putida.
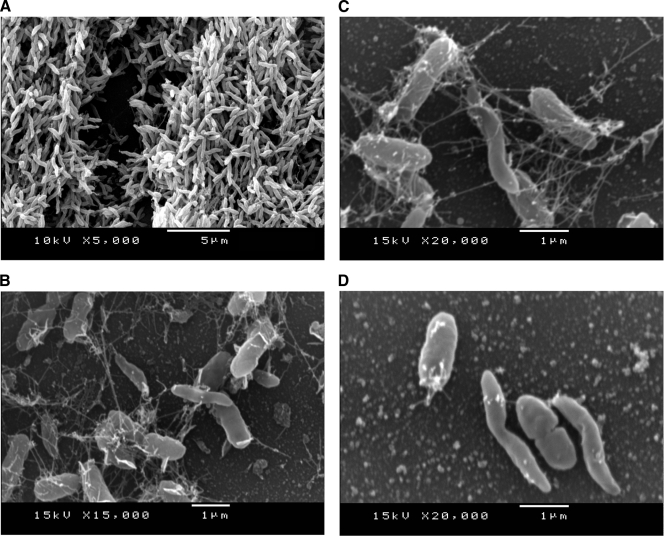
Grown under conditions of a microaerobic atmosphere, C. jejuni displayed its characteristic spiral morphology (Fig. 3A). Whereas it is known that C. jejuni cells undergo a morphological change from spiral to coccoid under normal oxygen tension (5), C. jejuni cells remained in the spiral morphology despite atmospheric oxygen tension when cocultured with P. putida (Fig. 3B to D). Moreover, C. jejuni and P. putida seem to interact by close contact and fiberlike structures, like a cobweb. The fiberlike structures appear to aggregate C. jejuni with P. putida (Fig. 3B to D).
FIG. 3.
Representative scanning electron microscopy. (A) SEM of C. jejuni DSM 4688 cultured under microaerobic conditions. (B and C) SEM of P. putida DSM 50198 and C. jejuni DSM 4688 cocultured under aerobic conditions for 32 h. C. jejuni cells are spiral shaped when cocultured with P. putida. (B to D) Cells of Pseudomonas and Campylobacter can be distinguished by the shape and thickness of the cells; fiberlike structures form cobwebs around the bacteria. (D) Both P. putida and C. jejuni seem to interact by close contact.
Swarming capability of C. jejuni isolates.
The swarming abilities of C. jejuni isolates grouped according to their previously demonstrated aerotolerance—survival for more than 48 h, survival but highly reduced CFU at 32 h, and survival for less than 32 h—were determined 24 and 48 h after plating. Whereas all isolates were able to swarm, no statistically significant difference between the groups was detected (i.e., box plots comprising the central 50% of samples as well as the 95% confidence interval [CI] for the median clearly overlapped).
DISCUSSION
C. jejuni is a major food-borne pathogen whose principle reservoir is in the intestinal tracts of warm-blooded animals. Colonization in food-producing animals has been studied and reaches up to 107 to 109 CFU per gram of intestinal content. Especially high rates of culturable Campylobacter spp. are found in the cecal and fecal contents of broilers reaching slaughter age (30), and thus chicken meat has been detected as the main source of human disease (34, 41). At slaughter, chicken meat can readily be contaminated by fecal contamination at the slaughter processing steps: scalding, defeathering, evisceration, and washing (25). Contamination of chicken meat with Campylobacter spp., determined by using cultural methods, ranges from 30 to 90% (4, 23, 29, 30). Thus, Campylobacter spp. can stay not only viable but also culturable on chicken meat surfaces despite their sensitivity to atmospheric oxygen tension. In general, Campylobacter species require 3 to 15% oxygen and 3 to 5% carbon dioxide for growth as they have a respiratory metabolism, resulting in the production of toxic reactive oxygen species, such as hydroxyl radical and hydrogen peroxide, that might lead to nucleic acid and protein injury (15, 28). Recent reports indicate that C. jejuni contains a range of enzymes involved in oxidative stress resistance. Another possible mechanism of dealing with high oxygen tension seems to be metabolic commensalism with aerobic microorganisms found on foods. This study confirms that theory by showing a beneficial effect for the aerobic survival of C. jejuni when incubated with well-adapted food-spoiling bacteria: Pseudomonas species. Several tested Pseudomonas species regularly found on chicken meat, like P. putida, P. fragi, P. fluorescens, and Pseudomonas chlororaphis in levels of up to 103 to 104 CFU/g (10, 21), were demonstrated to support growth and/or survival of C. jejuni in vitro under conditions of atmospheric oxygen tension.
