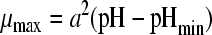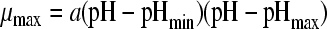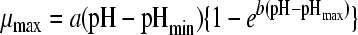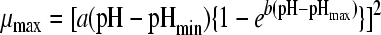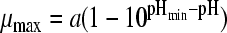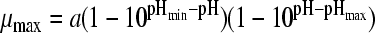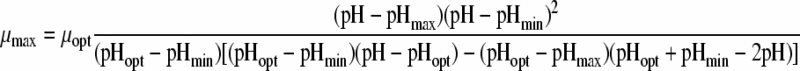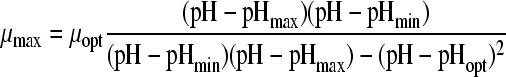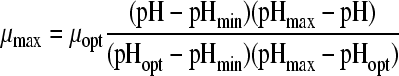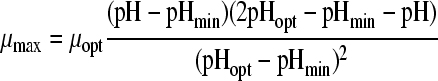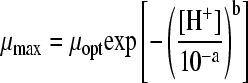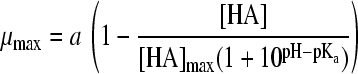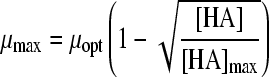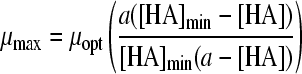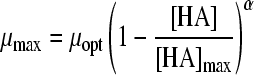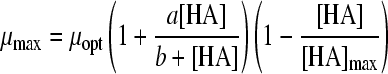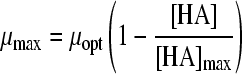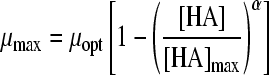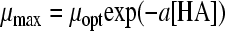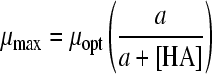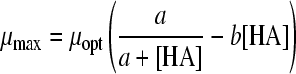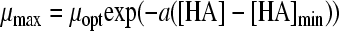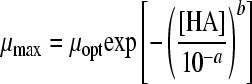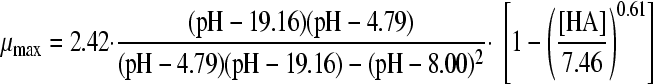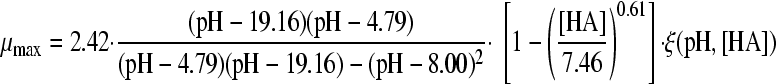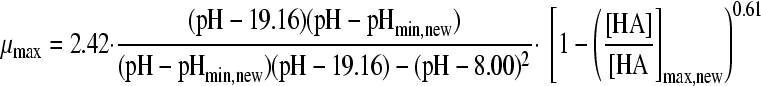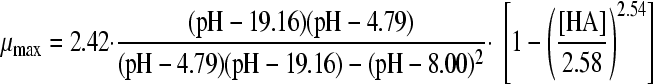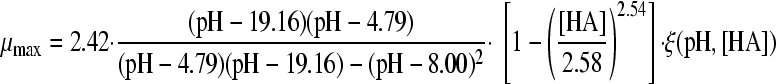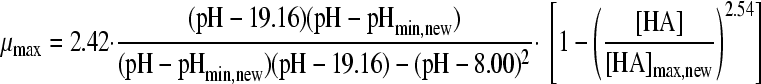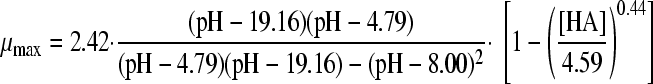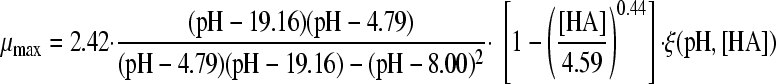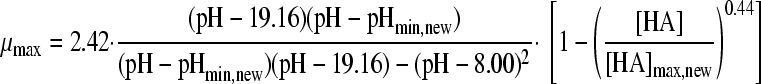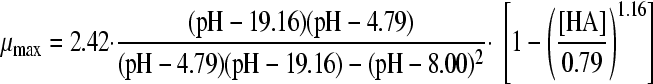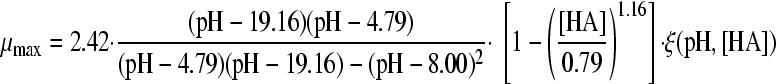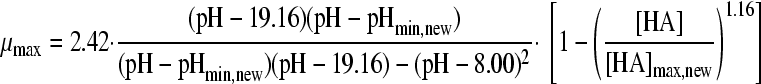Abstract
A combination of multiple hurdles to limit microbial growth is frequently applied in foods to achieve an overall level of protection. Quantification of hurdle technology aims at identifying synergistic or multiplicative effects and is still being developed. The gamma hypothesis states that inhibitory environmental factors aiming at limiting microbial growth rates combine in a multiplicative manner rather than synergistically. Its validity was tested here with respect to the use of pH and various concentrations of undissociated acids, i.e., acetic, lactic, propionic, and formic acids, to control growth of Bacillus cereus in brain heart infusion broth. The key growth parameter considered was the maximum specific growth rate, μmax, as observed by determination of optical density. A variety of models from the literature describing the effects of various pH values and undissociated acid concentrations on μmax were fitted to experimental data sets and compared based on a predefined set of selection criteria, and the best models were selected. The cardinal model developed by Rosso (for pH dependency) and the model developed by Luong (for undissociated acid) were found to provide the best fit and were combined in a gamma model with good predictive performance. The introduction of synergy factors into the models was not able to improve the quality of the prediction. On the contrary, inclusion of synergy factors led to an overestimation of the growth boundary, with the inherent possibility of leading to underestimation of the risk under the conditions tested in this research.
Consumers expect safe and sufficiently stable food within the given shelf life of a food product or component. Several growth-limiting factors, collectively referred to as hurdles, can be used to ensure food stability and safety. Examples of such hurdles are low pH, low water activity, or low temperature (12). Combining hurdles to achieve food stability and safety, known as hurdle technology, can be used to achieve an overall level of protection in food while minimizing impacts on food quality (20). When a combination of hurdles is used, generally the intensity of the hurdles may be lower, to exert a comparable preservative effect, than the intensity of those hurdles when used individually (20). Three classes of interaction can be defined when applying hurdle technology: “no interaction,” in which the effect of a combination is as expected from the response of the separate factors; “synergy,” in which the effect is greater than expected; and “antagonism,” in which the effect is less than expected (6).
Though the concept of hurdle technology is rather well established, the quantification of the combined impact of hurdles on growth of microorganisms is still being developed. One significant problem is that there are two opposite views of how antimicrobial factors combine. One view states that there are interactive effects between hurdles; when they are applied together, they give a protection significantly greater than that expected on the basis of the application of the individual hurdles (synergy). The alternative view considers that the combined effect may be complex but that there are no interactive effects culminating in synergy. The latter view is called the gamma hypothesis (41) and states that inhibitory environmental factors combine in a multiplicative manner to produce the observed overall microbial inhibition. A major benefit of models based on the gamma hypothesis is a reduction in experimental work, since growth rates and, as a result, growth boundaries can be estimated upon evaluating single hurdles rather than their various combinations. This benefit can only be realized, however, when the gamma hypothesis is valid for the combination of hurdles considered. If the hypothesis is not valid and interactive effects are present, growth boundaries are estimated wrongly, which might result in fail-safe predictions.