Whether C. jejuni or P. putida has a higher impact on the enhanced aerotolerance of C. jejuni is still unclear. Different C. jejuni strains coincubated with P. putida DSM 50198 exhibited prolonged aerobic survival to different extents (Fig. 2A); however, it also made a difference whether C. jejuni field isolates were coincubated with either P. putida FP56bII or DSM 50198 (Tables 2 and 3).
A very good supporter was found in P. putida FP56bII, isolated from chicken meat which was already naturally contaminated with Campylobacter bacteria. The contaminating Campylobacter strain was identified as C. coli FC56 (Table 1), and also this C. coli strain had the ability to survive for a prolonged period of time, more than 48 h in combination with FP56bII and other P. putida strains (data not shown). Without the support of Pseudomonas also, this C. coli strain failed to survive aerobic growth for more than 18 h when tested under the conditions described above.
Metabolic commensalism regarding oxygen depletion is known from bacterial communities building up dental plaques. Primary plaque-colonizing species tend to be facultative anaerobes, like streptococci, paving the way for fastidious microorganisms and obligate anaerobes by reducing oxygen tension (for a review, see reference 17). The strictly anaerobic bacterium Porphyromonas gingivalis can survive oxygen levels of 20% when grown together with Fusobacterium nucleatum (6). In this case, Porphyromonas gingivalis benefits from the NADH oxidase/peroxidase activity of F. nucleatum, which creates an optimum microenvironment by locally reducing oxygen levels.
Although some studies have identified mechanisms that enable C. jejuni to cope with reactive oxygen species, like hydrogen peroxide (2, 3, 31, 39), it might be a better way for C. jejuni to sense the environment for low oxygen tension, actively move to that site (11), and make close contact with strictly aerobic bacteria like members of the Pseudomonadaceae, thus coming to a microenvironment of lowered oxygen levels.
To make close contact to other bacterial cells as seen in the SEM pictures of C. jejuni laying side by side with P. putida (Fig. 3D), motility might be advantageous. It is known that the flagellated species of Campylobacter not only move by flagella but also use the flagella apparatus to colonize surfaces and epithelial cells (27). However, motility is also important for bacterium-bacterium metabolic interaction. The formation of microcolonies consisting of Burkholderia and Pseudomonas species, which are able to metabolically interact with each other, seems to benefit from motility of Pseudomonas cells (26). Very recently, it has been documented that oxygen sensing drives motility in microbial communities (8).
Nevertheless, the swarming ability of C. jejuni isolates tested in vitro was independent of the rate of aerobic survival in the presence of P. putida. Strains of C. jejuni surviving for less than 32 h had no different swarming activity than C. jejuni strains surviving for more than 42 h under conditions of atmospheric oxygen tension in coculture with P. putida. However, as motility of Campylobacter might be affected by quorum sensing (13), the in vitro conditions used might not allow for the essential signals that trigger this special movement.
Even though all C. jejuni strains (n = 145) tested had the ability to survive under conditions of atmospheric oxygen tension for a longer period in coculture with Pseudomonas spp. than without, some strains were even able to multiply in the first 18 h and to survive for more than 48 h, while others could survive only up to 30 h. By testing C. jejuni isolated from three different sources, chicken feces, chicken meat, and human isolates, we could identify a higher percentage of C. jejuni isolates from chicken meat that were able to benefit from cocultivation with Pseudomonas spp. more effectively than isolates from chicken feces. The biological relevance of this unexpected finding might point to a possible survival mechanism for C. jejuni on chicken meat. New studies have to be designed to determine whether Pseudomonas exhibits the same supporting activity for aerobic survival of C. jejuni on chicken meat.