Over the years, the gamma hypothesis has been confirmed by several studies (16, 17, 26, 34, 38) that concluded that the combined effect of hurdles on growth rates is multiplicative rather than synergistic. Contrarily, Rödel and Scheuer (30) concluded that interaction occurs when various hurdles are combined, stressing the occurrence of synergy. Both Le Marc et al. (21) and Augustin and Carlier (5) developed a synergy model to take account of synergy occurring when hurdles are combined. It is prudent to conclude that the effect of combinations of hurdles is best evaluated on a case-by-case basis in order to ensure appropriate utility of hurdle technology approaches in establishing food designs that are stable and safe.
This research aimed to validate or falsify the gamma hypothesis for two closely related hurdles often used in the food industry: the pH level and the undissociated acid concentration ([HA]). The approach chosen was to establish an overview of models for pH and undissociated acid from the literature. Based on predefined criteria, models were then selected to construct a new gamma model without synergy factors for the various hurdle combinations. The criteria were meant to enable evaluation of the fitting performance of all individual models to select the best-performing models for inclusion in the new gamma models. Finally, the validity of the gamma hypothesis was judged by comparing the predictive performance of the newly constructed gamma models with two gamma models, including a synergy factor reported in the literature. Bacillus cereus F4810/72, relevant for both food spoilage and poisoning (14, 19), was used as the model microorganism. Maximum specific growth rates were determined by optical density measurements combined with time to detection. This method was selected after thorough investigation of three different methods to obtain parameters for growth, as recently published (8).
MATERIALS AND METHODS
Bacterial strain and culture conditions.
B. cereus F4810/72, an emetic toxin producer, was originally isolated from human vomit (33). This strain is also known as B. cereus NCTC 11143, DSM 4312, and PAL 25 (36) (Health Protection Agency Culture Collections [www.nctc.org.uk]).
Preparation of the strain and the culture conditions were as previously described (8). In short, a loopful of microorganisms was inoculated in a 500-ml Erlenmeyer flask containing 100 ml brain heart infusion (BHI) broth and incubated for 16 h at 30°C with shaking at 200 rpm. The overnight culture was standardized by resuspending the concentrated bacterial suspension in 1 ml of 1% (wt/vol) peptone physiological salt solution (PPS). The cell suspension was diluted to an optical density at 600 nm (OD600) of 0.5 in a 1% PPS solution, corresponding to approximately 109 CFU/ml. This suspension was the standardized bacterial suspension used for further experiments.
BHI broth was prepared and autoclaved according to the manufacturer's instructions and adjusted to the appropriate pH (7 or 5.5) using sterile 0.5 M sulfuric acid (H2SO4) (Riedel-de Haën, Seelze, Germany).
Experimental design.
The maximum specific growth rate, μmax (h−1), was determined using the relative rate to detection (RRD) method by measuring time to detection (TTD) using the Bioscreen C analysis system (Oy Growth Curves AB Ltd., Helsinki, Finland) (8). This method was chosen since previous research showed that compared to other methods, it resulted in more data points close to the growth boundary where possible interactive effects are expected to be present (8).
The experiments were divided into three groups: 1, testing of the pH effect using strong acids (pKa < 1); 2, testing of the undissociated acid effect using weak acids (pKa > 1) in a buffer solution at a fixed pH of 5.5; and 3, testing of both pH and undissociated acid effects using weak acids. Table 1 provides an overview of the three groups of experiments, including the acids used (acetic acid [HAc], lactic acid [HLa], propionic acid [HPr], and formic acid [HFo]) and the ranges of pH values and undissociated acid concentrations ([HA]) tested.
TABLE 1.
Acids (solutes) used for experiments and key experimental data obtained
| Exptl group(s) | Acid | Chemical formula | Acid supplier | pKa | Tested pH range | No. of bins | No. of data points (no. excludeda) | Tested conc range (mM) | No. of bins | No. of data points (no. excludeda) | Conjugated salt | Salt supplier |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Sulfuric acid | H2SO4 | Riedel-de Haën, Seelze, Germany | <1 | 4.60-7.46 | 56 | 598 (2; 0) | |||||
| 2, 3 | Acetic acid (HAc) | C2H4O2 | Merck KGaA, Darmstadt, Germany | 4.75 | 5.11-7.0 | 36 | 399 (0; 1) | 0-10 | 16 | 596 (3; 1) | Potassium acetate (KAc) | Merck KGaA, Darmstadt, Germany |
| 2, 3 | Lactic acid (HLa) | C3H6O3 | Sigma-Aldrich Chemie GmbH, Steinheim, Germany | 3.85 | 4.86-7.11 | 49 | 496 (7; 0) | 0-5 | 16 | 400 (0; 0) | Potassium lactate (KLa) | PURAC Biochem, Gorinchem, Netherlands |
| 2, 3 | Propionic acid (HPr) | C3H6O2 | Merck KGaA, Darmstadt, Germany | 4.88 | 4.67-7.02 | 54 | 599 (0; 1) | 0-10 | 17 | 558 (0; 2) | Calcium propionate (CaPr) | Riedel-de Haën, Seelze, Germany |
| 2, 3 | Formic acid (HFo) | CH2O2 | Acros Organics, New Jersey | 3.75 | 4.49-7.00 | 39 | 399 (1; 0) | 0-8 | 21 | 599 (0; 1) | Potassium formate (KFo) | Sigma-Aldrich Chemie GmbH, Steinheim, Germany |
The first number is the number of data points excluded since at t = 0, the optical density (OD) values were above the detection limit of OD = 0.2. The second number is the number of data points excluded according to the criteria in Materials and Methods.
To test the effect of the undissociated acid concentration on μmax, the pH of the BHI broth was set to 5.5 using sulfuric acid. This value was chosen for being approximately half a pH unit higher than the minimal pH to be able to observe either an increase or a decrease in the μmax. The ratio between the dissociated and undissociated forms at the set pH was calculated using the Henderson-Hasselbalch equation (equation 1).
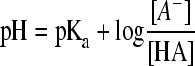 |
(1) |
where pH is the preset pH of the medium using sulfuric acid, pKa is the acid dissociation constant (acid dependent; see Table 1), [A−] is the concentration of anions, and [HA] is the concentration of the undissociated acid, which at a fixed pH has a fixed ratio. For each type of weak acid, a stock solution was prepared at pH 5.5 by adding the acid and its conjugated salt ion in the correct ratio (Table 1).
Effect of pH and undissociated acid concentrations on μmax.
For every experiment of groups 1 and 3, 20 bottles containing 50 ml BHI broth (pH 7) were pH adjusted with the selected acid, either H2SO4, HAc, HLa, HPr, or HFo. After the pH of the BHI had been set, the liquids were filter sterilized (Steritop/Steriflip; Milipore Corporation, Massachusetts). The standardized bacterial suspension was diluted 10,000-fold in pH-adjusted BHI, using a new dilution series for every pH value, aiming at an initial cell concentration of approximately 104 CFU/ml. This resulted in 20 inoculated test tubes, 1 for every pH value to be tested. The content of the tube of pH 7 was spiral plated on BHI agar plates for enumeration.
Honeycomb plates were filled and incubated as previously described (8). In short, wells were filled with 150 μl of pH-adjusted BHI, filling five wells per pH value. Every pH value was investigated in a duplicate plate. Every first well of a pH series was inoculated with 150 μl of a particular target pH value-adjusted bacterial culture, and after mixing, half of the content was transferred to the nearest well of the same pH, continuing in this manner up to the fifth well. Both honeycomb plates were incubated in a Bioscreen C system at 30°C for 3 days with continuous shaking at the medium setting. The OD600 was measured every 10 min. The OD600 data obtained from the Bioscreen system were imported into the Microsoft Excel software program for data capturing. Wells with an initial OD600 above 0.2 were removed from the data set since they were likely to have an incidental too-high inoculum level (8). For all relevant data series, the time to detection (TTD), defined as the time (h) to reach an OD600 of 0.2, was determined. For wells not reaching an OD600 of 0.2 within the time frame of the experiment, viability of bacteria was determined, and if no viable bacteria were detected, the μmax value was set to 0 h−1. For all acids to be tested, the experiment was repeated at least once. In case two experiments did not give enough information about the exact growth boundary, the experiment was repeated once more.
The TTD determined for every test condition (TTDi) was related to the TTD under optimal conditions (TTDopt), in this case at pH 7. Subsequently, the specific growth rate (μmax,i) was calculated according to equation 2.
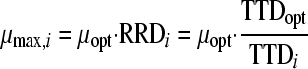 |
(2) |
The μopt value was estimated independently by plate counting and subsequent fitting of the Gompertz model to the counts (8). In assessing TTDopt and TTDi for use in equation 2, care was taken to always start with equal inoculum levels.
To test the effect of the undissociated acid concentration for the weak acids (experiments of group 2), a buffer solution was prepared. Ten bottles containing 50 ml of standardized BHI were adjusted to pH 5.5 using sulfuric acid. The acid and the conjugated salt were added to the pH-adjusted BHI in the right ratio according to equation 1, whereupon the medium was filter sterilized. The concentration ranges of the acids tested are presented in Table 1. The standardized bacterial suspension was diluted 10,000-fold in adjusted BHI, one for every undissociated acid concentration condition to be tested. The dilution with no undissociated acid present was spiral plated on BHI agar plates in duplicate for enumeration. Filling of the plates, running of the Bioscreen system, and capturing and processing of data were as described for the experiments of groups 1 and 3, except for the number of replicates within the experiments, which was four instead of two. Every experiment was repeated at least once and twice in cases where the growth boundary could not be determined from the previous experiments. To determine μmax values, equation 2 was used. TTDopt was defined as the test condition with no undissociated acid present.
For the three groups of experiments, the growth rate curves were studied in more detail. Generally, the μmax values per replicate of tested pH value or undissociated acid concentration were in the same order of magnitude, but sometimes no growth was measured for some of the replicates, while the other points showed considerable μmax values. For these cases, the data points per test conditions were divided into two groups: growth and no growth. The number of data points per group was determined, and the ratio between the numbers in the smallest and largest groups was calculated. In cases where the number of data points in the smallest group was more than 10%, the values in this group were considered to be representative for the experimental condition and were included for further analysis. In cases where the contribution was less than 10%, the 99% confidence interval of the μmax values of the largest group was calculated. If the μmax values of the smallest group were within the 99% confidence interval, the values were also included for further research; otherwise, they were excluded from further data analysis.
Model selection and performance.
Three criteria were used to select the best-fitting models: (i) the mean square error (MSE) value for the model fit should be below 0.01 to ensure a high level of fit; (ii) the standard deviations for individual model parameters should be smaller than the parameter estimates themselves, since standard deviations greater than the respective parameter estimate indicate large variation; and (iii) the model parameters should preferably have biological significance.
Secondary models for growth rate, which actually included or could be amended to include a pH term or an undissociated acid term, were selected from the literature. The pH models are summarized in Table 2, and the undissociated acid models are summarized in Table 3 . The names of the parameters were standardized to improve transparency and comparability throughout this research. All outcomes of the models were expressed as μmax values. The pH models were fitted to the μmax data of group 1, and the undissociated acid models were fitted to the μmax data of group 2. Model performance (MSE values) and parameter estimates for the two types of models are included in Tables 2 and 3, respectively.
TABLE 2.
Singular models describing the maximum specific growth rate (h−1) as a function of pH, their fitting performance (indicated by the MSE value), and the optimal parameter estimates with standard deviations when fitted to the experimental data set obtained with B. cereus F4810/72
| Model no.a | Model | Reference | No. of parameters | MSE | μopt (SE) | a (SE) | b (SE) | pHmin (SE) | pHmax (SE) | pHopt (SE) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
|
1 | 2 | 0.0765 | 1.03 (0.007) | 4.39 (0.020) | |||||
| 2 |
|
37 | 3 | 0.0074 | −0.52 (0.007) | 4.72 (0.004) | 9.02 (0.031) | ||||
| 3 |
|
37 | 4 | 0.0075 | 79.73 (446.78) | 0.01 (0.038) | 4.71 (0.005) | 9.00 (0.107) | |||
| 4 |
|
29 | 4 | 0.0257 | 152.59 (5381) | 0.002 (0.060) | 4.29 (0.027) | 9.23 (0.172) | |||
| 5 |
|
28 | 2 | 0.0208 | 2.21 (0.009) | 4.86 (0.003) | |||||
| 6 |
|
28 | 3 | 0.0208 | 2.21 (0.013) | 4.86 (0.00) | 16.11 (4.2*106) | ||||
| 7 |
|
32 | 4 | 0.0075 | 2.41 (0.007) | 4.73 (27.384) | 9.01 (0.033) | 6.87 (0.015) | |||
| 8 |
|
23, 31 | 4 | 0.0023 | 2.49 (0.018) | 4.79 (0.002) | 19.16 (1.252) | 8.00 (0.135) | |||
| 9 |
|
40 | 4 | 0.0074 | 2.38 (71884) | 4.72 (0.004) | 9.02 (0.031) | 7.10 (298016) | |||
| 10 |
|
40 | 3 | 0.0074 | 2.41 (0.007) | 4.72 (0.004) | 6.87 (0.015) | ||||
| 11 |
|
16 | 3 | 0.0097 | 2.35 (0.010) | 5.15 (0.004) | 1.08 (0.016) |
Boldface indicates model selected for further analysis.
TABLE 3.