All isolates from human clinical samples used in this study showed a high level of ability to survive in coculture with Pseudomonas species for a prolonged period of time. Therefore, it is notable that Campylobacter species maintain these characteristics during the passage through the human host, suggesting it to be a highly important feature for the pathway of human infection.
Acknowledgments
This study was in part funded by European research project PoultryFlorGut contract no. FOOD-CT-2005-007076 in FP6.
We are grateful to acknowledge Gebhard Feierl for providing C. jejuni human isolates and Karin Siebert-Gulle and Erich Schopf for technical assistance.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 2.Atack, J. M., P. Harvey, M. A. Jones, and D. J. Kelly. 2008. The Campylobacter jejuni thiol peroxidases Tpx and Bcp both contribute to aerotolerance and peroxide-mediated stress resistance but have distinct substrate specificities. J. Bacteriol. 190:5279-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baillon, M. L., A. H. van Vliet, J. M. Ketley, C. Constantinidou, and C. W. Penn. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrang, M. E., J. S. Bailey, S. F. Altekruse, B. Patel, W. K. Shaw, Jr., R. J. Meinersmann, and P. J. Fedorka-Cray. 2007. Prevalence and numbers of Campylobacter on broiler carcasses collected at rehang and postchill in 20 U.S. processing plants. J. Food Prot. 70:1556-1560. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, S. N., E. R. Slater, A. H. L. Chamberlain, and M. R. Adams. 1994. Production and viability of coccoid forms of Campylobacter jejuni. J. Appl. Bacteriol. 77:303-307. [DOI] [PubMed] [Google Scholar]
- 6.Diaz, P. I., P. S. Zilm, and A. H. Rogers. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology 148:467-472. [DOI] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay, B. J., and G. F. Esteban. 2009. Oxygen sensing drives predictable migrations in a microbial community. Environ. Microbiol. 11:81-85. [DOI] [PubMed] [Google Scholar]
- 9.Fravalo, P., M. J. Laisney, M. O. Gillard, G. Salvat, and M. Chemaly. 2009. Campylobacter transfer from naturally contaminated chicken thighs to cutting boards is inversely related to initial load. J. Food Prot. 72:1836-1840. [DOI] [PubMed] [Google Scholar]
- 10.Ghafir, Y., B. China, K. Dierick, L. De Zutter, and G. Daube. 2008. Hygiene indicator microorganisms for selected pathogens on beef, pork, and poultry meats in Belgium. J. Food Prot. 71:35-45. [DOI] [PubMed] [Google Scholar]
- 11.Guerry, P. 2007. Campylobacter flagella: not just for motility. Trends Microbiol. 15:456-461. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, M.-N., and G. M. Ederer. 1975. Rapid hippurate hydrolysis method for presumptive identification of group B streptococci. J. Clin. Microbiol. 1:114-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon, B., K. Itoh, N. Misawa, and S. Ryu. 2003. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol. Immunol. 47:833-839. [DOI] [PubMed] [Google Scholar]
- 14.Kalischuk, L. D., G. D. Inglis, and A. G. Buret. 2009. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathog. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly, D. J. 2008. Complexity and versatility in the physiology and metabolism of Campylobacter jejuni, p. 41-61. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 16.Kielwein, G. 1969. Ein Nährboden zur selektiven Züchtung von Pseudomonaden und Aeromonaden. Arch. Lebensmittelhyg. 20:131-133. [Google Scholar]
- 17.Kuramitsu, H. K., X. He, R. Lux, M. H. Anderson, and W. Shi. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linton, D., S. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luber, P., and E. Bartelt. 2007. Enumeration of Campylobacter spp. on the surface and within chicken breast fillets. J. Appl. Microbiol. 102:313-318. [DOI] [PubMed] [Google Scholar]
- 20.Mazmanian, S. K., J. K. Round, and D. L. Kasper. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620-625. [DOI] [PubMed] [Google Scholar]
- 21.Mead, G. C., W. R. Hudson, and M. H. Hinton. 1993. Microbiological survey of five poultry processing plants in the UK. Br. Poult. Sci. 34:497-503. [DOI] [PubMed] [Google Scholar]
- 22.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meldrum, R. J., R. M. Smith, and I. G. Wilson. 2006. Three-year surveillance program examining the prevalence of Campylobacter and Salmonella in whole retail raw chicken. J. Food Prot. 69:928-931. [DOI] [PubMed] [Google Scholar]
- 24.Montville, T. J., and K. R. Matthews. 2007. Growth, survival and death of microbes in foods, p. 3-22. In M. P. Doyle and L. R. Beuchat (ed.), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC.