Singular models describing the effect of undissociated acid on the maximum specific growth rate (h−1), fitting performance (indicated by MSE value), and optimal parameter estimates with their standard deviations when fitted to the experimental data set obtained with B. cereus F4810/72
| Model no.a | Model | Reference | No. of parameters | Acids | MSE | MSE total, 4 acids | μopt (SE) | [HA]max (SE) | a value (SE) | b value (SE) | α value (SE) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 |
|
28 | 2 | HAc | 0.0732 | 0.262 | 4.00 (1.139) | 1.00 (0.021) | ||||
| HLa | 0.0657 | 2.00 (4.370) | 1.06 (0.032) | |||||||||
| HPr | 0.0579 | 2.00 (0.517) | 1.04 (0.030) | |||||||||
| HFo | 0.0652 | 0.40 (1.052) | 1.06 (0.029) | |||||||||
| 13 |
|
20 | 2 | HAc | 0.0028 | 0.1143 | 1.62 (0.005) | 8.24 (0.049) | ||||
| HLa | 0.0840 | 1.91 (0.038) | 3.89 (0.110) | |||||||||
| HPr | 0.0040 | 1.48 (0.007) | 8.89 (0.054) | |||||||||
| HFo | 0.0235 | 1.73 (0.017) | 1.02 (0.014) | |||||||||
| 14 |
|
13 | 3 | HAc | 0.0006 | 0.0310 | 1.51 (0.003) | 8.65 (0.051) | −4.93 (0.067) | |||
| HLa | 0.0058 | 1.55 (0.009) | 2.52 (0.006) | 3.56 (0.048) | ||||||||
| HPr | 0.0034 | 1.51 (0.007) | 10.17 (0.128) | −3.32 (0.091) | ||||||||
| HFo | 0.0212 | 1.59 (0.017) | 0.95 (0.015) | 19.73 (23.760) | ||||||||
| 15 |
|
22 | 3 | HAc | 0.0014 | 0.0161 | 1.48 (0.004) | 15.68 (0.693) | 4.24 (0.223) | |||
| HLa | 0.0066 | 1.56 (0.010) | 2.28 (0.007) | 0.34 (0.010) | ||||||||
| HPr | 0.0045 | 1.50 (0.008) | 2.78 × 108 (2.5 × 108) | 9.40 × 107 (2.5 × 108) | ||||||||
| HFo | 0.0036 | 1.55 (0.007) | 0.77 (0.005) | 0.87 (0.017) | ||||||||
| 16 |
|
27 | 4 | HAc | 0.0004 | 0.0119 | 1.52 (0.003) | 8.20 (0.037) | −0.70 (0.016) | 2.95 (0.112) | ||
| HLa | 0.0050 | 1.50 (0.011) | 2.53 (0.007) | −2.38 (0.405) | −5.98 (0.756) | |||||||
| HPr | 0.0030 | 1.51 (0.007) | 9.39 (0.097) | −0.70 (0.030) | 0.84 (0.084) | |||||||
| HFo | 0.0035 | 1.54 (0.008) | 0.78 (0.006) | 67.38 (12797) | 275.97 (52475) | |||||||
| 17 |
|
11 | 2 | HAc | 0.0083 | 0.0587 | 1.36 (0.006) | 6.45 (0.050) | ||||
| HLa | 0.0291 | 1.75 (0.020) | 2.99 (0.035) | |||||||||
| HPr | 0.0175 | 1.33 (0.014) | 6.46 (0.087) | |||||||||
| HFo | 0.0038 | 1.57 (0.006) | 0.81 (0.003) | |||||||||
| 18 |
|
24 | 3 | HAc | 0.0014 | 0.0132 | 1.54 (0.005) | 7.46 (0.045) | 0.61 (0.006) | |||
| HLa | 0.0050 | 1.49 (0.008) | 2.58 (0.055) | 2.54 (0.007) | ||||||||
| HPr | 0.0035 | 1.51 (0.008) | 4.59 (0.034) | 0.44 (0.007) | ||||||||
| HFo | 0.0033 | 1.53 (0.007) | 0.79 (0.004) | 1.16 (0.018) | ||||||||
| 19 |
|
2 | 2 | HAc | 0.0022 | 0.1090 | 1.52 (0.004) | 0.32 (0.002) | ||||
| HLa | 0.0803 | 1.79 (0.039) | 0.59 (0.020) | |||||||||
| HPr | 0.0046 | 1.50 (0.008) | 0.34 (0.004) | |||||||||
| HFo | 0.0219 | 1.66 (0.017) | 2.37 (0.043) | |||||||||
| 20 |
|
27 | 2 | HAc | 0.0179 | 0.2249 | 1.62 (0.015) | 1.63 (0.044) | ||||
| HLa | 0.1359 | 1.72 (0.056) | 0.99 (0.076) | |||||||||
| HPr | 0.0173 | 1.55 (0.017) | 1.20 (0.037) | |||||||||
| HFo | 0.0538 | 1.64 (0.029) | 0.22 (0.010) | |||||||||
| 21 |
|
27 | 3 | HAc | 0.0011 | 0.1226 | 1.53 (0.004) | 2.80 (0.029) | 0.03 (0.0003) | |||
| HLa | 0.0638 | 1.69 (0.036) | 3.13 (0.414) | 0.12 (0.010) | ||||||||
| HPr | 0.0071 | 1.53 (0.011) | 1.75 (0.043) | 0.01 (0.000) | ||||||||
| HFo | 0.0506 | 1.63 (0.028) | 0.24 (0.011) | 0.01 (0.002) | ||||||||
| 22 |
|
39 | 3 | HAc | 0.0003 | 0.1580 | 0.13 (3016) | 8.00 (75794) | 0.31 (0.001) | |||
| HLa | 0.0207 | 0.95 | 2.00 | 0.25 (0.011) | ||||||||
| HPr | 0.1280 | 1.48 (3.7 × 106) | 0.34 (2.0 × 107) | 0.07 (2.2 × 106) | ||||||||
| HFo | 0.0090 | 0.37 | 0.75 | 1.96 (0.028) | ||||||||
| 23 |
|
16 | 3 | HAc | 0.0019 | 0.0175 | 1.49 (0.005) | −0.50 (0.003) | 1.08 (0.010) | |||
| HLa | 0.0071 | 1.44 (0.009) | −0.32 (0.002) | 4.67 (0.127) | ||||||||
| HPr | 0.0044 | 1.51 (0.009) | −0.47 (0.005) | 0.98 (0.015) | ||||||||
| HFo | 0.0041 | 1.49 (0.007) | 0.30 (0.002) | 2.16 (0.035) |
Boldface indicates model selected for further analysis.
The models for pH and undissociated acid concentration that were selected on the basis of the three criteria stated above were tested against the best performing model with one parameter less, using an F test to evaluate if the reduction of one parameter was still statistically acceptable (10). The experimental f value was tested against the 95% confidence F table value (F∞1 = 3.84). If the f value was smaller than the F table value, the F test was accepted and the model with the least number of parameters was accepted.
Evaluating gamma hypothesis validity.
The selected best-fitting models for pH and undissociated acid concentration were combined in a gamma model according to equation 3 (41):
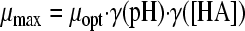 |
(3A) |
with
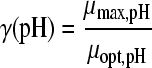 |
(3B) |
and
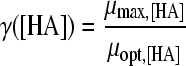 |
(3C) |
where μmax is the maximum specific growth rate under the tested condition, μopt and μopt,pH are the maximum specific growth rates in medium with pH 7 as determined by a plate count (2.42 h−1), and μopt,[HA] is the maximum specific growth rate when no undissociated acid is present in buffer solution with pH 5.5, as determined from the pH-growth rate curve for H2SO4 fitted with the model of Rosso et al. (31) (1.51 h−1); all maximum growth rates were obtained from the pH-growth rate curve and the best-fitting model of this curve. Parameter estimates derived by fitting of single models were incorporated into the gamma model, and predictions were made using the optimal fit of the growth rate curves for the combined effect of pH and undissociated acid. In total, four sets of predictions were made, one for each acid tested. These predictions were compared to the experimental data of the acids of group 3, where both pH and undissociated acid effects were present. The ratio between the dissociated and undissociated forms of the acid could be calculated from equation 3, since the pH of the broth was measured while adding the acid. The concentrations of the dissociated and undissociated acids could be calculated, since the amount and concentration of the acid added to the broth were known. The differences between predictions and experimental data were expressed as MSE values to be able to compare between the acids and, if necessary, between models.
Two gamma models, including a synergy factor, were found in the literature and compared with the newly composed gamma models combining pH and undissociated acid effects. The first synergy model was described by Le Marc et al. (21) (equation 4):
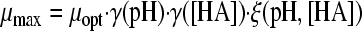 |
(4) |
in which “ξ” is the synergy factor. The second synergy model was the model of Augustin and Carlier (4, 5). This model does not include a synergy factor, but the different inhibitory factors were corrected independently for synergy by estimating new minimal growth values, which were again used in the nonsynergistic gamma model according to equations 5A, B, and C:
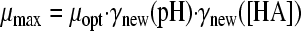 |
(5A) |
with
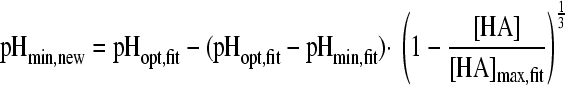 |
(5B) |
and
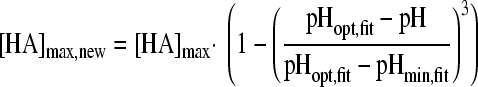 |
(5C) |
RESULTS
Effect of pH and undissociated acid concentrations on μmax.
Three groups of experiments were conducted to investigate the effect of pH (group 1), the effect of undissociated acid (group 2), and the combined effect of pH and undissociated acid (group 3). For every group of experiments and for every acid evaluated, Table 1 shows the pH range tested, the number of pH groups (bins), the number of data points in the data set generated, and the number of data points removed from the initial data set (for reasons described in Materials and Methods).
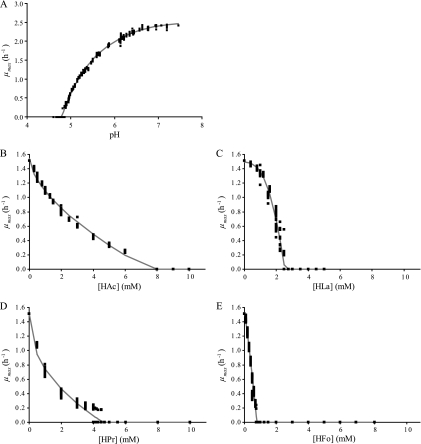
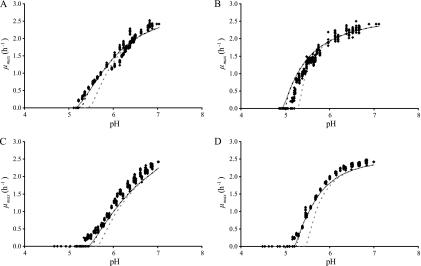
Using H2SO4 to set the pH, the lower pH boundary was pH 4.8. The experimental data for this first group of experiments are displayed in Fig. 1A. The results for the undissociated effect of HAc, HLa, HPr, and HFo in a pH 5.5 buffer solution are shown in Fig. 1B to E. For these, the measured growth boundaries were 8, 2.5, 4.7, and 0.80 mM undissociated acid, respectively.
FIG. 1.
Experimental data (squares) for the effect on μmax of pH, set using H2SO4 (A), or of the undissociated acid concentration ([HA], using HAc (B), HLa (C), HPr (D), or HFo (E); the solid gray line shows the fit of the best-fitting model for pH (model 8) and [HA] (model 18) to the data sets.
Selection of the best-fitting model to describe growth rate as a function of pH.
Eleven pH models, as displayed in Table 2, were fitted to the pH-growth rate curve of B. cereus F4810/72 cultured in BHI adjusted for pH by the addition of H2SO4. The fitting performance (expressed as MSE values) and the parameter estimates with their standard deviations are represented in Table 2, as well. Five models, namely, models 3, 4, 6, 7, and 9, were rejected, since at least one of the standard deviations of their estimated parameters was larger than the parameter estimate itself. Models 1 and 5 were not considered further, since the MSE values exceeded 0.01. Models 2 and 11 were rejected based on the inclusion of parameters without an evident biological relevance, while, additionally, model 11 was not able to fit the growth boundary since it is an asymptotic model. Of the two remaining models, models 8 and 10, model 8 of Rosso et al. (31) was considered the best model since it had the best-fitting performance (MSE = 0.0023) and included parameters that were biological relevant. Model 10 of Zwietering et al. (41), with one parameter less, met all criteria but had a slightly lower fitting performance (MSE = 0.0074). The F test was applied to investigate if the difference in MSE values between model 8 and model 10 was significant. Since f = 1,364.68 exceeds
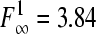 |
, model 8 with four parameters was judged to be the best model to describe the behavior of B. cereus cultured in BHI acidified by H2SO4.
Selection of the best-fitting model to describe growth in the presence of undissociated acid.
Twelve undissociated acid models (Table 3) were fitted to the undissociated acid-growth rate curves of B. cereus F4810/72 cultured in pH 5.5 buffered BHI with various concentrations of undissociated acid. The models were fitted to all four data sets: HAc, HLa, HPr, and HFo. Table 3 shows the MSE values and the parameter estimates and their standard deviations for the various models applied to all four acids. Models 12, 14, 15, 16, and 22 were rejected since the standard deviation of one or more parameters exceeded the estimated value of that parameter for at least one acid. Models 13, 17, 19, 20, and 21 were rejected since for at least one acid the MSE value exceeded the MSE value criterion of 0.01. Model 18 of Luong (MSE = 0.0132) (24) and model 23 of Lambert and Bidlas (MSE = 0.0175) (16) were the models remaining, although both include a parameter with no evident biological significance. Both models include three parameters. Model 18 of Luong was selected as the best-fitting model since it had the lowest MSE value. Although model 16 of Pasos et al. (27) had been rejected, the criteria for rejection were tested by comparing model 18 to model 16, which had a lower MSE value (MSE = 0.0119) than model 18 but a standard deviation exceeding the parameter estimate for HFo. Model 16 also contains one more parameter than model 18. The two models were tested for all four acid data sets with the F test (F∞1 = 3.84). The ƒ values for the HAc, HLa, HPr, and HFo data sets were 424, 1, 8, and −35, respectively. This indicates that for the HLa and HFo datasets, model 18 fits best, whereas for HAc and HPr, model 16 is the best to use. Model 18 of Luong was also compared with an F test to model 17 of Ghose and Tyagi (11), which was the best model with two parameters for all data sets. The sum of the MSE values for model 17 was the lowest of the two parameter models tested, but this model was initially rejected, since the MSE values exceeded the criterion of 0.01. The ƒ values were 494, 330, 445, and 79 for the respective acids, indicating that the reduced model with two parameters would not be the best choice for fitting the data, as was already concluded from the high MSE values.
Evaluating gamma hypothesis validity.
Models 8 and 18 were combined in a gamma model according to equation 3 to form the models 24A, 25A, 26A, and 27A, as presented in Table 4. In addition, the synergy models of Le Marc et al. (equation 4; Table 4, models B) (21) and Augustin and Carlier (equation 5, Table 4, models C) (4, 5) were combined into predictive models. With these three equations, the growth of B. cereus in BHI broth in the presence of weak acid was predicted, as was the growth boundary. Figure 2A, B, C, and D show the predictions of the three models for HAc, HLa, HPr, and HFo, respectively, next to the experimental data. For acetic acid and formic acid, the shape of the curves and the growth boundaries were predicted very well by the gamma model. For all four acids, the predictions of the models of μmax were lower than the experimental data for the whole curve. For lactic acid, however, the growth boundary was underestimated by 0.25 pH units, while the data points at pH 5.5 or higher were predicted correctly by the gamma model. The model of Le Marc et al. underestimated the boundary by 0.15 pH unit, and the model of Augustin and Carlier overestimated the boundary by 0.15. For all acids, the predictions by Le Marc et al. were very close to those of the gamma model, while the model of Augustin and Carlier consequently underestimated growth rates in the lower half of the pH range.