- 25.Nauta, M., A. Hill, H. Rosenquist, S. Brynestad, A. Fetsch, P. van der Logt, A. Fazil, B. Christensen, E. Katsma, B. Borck, and A. Havelaar. 2009. A comparison of risk assessments on Campylobacter in broiler meat. Int. J. Food Microbiol. 129:107-123. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen, A. T., T. Tolker-Nielsen, K. B. Barken, and S. Molin. 2000. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 27.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interaction. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 28.Park, S. F. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74:177-188. [DOI] [PubMed] [Google Scholar]
- 29.Pointon, A., M. Sexton, P. Dowsett, T. Saputra, A. Kiermeier, M. Lorimer, G. Holds, G. Arnold, D. Davos, B. Combs, S. Fabiansson, G. Raven, H. McKenzie, A. Chapman, and J. Sumner. 2008. A baseline survey of the microbiological quality of chicken portions and carcasses at retail in two Australian states (2005 to 2006). J. Food Prot. 71:1123-1134. [DOI] [PubMed] [Google Scholar]
- 30.Reich, F., V. Atanassova, E. Haunhorst, and G. Klein. 2008. The effects of Campylobacter numbers in caeca on the contamination of broiler carcasses with Campylobacter. Int. J. Food Microbiol. 127:116-120. [DOI] [PubMed] [Google Scholar]
- 31.Reuter, M., A. Mallett, B. M. Pearson, and A. H. van Vliet. 2010. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76:2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachs, L. 1992. Angewandte Statistik, 7th ed. Springer, Berlin, Germany.
- 33.Sambrook, J. F., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sheppard, S. K., J. F. Dallas, N. J. Strachan, M. MacRae, N. D. McCarthy, D. J. Wilson, F. J. Gormley, D. Falush, I. D. Ogden, M. C. Maiden, and K. J. Forbes. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snelling, W. J., J. P. McKenna, D. M. Lecky, and J. S. Dooley. 2005. Survival of Campylobacter jejuni in waterborne protozoa. Appl. Environ. Microbiol. 71:5560-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern, N. J., E. A. Svetoch, B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. D. Pokhilenko, V. P. Levchuk, O. E. Svetoch, and B. S. Seal. 2006. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 50:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stern, N. J., J. Reiersen, R. Lowman, J. R. Bisaillon, V. Fridriksdottir, E. Gunnarsson, K. L. Hiett, and the Campy-on-Ice Consortium. 2005. Occurrence of Campylobacter spp. in cecal contents among commercial broilers in Iceland. Foodborne Pathog. Dis. 2:82-89. [DOI] [PubMed] [Google Scholar]
- 38.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 39.Trachoo, N., J. F. Frank, and N. J. Stern. 2002. Survival of Campylobacter jejuni in biofilms isolated from chicken houses. J. Food Prot. 65:1110-1116. [DOI] [PubMed] [Google Scholar]
- 40.Wegmüller, B., J. Luethy, and U. Candrian. 1993. Direct polymerase chain reaction detection of Campylobacter jejuni and Campylobacter coli in raw milk and dairy products. Appl. Environ. Microbiol. 59:2161-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, D. J., E. Gabriel, A. J. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, P. Fearnhead, C. A. Hart, and P. J. Diggle. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]