TABLE 4.
MSE values for predictions obtained with the nonsynergistic gamma model, model of Le Marc et al., and model of Augustin and Carlier for HAc, HLa, HPr, and Hfo
| Model IDa | Acid | Model | MSE | Boundary pH | |
|---|---|---|---|---|---|
| 24 | |||||
| A | HAc |
|
0.0181 | 5.19 | |
| B | HAc |
|
0.0226 | 5.30 | |
| C | HAc |
|
0.0609 | 5.54 | |
| 25 | |||||
| A | HLa |
|
0.0925 | 4.95 | |
| B | HLa |
|
0.0889 | 5.02 | |
| C | HLa |
|
0.0745 | 5.31 | |
| 26 | |||||
| A | HPr |
|
0.0355 | 5.50 | |
| B | HPr |
|
0.0384 | 5.56 | |
| C | HPr |
|
0.0814 | 5.66 | |
| 27 | |||||
| A | HFo |
|
0.0127 | 5.25 | |
| B | HFo |
|
0.0149 | 5.30 | |
| C | HFo |
|
0.0919 | 5.46 |
FIG. 2.
Experimental data (diamonds) and predictions using model 8 and model 18 for the combined effect of pH and the undissociated acid concentration on μmax without (solid gray line) or with (dashed black line) interaction factor and predictions using the model of Augustin (dashed gray line) for HAc (A), HLa (B), HPr (C), or HFo (D).
As shown in Table 4 and Fig. 2, the addition of a synergy factor according to the model of Le Marc et al. does not reduce the MSE values between predictions and experiments. On the contrary, in most cases the MSE values increased compared to those for the gamma model. For lactic acid, however, the MSE value decreased due to the addition of the synergy factors, and the growth boundary was more closely approached. The proposed model of Augustin and Carlier was also able to reduce the MSE value between predictions and experiments for the HLa data, but on the other hand the growth boundary was overestimated. For the other three acids, both synergy models lead to overestimation of the minimal growth boundaries and an increase in MSE values compared to the gamma model.
DISCUSSION
Model criteria and selection.
The model selection applied started by reviewing literature for pH models and undissociated acid models. This resulted in a selection of 11 pH models and 12 undissociated acid models. Few articles present an extended overview of available models (27, 35), and for this reason, this review and the selection procedure might be of value for predictive microbiology studies.
For all models, the parameters were standardized, and all models were expressed in the form μmax = μopt·γ(pH) or μmax = μopt·γ([HA]). Also, the RRD models (model 11 and model 23) of Lambert and Bidlas (16) were expressed as such to be able to compare MSE values between experiments and to allow fitting performance to be expressed on the same scale.
Three selection criteria were proposed for model selection. First, MSE values should be below 0.01. This value was proposed on the basis of multiple visual inspections of the fitting performance of different models with respect to the experimental data. Moreover, the MSE value of 0.01 allowed some difference between experiments and model fitting performance while significantly reducing the number of models to be considered in gamma hypothesis validation. Second, standard deviations for any parameter had to be smaller than the parameter estimates. A large standard deviation indicates that a large variation in the estimated parameter is possible, meaning that parameter estimates can be zero, which indicates overparameterization of the model. The third criterion was the inclusion of parameters without biological significance; this should be kept to a minimum. A set of criteria should be preferred over a single criterion since several aspects of model fit can be considered. The proposed criterion for the MSE value eliminated, for example, linear models, like model 1, which did not fit the experimental data well, whereas the model would not be eliminated based only on the standard deviations of parameter estimates.
The criteria were further evaluated for relevance by comparing the undissociated acid concentration selected model 18 (MSEtot = 0.0132), with three parameters, to model 17 (MSEtot = 0.0587), with two parameters, which just exceeded the sum total MSE value of 0.04. Comparison by an F test showed that for all four acids, model 17 was not to be selected as fitting best, meaning that the proposed criteria (MSE = 0.01) are adequate selection criteria and do not need to be less stringent. Concerning the criterion that standard deviations should not exceed parameter estimates, this was tested by comparing model 18 to model 16, with four parameters, in the case of HFo, where the standard deviation exceeded the parameter estimate for model 16. Model 16 was found to not be acceptable for HAc and HPr, while it was acceptable for HLa and HFo. However, in the case of HFo, the standard deviations exceeded the parameter estimates and two parameters without evident biological significance are included. Consequently, it was concluded that model 16 was not the best choice. The third criterion, that parameters preferably have biological meaning, is in principle met for the chosen model 8. However, although the maximum-pH parameter (pHmax) has biological meaning, the estimated value is unrealistic, caused by the fact that no experiments were performed at pH values above 7.5. Fixing pHmax in model 8 at a pH of 11 resulted in a fit of the data with an MSE value of 0.0033. Exclusion of this parameter by choosing model 10 with a slightly higher MSE value also solved the problem of an unrealistic pHmax, but model 8 still had better performance, as shown in Results. Apparently the pHmax value is a valuable shape parameter, necessary to make a good fit of the data.
Undoubtedly, proper and thorough selection of models for describing combined pH and undissociated acid effects needs to rely on selection of the models that are best able to accurately describe the individual hurdles correctly or most optimally, since the model choice may well impact on conclusions drawn about possible synergy between the hurdles.
Evaluation of gamma hypothesis validity.
To falsify the hypothesis that no interaction between the two hurdles occurs at the growth boundary, as suggested by Bidlas and Lambert (7), two models assuming synergy between factors were compared to the newly constructed gamma models for the four acids combined with pH effect. Considering the acids HAc, HPr, and HFo, the synergy models of Le Marc et al. and Augustin and Carlier did not reduce the MSE values, so the predictions were not better than those obtained with the gamma model without an interaction factor. For HPr, all three models underestimated growth, suggesting that pH and the concentration of undissociated propionic acid exhibit a slight antagonistic effect when applied in combination. The growth boundary, however, was predicted relatively well by the gamma model, and the inclusion of a synergy factor did not improve the quality of the prediction. It has to be noted that both synergy models are developed to study growth kinetics of Listeria. The use of the synergy models to study growth kinetics of B. cereus might therefore lead to nonoptimal predictions of growth behavior.
For the lactic acid data set as shown in Fig. 2B, the gamma model performed less well than for the other acids, mainly because the growth boundary was not properly estimated. New parameter estimates for the effect of undissociated lactic acid were made based on a data set with different concentrations of undissociated acid in a buffered BHI solution at pH 5.7 (data not shown). These new parameter estimates (and standard deviations) (μopt = 1.7 [0.011]; [HA]max = 2.57 [0.016];, α = 2.21 [0.068]) were incorporated in the gamma model, and a new prediction of the HLa experiment for the two hurdles was made. The MSE value between the prediction and the experiment was 0.0790, somewhat lower than that for model performance using the original parameter estimates at pH 5.5 (MSE = 0.0925), but the growth boundary was still underestimated by 0.2 pH unit with the new parameter estimates. The observed improvement in model performance, as judged from the decreased MSE, may be due mainly to better predictability of experimental data generated at somewhat higher pH values rather than by a shift in the growth boundary. For lactic acid, the use of model 16 in the combined gamma model (which had been rejected based on an F test) did not result in a better estimate of the growth boundary (MSE = 0.1367) either. As a consequence, it cannot be ruled out that for HLa, interaction between hurdles might play a role close to the growth boundary. For the HLa data, the synergy models gave slightly lower MSE values than the noninteraction models, but visual inspection of the relevant pH-growth rate curves showed that the growth boundary was not predicted correctly. The model of Augustin and Carlier predicted more interaction than did the model of Le Marc et al.
According to Augustin and Carlier (5), their interaction model improves the fail-safe growth predicted by 1.4% and the fail-dangerous no-growth by 9%. There may be more than one reason why different studies draw different conclusions regarding synergy. While the current study would not support a synergistic effect with the factors investigated, Augustin and Carlier (4) set up a model to specifically take into account interactive effects, based on a large quantity of growth data retrieved from the literature (4). It may be that due to the particular origin of their data set, in which there is more likely spreading of the data, especially around the growth boundary, the results are not comparable to the data set generated in the investigation described here. Conceivably, in a situation of data spreading, assuming interaction between factors and use of a synergistic model may improve the prediction of growth data and of the growth boundary.
The possibility of predicting growth behavior of microorganisms with multiple inhibitory factors by the use of models without interaction was found to be applicable for several microorganisms for different combined hurdles (16, 17, 26, 34, 38). Other studies concluded there is indeed synergy when using a mixture of nisin and lactates, salts, or lysozyme and nisin for different bacteria and yeast (3, 9, 25). This synergy is supposed to occur due to metal cations, which have an effect on intracellular ATP levels, which results in higher sensitivity of cells to other hurdles (25). Importantly, as stressed by Lambert and Lambert (18), when investigating the effect of combinations of antimicrobial factors on microorganisms, it is important to understand whether or not the individual factors exert the same inhibitory impact at the same concentration, i.e., whether the factors have an identical response. Mixing antimicrobials with different dose-response relationships may result in apparently synergistic impacts without actually being based on synergistic interactions. Next to insight into the response effects of individual inhibitory factors that are to be applied in combination, it is important to understand the mechanisms of action of the individual factors (6). Lambert and Stratford (15) studied mechanisms of microbial inhibition and responses for weak-acid preservatives and concluded that inhibition depends more on the degree to which individual preservatives are concentrated within the cell rather than on the undissociated acid concentration. Physiological experiments exploring the mechanism of action of HLa on the B. cereus strain investigated in our current study may provide insight into why, with this acid only, there was a notable difference between predictions and experimental data, in particular an overestimation of the growth boundary. As suggested in the literature and tested for Listeria monocytogenes, this synergistic effect for combinations including lactate might be due to metal cations, which have an effect on intracellular ATP levels, which results in higher sensitivity of cells to other hurdles (25). It is interesting to investigate if this hypothesis is also valid for B. cereus, which might explain why, with this acid only, there is an overestimation of the growth boundary.
In conclusion, the nonsynergistic gamma model built in this study with the two best-fitting models for individual factors was sufficiently capable of describing the combined effect of these factors, pH and undissociated acid concentration, on the growth rate of the test microorganism. Synergy between these two factors thus could not be proven, except maybe for the use of lactic acid. This investigation also established an extensive overview of models for predicting effects of either pH or undissociated acid on growth of microorganisms as reported in the literature. The model selection criteria used were found to be practical to identify those models that best fit the data sets. For describing the effects of pH and of the undissociated acid concentration, the models of Rosso et al. and of Luong, respectively (24, 31) were found to be the best-fitting models. The model of Luong could be used for all four acids investigated (i.e., acetic, lactic, propionic, and formic) regardless of the shape of the undissociated acid curve. When using these models in a gamma model for predicting the combined effect of pH and the undissociated acid concentration, it was found to be unnecessary to include a synergy factor in the model, since in three out of four cases, addition of this factor to the nonsynergistic model did not reduce MSE values between predictions and experiments. For lactic acid, the MSE value was reduced by the use of a synergistic model, but the growth boundary was still not estimated correctly. In general, both synergy models that were investigated were found to shift the growth boundary to a higher pH due to assumed interaction between the factors. Users of predictive models, notably those in the food industry that use them in establishing safe product and process designs, should be cautioned against the use of synergistic models in situations where there is no synergy. Evidently, when models erroneously predict an upward shift of a growth boundary to a higher pH, for instance, their use may possibly lead to unsafe food designs or other situations compromising food safety.
Acknowledgments
This work was financially supported by Nestlé Research Centre, Lausanne, Switzerland.
Fan Wang and Linlin Fan are gratefully acknowledged for skillful experimental work during this research.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Adams, M. R., C. L. Little, and M. C. Easter. 1991. Modelling the effect of pH, acidulant and temperature on the growth rate of Yersinia enterocolitica. J. Appl. Bacteriol. 71:65-71. [PubMed] [Google Scholar]
- 2.Aiba, S., M. Shoda, and M. Nagatani. 1968. Kinetics of product inhibition in alcohol fermentation. Biotechnol. Bioeng. 10:845-864. [DOI] [PubMed] [Google Scholar]
- 3.Almagro, A., C. Prista, S. Castro, C. Quintas, A. Madeira-Lopes, J. Ramos, and M. C. Loureiro-Dias. 2000. Effects of salts on Debaryomyces hansenii and Saccharomyces cerevisiae under stress conditions. Int. J. Food Microbiol. 56:191-197. [DOI] [PubMed] [Google Scholar]
- 4.Augustin, J.-C., and V. Carlier. 2000. Mathematical modelling of the growth rate and lag time for Listeria monocytogenes. Int. J. Food Microbiol. 56:29-51. [DOI] [PubMed] [Google Scholar]
- 5.Augustin, J.-C., and V. Carlier. 2000. Modelling the growth rate of Listeria monocytogenes with a multiplicative type model including interactions between environmental factors. Int. J. Food Microbiol. 56:53-70. [DOI] [PubMed] [Google Scholar]
- 6.Berenbaum, M. C. 1989. What is synergy? Pharmacol. Rev. 41:93-141. [PubMed] [Google Scholar]
- 7.Bidlas, E., and R. J. W. Lambert. 2008. Quantification of hurdles: predicting the combination of effects—interaction vs. non-interaction. Int. J. Food Microbiol. 128:78-88. [DOI] [PubMed] [Google Scholar]
- 8.Biesta-Peters, E. G., M. W. Reij, H. Joosten, L. G. M. Gorris, and M. H. Zwietering. 2010. Comparison of two optical density methods and a plate count method for estimation of growth parameters of Bacillus cereus. Appl. Environ. Microbiol. 76:1399-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, W., and R. E. W. Hancock. 2000. Action of lysozyme and nisin mixtures against lactic acid bacteria. Int. J. Food Microbiol. 60:25-32. [DOI] [PubMed] [Google Scholar]
- 10.den Besten, H. M. W., M. Mataragas, R. Moezelaar, T. Abee, and M. H. Zwietering. 2006. Quantification of the effects of salt stress and physiological state on thermotolerance of Bacillus cereus ATCC 10987 and ATCC 14579. Appl. Environ. Microbiol. 72:5884-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghose, T. K., and R. D. Tyagi. 1979. Rapid ethanol fermentation of cellulose hydrolysate. I. Batch versus continuous systems. Biotechnol. Bioeng. 21:1387-1400. [Google Scholar]
- 12.Gould, G. W. 1996. Methods for preservation and extension of shelf life. Int. J. Food Microbiol. 33:51-64. [DOI] [PubMed] [Google Scholar]
- 13.Houtsma, P. C., B. J. M. Kusters, J. C. de Wit, F. M. Rombouts, and M. H. Zwietering. 1994. Modelling growth rates of Listeria innocua as a function of lactate concentration. Int. J. Food Microbiol. 24:113-123. [DOI] [PubMed] [Google Scholar]
- 14.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microb. Infect. 2:189-198. [DOI] [PubMed] [Google Scholar]
- 15.Lambert, R. J., and M. Stratford. 1999. Weak-acid preservatives: modelling microbial inhibition and response. J. Appl. Microbiol. 86:157-164. [DOI] [PubMed] [Google Scholar]
- 16.Lambert, R. J. W., and E. Bidlas. 2007. An investigation of the Gamma hypothesis: a predictive modelling study of the effect of combined inhibitors (salt, pH and weak acids) on the growth of Aeromonas hydrophila. Int. J. Food Microbiol. 115:12-28. [DOI] [PubMed] [Google Scholar]
- 17.Lambert, R. J. W., and E. Bidlas. 2007. A study of the Gamma hypothesis: predictive modelling of the growth and inhibition of Enterobacter sakazakii. Int. J. Food Microbiol. 115:204-213. [DOI] [PubMed] [Google Scholar]
- 18.Lambert, R. J. W., and R. Lambert. 2003. A model for the efficacy of combined inhibitors. J. Appl. Microbiol. 95:734-743. [DOI] [PubMed] [Google Scholar]
- 19.Larsen, H. D., and K. Jørgensen. 1999. Growth of Bacillus cereus in pasteurized milk products. Int. J. Food Microbiol. 46:173-176. [DOI] [PubMed] [Google Scholar]
- 20.Leistner, L., and L. G. M. Gorris. 1995. Food preservation by hurdle technology. Trends Food Sci. Technol. 6:41-46. [Google Scholar]
- 21.Le Marc, Y., V. Huchet, C. M. Bourgeois, J. P. Guyonnet, P. Mafart, and D. Thuault. 2002. Modelling the growth kinetics of Listeria as a function of temperature, pH and organic acid concentration. Int. J. Food Microbiol. 73:219-237. [DOI] [PubMed] [Google Scholar]
- 22.Levenspiel, O. 1980. The Monod equation: a revisit and a generalization to product inhibition situations. Biotechnol. Bioeng. 22:1671-1688. [Google Scholar]
- 23.Lobry, J. R., L. Rosso, and J. P. Flandrois. 1991. A FORTRAN subroutine for the determination of parameter confidence limits in non-linear models. Binary 3:86-93. [Google Scholar]
- 24.Luong, J. H. T. 1985. Kinetics of ethanol inhibition in alcohol fermentation. Biotechnol. Bioeng. 27:280-285. [DOI] [PubMed] [Google Scholar]
- 25.McEntire, J. C., T. J. Montville, and M. L. Chikindas. 2003. Synergy between nisin and select lactates against Listeria monocytogenes is due to the metal cations. J. Food Prot. 66:1631-1636. [DOI] [PubMed] [Google Scholar]
- 26.McMeekin, T. A., K. Presser, D. A. Ratkowsky, T. Ross, M. Salter, and S. Tienungoon. 2000. Quantifying the hurdle concept by modelling the bacterial growth/no growth interface. Int. J. Food Microbiol. 55:93-98. [DOI] [PubMed] [Google Scholar]
- 27.Pasos, F. V., H. P. Fleming, D. F. Ollis, D. F. Hassan, and R. M. Felder. 1993. Modeling the specific growth rate of Lactobacillus plantarum in cucumber extract. Appl. Microbiol. Biotechnol. 40:143-150. [Google Scholar]
- 28.Presser, K. A., D. A. Ratkowsky, and T. Ross. 1997. Modelling the growth rate of Escherichia coli as a function of pH and lactic acid concentration. Appl. Environ. Microbiol. 63:2355-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratkowsky, D. A., R. K. Lowry, T. A. McMeekin, A. N. Stokes, and R. E. Chandler. 1983. Model for bacterial culture growth rate throughout the entire biokinetic temperature range. J. Bacteriol. 154:1222-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rödel, W., and R. Scheuer. 2007. Recent results on the hurdle technology—measuring of combined hurdles. Mitteilungsblatt Fleischforschung Kulmbach 46:3-10. [Google Scholar]
- 31.Rosso, L., J. R. Lobry, S. Bajard, and J. P. Flandrois. 1995. Convenient model to describe the combined effects of temperature and pH on microbial growth. Appl. Environ. Microbiol. 61:610-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosso, L., J. R. Lobry, and J. P. Flandrois. 1993. An unexpected correlation between cardinal temperatures of microbial growth highlighted by a new model. J. Theor. Biol. 162:447-463. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, A. J., and R. J. Gilbert. 1975. Bacillus cereus food poisoning: a provisional serotyping scheme. J. Med. Microbiol. 8:543-550. [DOI] [PubMed] [Google Scholar]
- 34.Te Giffel, M. C., and M. H. Zwietering. 1999. Validation of predictive models describing the growth of Listeria monocytogenes. Int. J. Food Microbiol. 46:135-149. [DOI] [PubMed] [Google Scholar]
- 35.Vereecken, K. M., F. Devlieghere, A. Bockstaele, J. Debevere, and J. F. van Impe. 2003. A model for lactic acid-induced inhibition of Yersinia enterocolitica in mono- and coculture with Lactobacillus sakei. Food Microbiol. 20:701-713. [Google Scholar]
- 36.Wijnands, L. M., J. B. Dufrenne, and F. M. van Leusden. 2005. Bacillus cereus: characteristics, behaviour in the gastro-intestinal tract, and interaction with Caco-2 cells. RIVM report 250912003. RIVM, Bilthoven, Netherlands.
- 37.Wijtzes, T., J. C. de Wit, J. H. J. Huis in 't Veld, K. van 't Riet, and M. H. Zwietering. 1995. Modelling bacterial growth of Lactobacillus curvatus as a function of acidity and temperature. Appl. Environ. Microbiol. 61:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wijtzes, T., F. M. Rombouts, M. L. T. Kant-Muermans, K. van 't Riet, and M. H. Zwietering. 2001. Development and validation of a combined temperature, water activity, pH model for bacterial growth rate of Lactobacillus curvatus. Int. J. Food Microbiol. 63:57-64. [DOI] [PubMed] [Google Scholar]
- 39.Yeh, P. L.-H., R. K. Bajpai, and E. L. Iannotti. 1991. An improved kinetic model for lactic acid fermentation. J. Ferment. Bioeng. 71:75-77. [Google Scholar]
- 40.Zwietering, M. H., J. C. de Wit, and S. Notermans. 1996. Application of predictive microbiology to estimate the numbers of Bacillus cereus in pasteurised milk at the point of consumption. Int. J. Food Microbiol. 30:55-70. [DOI] [PubMed] [Google Scholar]
- 41.Zwietering, M. H., T. Wijtzes, J. C. de Wit, and K. van 't Riet. 1992. A decision support system for prediction of the microbial spoilage in foods. J. Food Prot. 55:973-979. [DOI] [PubMed] [Google Scholar]